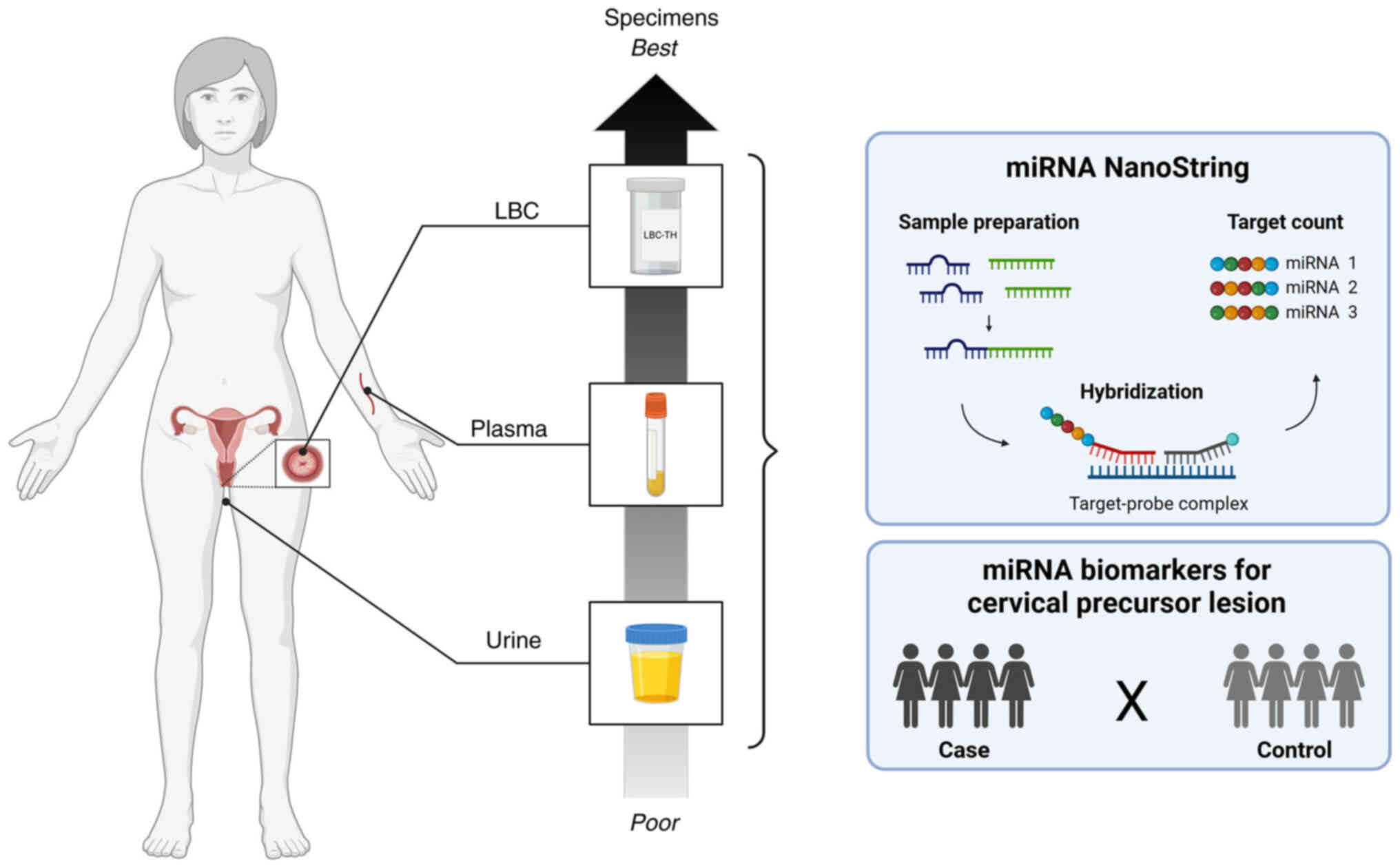
Introduction
Cervical cancer (CC) ranks as the fourth most common
cancer worldwide (1). Persistent
infection of high-risk (hr)-human papillomavirus (HPV) is the
principal cause of CC, initiating carcinogenic processes that
involve the accumulation of genetic and epigenetic changes
(2).
Liquid biopsy (LB) has emerged as an innovative
cancer diagnosis and prognosis approach, offering a non-invasive
alternative to traditional tissue biopsies (3). By detecting circulating tumor
components such as circulating tumor cells, cell-free DNA,
cell-free RNA and exosomes in biological fluids (blood, urine,
feces and saliva), LB has demonstrated utility in early cancer
detection, personalized treatment decisions and prognostic
assessment (3,4). In CC, LB enables the detection of
molecular alterations associated with precursor lesions and disease
progression, potentially before morphological changes are evident
(5).
Identifying molecular markers at an early stage is
crucial to improving screening strategies and clinical outcomes
(6). Among the sample types used in
this context, liquid-based cytology (LBC), which contains
exfoliated cervical cells, is widely employed for conventional
cytological screening (Pap smear) (7). Whilst it is not traditionally
considered a LB, LBC offers a minimally invasive means to collect
cellular material and has proven useful for molecular analyses
(8).
Plasma is the most utilized specimen in LB research
for detecting biomarkers in several types of cancers due to its
easy accessibility (9). Although
several studies have focused on identifying biomarkers in LBC or
blood (9,10), urine can be easily self-collected
recurrently in relatively large volumes and it has greater patient
compliance, suggesting that it could be a promising non-invasive
alternative for CC screening and monitoring (11). However, technical limitations, such
as low RNA yield, concentration variability and enzymatic
degradation, may affect its analytical performance (12,13).
Given its non-invasive nature, LB using these types of specimens
allows the detection of molecular biomarkers, such as HPV DNA
(13–15), DNA methylation (12), circulating tumor cells (16) or dysregulated microRNAs
(miRNAs/miRs) (10,11), which may indicate cervical
carcinogenesis before the progression to high-grade lesions or
invasive cancer (5).
miRNAs are small non-coding RNAs that serve
essential roles in regulating gene expression at the
post-transcriptional level by inhibiting mRNA translation or
promoting mRNA degradation (17).
They are stable and easily detected in LB samples, and certain
studies have demonstrated their role in several biological pathways
in CC (18,19). miRNAs identified in LBC (10), plasma (20,21) or
urine (11) samples have been
directly associated with the diagnosis, treatment monitoring and
progression of CC and its precursor lesions. However, most studies
have explored the expression of miRNAs in a single specimen using
reverse transcription-quantitative PCR (11,20,21),
which has limitations such as low throughput, amplification bias
and complex normalization steps (22). This highlights the potential of
high-throughput digital platforms, such as the NanoString
nCounter® Analysis System, which enables comprehensive
miRNA profiling without amplification. It also offers high
sensitivity, even from degraded samples, such as those obtained
from formalin-fixed paraffin-embedded tissues. Such technologies
simultaneously facilitate detection of multiple targets, making
them valuable for large-scale biomarker studies (22). To date, only one study has employed
NanoString technology in this context, to the best of our knowledge
(10).
Therefore, the aim of the present study was to
identify the most suitable specimen for miRNA-based detection of CC
precursor lesions using NanoString nCounter technology to compare
miRNA profiles across three specimen types, LBC, plasma and urine,
from women with and without high-grade cervical intraepithelial
neoplasia (CIN).
Materials and methods
Study population and sample
collection
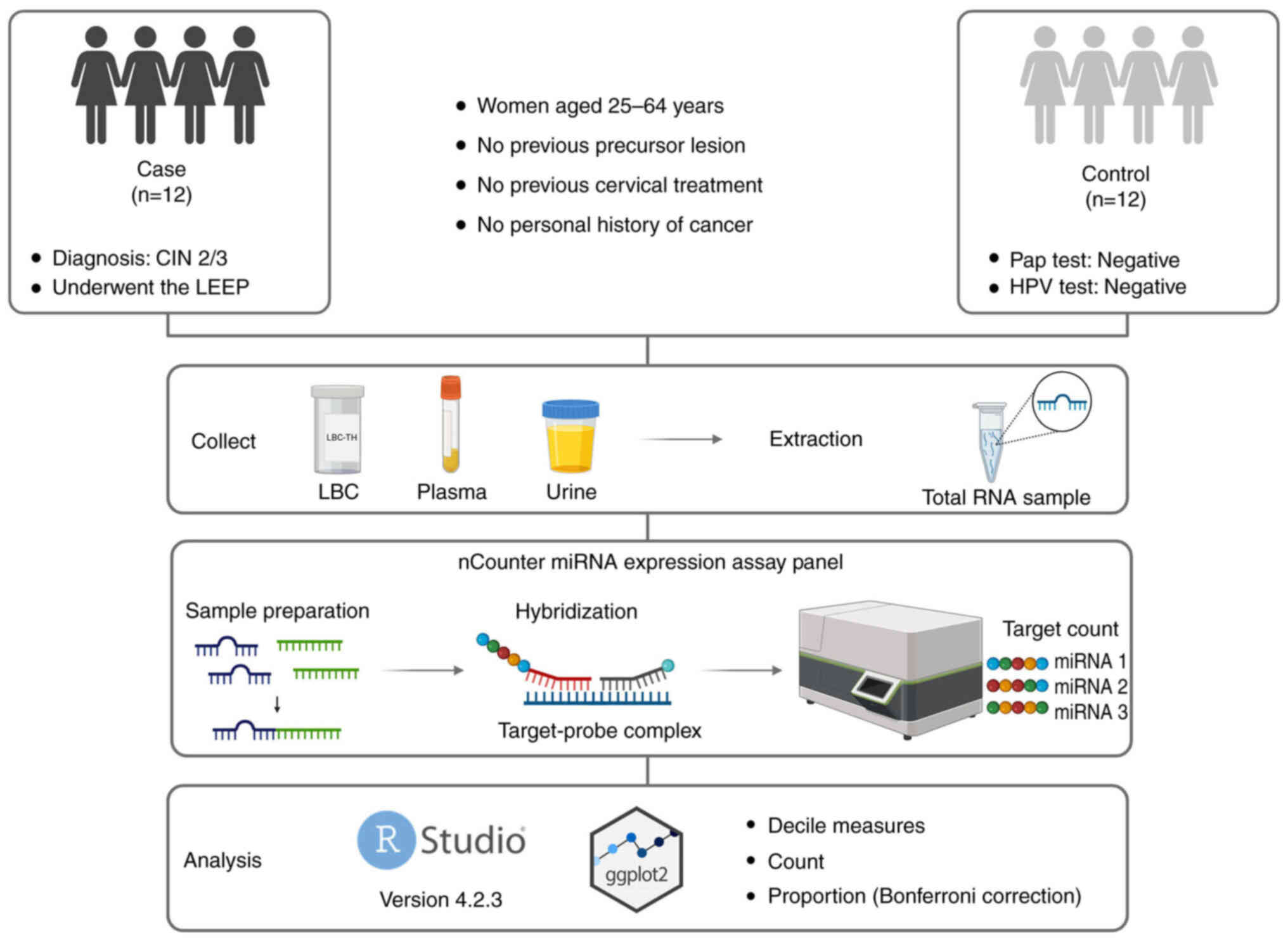
An overview of the methods and key steps of the
present study are presented in Fig.
1. The present study included 24 women, with paired LBC, plasma
and urine samples collected from each participant at the Prevention
Department of Barretos Cancer Hospital, Barretos, Brazil from June
2021 to November 2022. The present study was approved by the
Research Ethics Committee of the Barretos Cancer Hospital
(Barretos, Brazil; approval no. 3.926.525). All patients signed an
informed consent form for their samples to be used in the
research.
Eligible participants were between 25 and 64 years
old, with no history of cervical precursor lesions, no previous
cervical procedures or prior cancer diagnoses. The case group
included 12 women with histologically confirmed high-grade cervical
intraepithelial lesion (CIN 2/3) (23), confirmed by histopathological
examination of the excised tissue obtained through the Loop
Electrosurgical Excision Procedure (LEEP). All patients in the case
group were tested for hr-HPV. The control group also comprised 12
women, selected based on negative results for both Pap smear
cytology and high-risk HPV testing, performed using the Cobas ×480™
system (Roche Diagnostics) (24).
Patients were matched by age (±2 years). The epidemiological and
clinicopathological characteristics of the study participants are
summarized in Table I.
 | Table I.Participant characteristics and
characteristics in case and control groups. |
Table I.
Participant characteristics and
characteristics in case and control groups.
| Characteristic | Case group
(n=12) | Control group
(n=12) |
|---|
| Age, years |
|
|
| Mean
(SD) | 32.41 (7.02) | 32.91 (6.87) |
| Median
(min-max) | 29.00 (25–48) | 30.50 (25–48) |
| Smoking |
|
|
|
Yes | 1 (8.33) | 0 (0.00) |
| No | 10 (83.33) | 11 (91.67) |
| Former
smoker | 1 (8.33) | 1 (8.33) |
| Alcohol
consumption |
|
|
|
Yes | 10 (83.33) | 6 (50.00) |
| No | 1 (8.33) | 2 (16.67) |
| Former
consumption | 1 (8.33) | 4 (33.33) |
| Menopausal
status |
|
|
|
Pre-menopause | 11 (91.67) | 11 (91.67) |
|
Post-menopause | 1 (8.33) | 1 (8.33) |
| Use of hormonal
contraceptives |
|
|
|
Never | 0 (0.00) | 2 (16.67) |
|
Currently | 6 (50.00) | 5 (41.67) |
|
Former | 6 (50.00) | 5 (41.67) |
| HPV vaccine |
|
|
|
Yes | 0 (0.00) | 1 (8.33) |
| No | 12 (100.00) | 11 (91.67) |
| Cytology |
|
|
|
ASC-US | 1 (8.33) | 0 (0.00) |
|
ASC-H | 0 (0.00) | 0 (0.00) |
|
LSIL | 0 (0.00) | 0 (0.00) |
|
HSIL | 11 (91.67) | 0 (0.00) |
|
Negative | 0 (0.00) | 12 (100.00) |
| Consolidated
diagnosisa |
|
|
| CIN
2 | 1 (8.33) | 0 (0.00) |
| CIN
2/3 | 2 (16.67) | 0 (0.00) |
| CIN
3 | 9 (75.00) | 0 (0.00) |
| HPV status |
|
|
| HPV
16 | 5 (41.67) | 0 (0.00) |
| HPV
18 | 1 (8.33) | 0 (0.00) |
| Other
hr-HPV types | 6 (50.00) | 0 (0.00) |
LBC cervical samples were collected from each
participant immediately before the LEEP (case group) or Pap test
(control group). Samples from cervical cell scraping were collected
using the Cervex-Brush® (cat. no. 36825G; Rovers Medical
Devices B.V.). A total of two cervical smear samples were collected
from each participant: One sample was preserved in BD
SurePath™ Preservative Fluid (cat. no. 491337; Becton,
Dickinson and Company) for morphological and HPV tests, and the
other sample was preserved in ThinPrep® Pap test (cat.
no. 70097-001; Hologic, Inc.) for miRNA analysis. Blood samples for
plasma were collected in anticoagulant tubes, whilst urine samples
were collected without any restrictions on time or preparation. All
fluids were stored at Barretos Cancer Hospital Biobank (25).
Biological fluids RNA isolation
LBC, plasma and urine total RNA isolation were
performed using the miRNeasy Mini Kit (cat. no. 217004; Qiagen
GmbH), miRNeasy Serum/Plasma Kit (cat. no. 217184; Qiagen GmbH) and
Urine Cell-Free Circulating RNA Purification Mini Kit (cat. no.
56900; Norgen Biotek Corp.), respectively, according to the
manufacturer's protocol. The total RNA extracted was stored at
−80°C until use. The total RNA concentration and purity of each
sample were measured using the NanoDrop™ 2000/2000c
Spectrophotometer (Thermo Fisher Scientific, Inc.).
nCounter analysis assays
miRNA expression profiling was performed using the
nCounter miRNA Expression Assay Kit (cat. no. CSO-MIR3-12;
NanoString; Bruker Spatial Biology, Inc.), according to the
manufacturer's protocol, using the nCounter Human v3 miRNA that
evaluates 798 miRNAs. In summary, 100 ng total RNA was subjected to
tag binding and hybridization with the Reporter CodeSet and Capture
ProbeSet for 24 h at 65°C. Subsequently, the samples were processed
using the NanoString PrepStation and immobilized on the nCounter
cartridge, which was transferred to the nCounter Digital Analyzer
for image capture (555 field-of-views) and data acquisition.
Statistical analysis
Statistical analyses were performed using R version
4.2.3 in RStudio 2023 (26), with
the ggplot2 library (27) for
constructing graphical analyses. Normalization was performed using
the NanoStringNorm R package (version 1.2.1) (28). Background correction and positive
control normalization were not applied. Sample content
normalization was based on the geometric mean of housekeeping
miRNAs, which were selected for their low coefficient of variation
(CV) across all samples, as detailed in Table SI. Log transformation was applied
to the normalized values. The CV was calculated as the ratio of the
standard deviation (SD) to the mean (CV=SD/mean), providing a
dimensionless measure of dispersion across datasets with different
scales. A reduction in CV after normalization indicated effective
removal of technical noise and improved reproducibility. As CV can
be sensitive to small means, the SD was also assessed as an
alternative dispersion metric (29).
For data exploration, miRNA deciles, counts and
proportions were evaluated. Low-count miRNAs were defined as those
with <10 counts in >50% of samples. Bonferroni correction was
applied to compare the proportions of low-count miRNAs between case
and control groups within each specimen type, as well as across
different specimen types.
As no differential expression analysis was
performed, and the aim was to assess general detection capacity
rather than evaluate specific miRNAs, statistical comparisons of
individual miRNAs between groups were not performed. Therefore, no
multiple testing correction was applied.
Results
NanoString technology comparison of
raw data counts
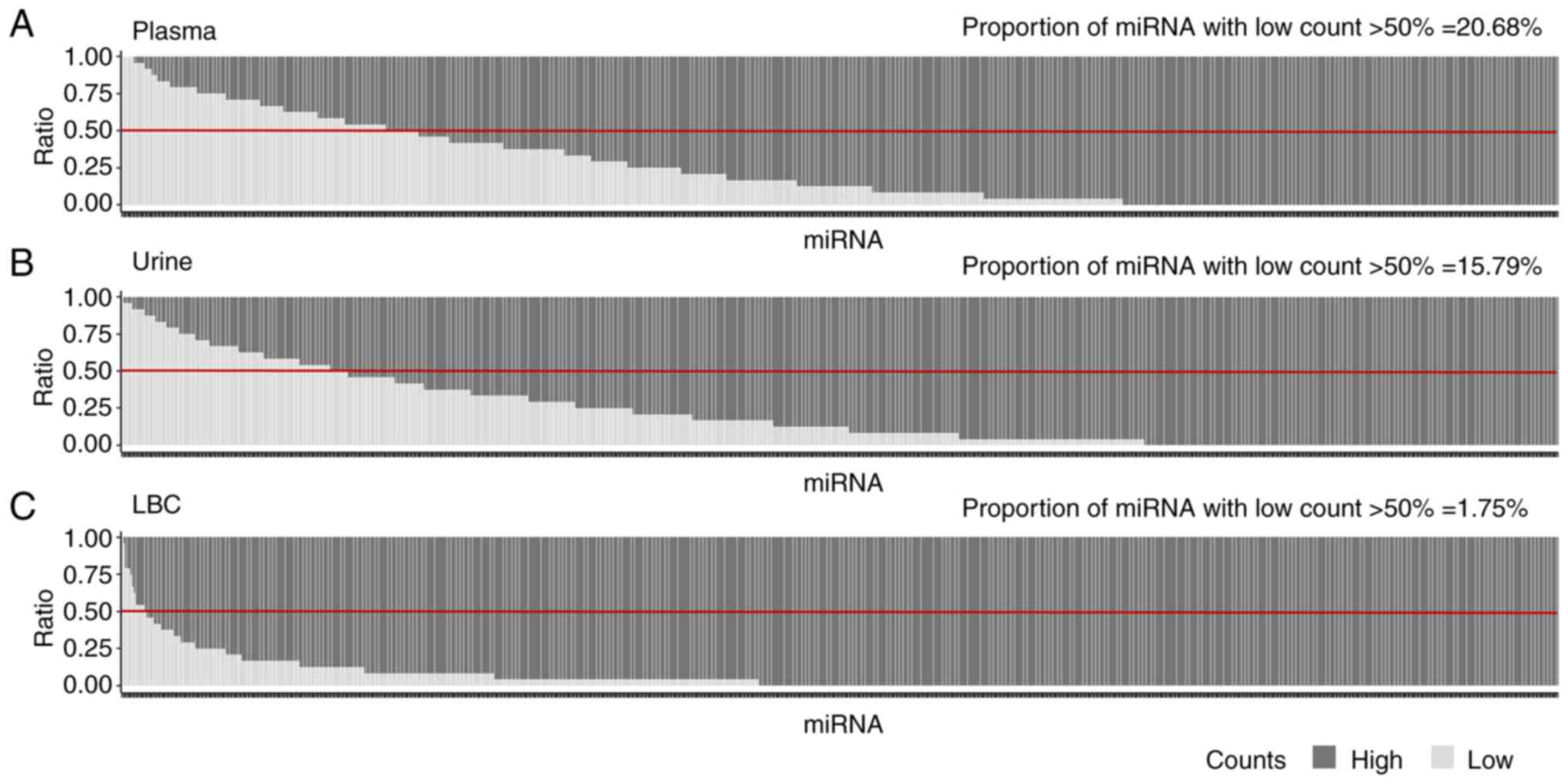
The distribution of the counts of 798 miRNAs across
LBC, plasma and urine samples was analyzed using the raw data.
Median miRNA counts were similar between the case and control
groups (Table SII, Table SIII, Table SIV).
In the raw data analysis, a cutoff was established
using the NanoStringNorm package to distinguish between low and
high counts. Counts <10 were defined as low, whilst those ≥10
were considered high. Initially, raw data counts were compared in
three specimens without stratifying by clinical group. Due to the
characteristics of the raw data, statistical tests were not applied
and the analysis remained descriptive. The detected miRNA counts in
plasma, urine and LBC were analyzed, which demonstrated that LBC
exhibited proportionally higher levels of detected miRNAs than
plasma and urine (Fig. 2). These
results suggest a greater abundance of miRNAs in LBC compared with
the other specimens. By contrast, the comparison between plasma and
urine revealed no notable difference in the proportions of high and
low counts of miRNAs, suggesting comparable levels of miRNA
detection between these two fluids.
The technical threshold was set at counts of <10,
which were defined as low. A miRNA was considered to have a low
count if its expression was <10 in >50% of samples. According
to this analysis, LBC revealed a lower percentage of detected
miRNAs with low counts (1.75%) compared with plasma (20.68%) and
urine (15.79%). This indicates a higher overall detection
efficiency of miRNAs in LBC compared with the other fluids.
Although plasma and urine demonstrated similar proportions of
low-count miRNAs, this indicates comparable levels of detectable
miRNAs in these fluids. However, further analysis is needed to
determine if similar miRNA species are being captured.
Comparison of raw data counts between
case and control groups
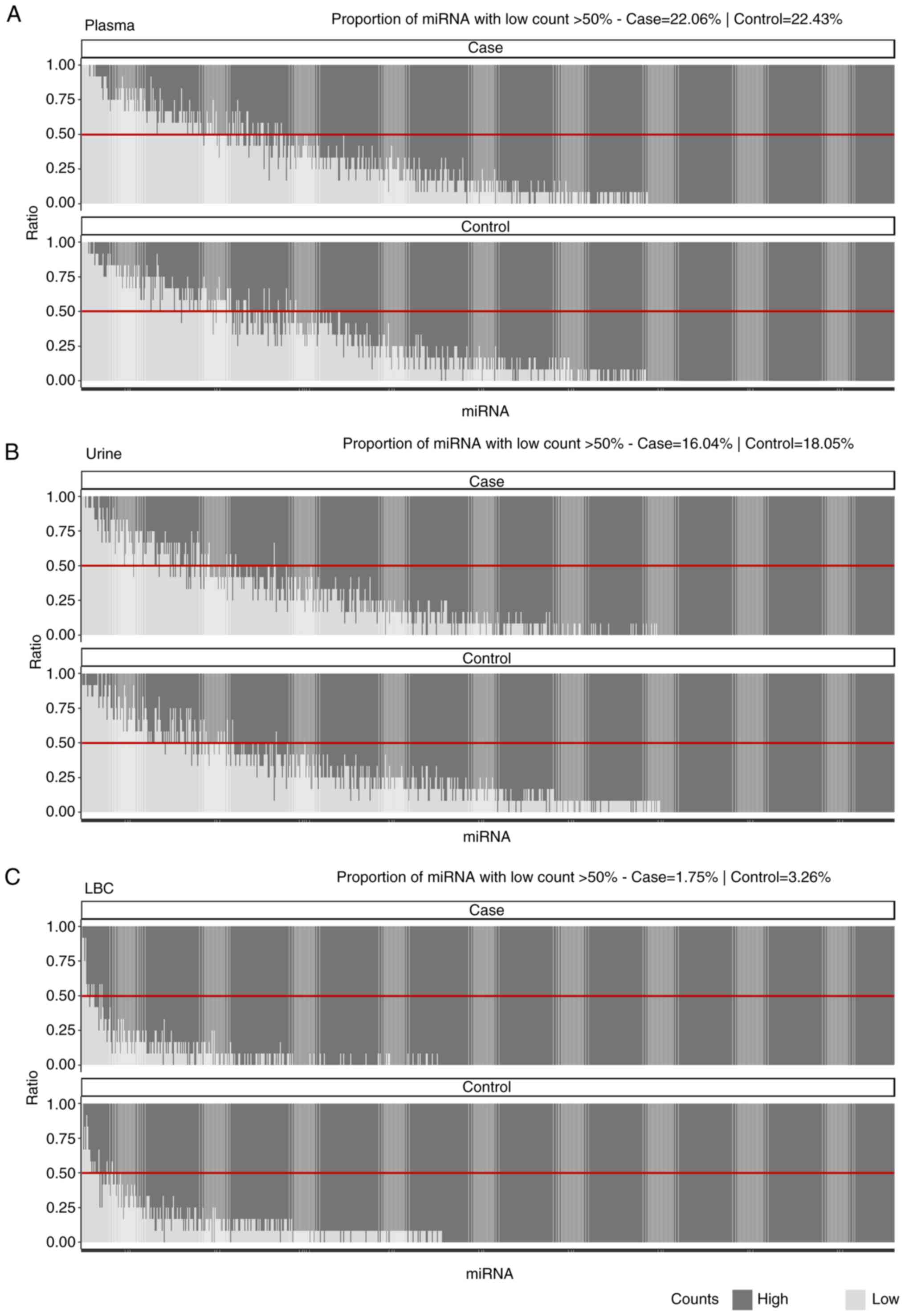
Additionally, a comparative assessment of raw data
counts between the case and control groups for each specimen type
was performed (Fig. 3). The results
demonstrated differences in the proportions of low-count miRNAs
between the case and control groups in LBC (case, 1.75%; control,
3.26%) and urine (case, 16.04%; control, 18.05%). This suggests
that certain miRNAs presented a higher abundance in the case group
compared with in the control group. However, in plasma, the
proportions were similar between the case (22.06%) and control
(22.43%) groups, indicating no difference in low-count miRNAs for
this specimen type.
Comparison of normalized data
counts
The distribution of the counts of 798 miRNAs across
LBC, plasma and urine samples was analyzed, using the normalized
data. Median miRNA counts were similar between the case and control
groups (Table SIV, Table SV, Table SVI, Table SVII) and box plots demonstrated
variability across the sample types (Fig. S1).
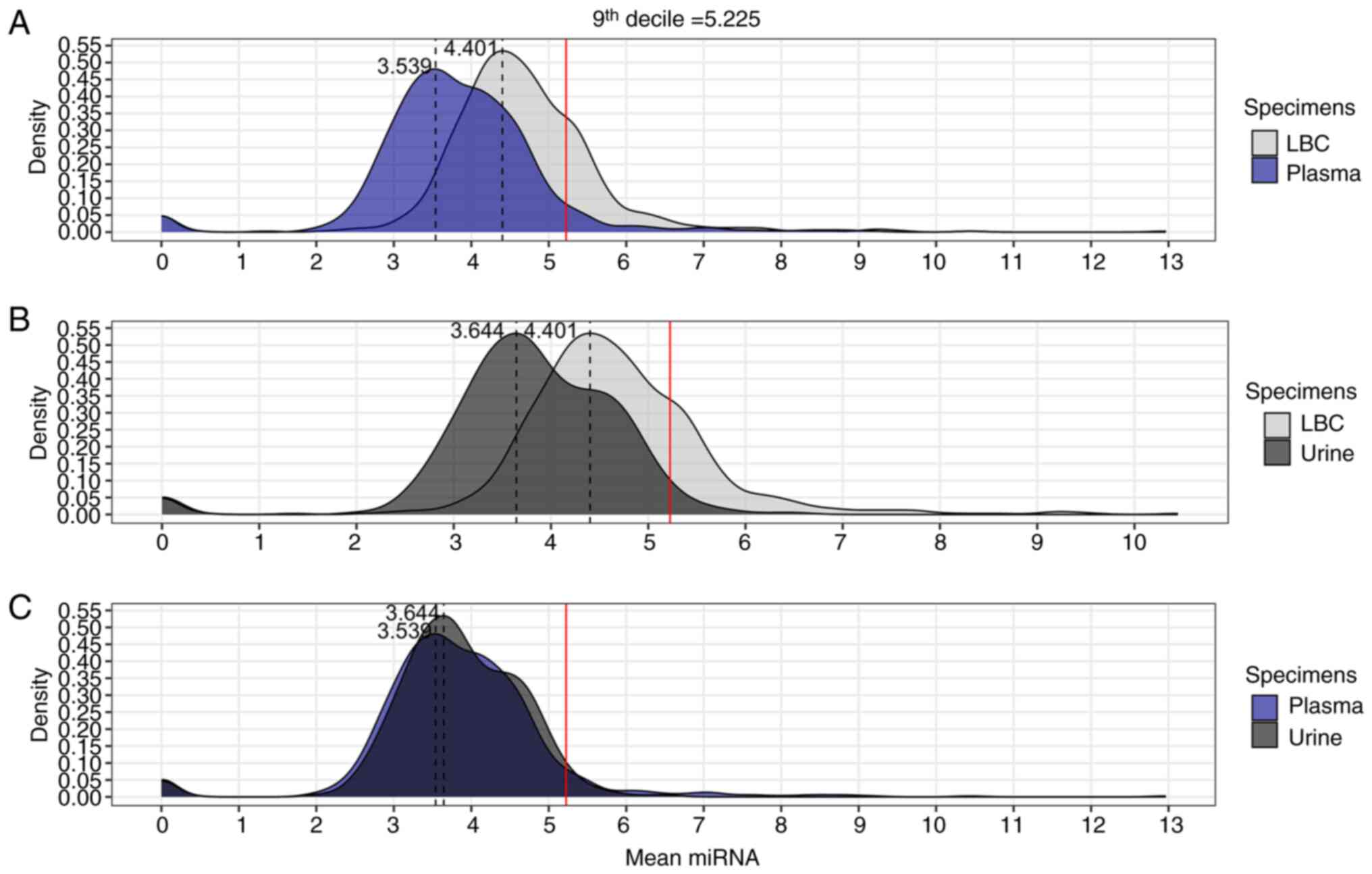
Subsequently, the mean normalized counts of each
detected miRNA were compared across the three specimen types using
the normalized data (Fig. 4).
Density plots were generated to visualize the distribution of mean
miRNA counts in LBC, plasma and urine (Fig. 4). The 9th decile (5.225) was used as
a reference to identify regions with extreme values of mean miRNA
counts, allowing for a clear comparative analysis of miRNA count
distributions between the different specimens. This approach
highlighted differences and extreme values in the density plots,
facilitating the visualization of distinctive characteristics
between the analyzed fluids. Additionally, the proportions of miRNA
counts above the 9th decile were calculated for each specimen: LBC
vs. plasma, LBC vs. urine, and plasma vs. urine (Table II). The comparisons revealed that
LBC was superior to the other specimens, with a significantly
higher concentration of miRNAs with counts >9th decile (21.3%),
compared with plasma (5.89%) (P<0.01; Fig. 4A; Table
II) and urine (2.88%) (P<0.01; Fig. 4B; Table
II). Furthermore, the difference in miRNAs with high counts
>9th decile between plasma (5.89%) and urine (2.88%) was
significant (P<0.01; Fig. 4C;
Table II).
 | Table II.Comparison of the proportions of
microRNA counts >9th decile between liquid-based cytology,
plasma and urine samples. |
Table II.
Comparison of the proportions of
microRNA counts >9th decile between liquid-based cytology,
plasma and urine samples.
| A, LBC vs.
plasma |
|---|
|
|---|
| Specimen | Count | Proportion, % | P-value |
|---|
| LBC | 170/798 | 21.3 | <0.01 |
| Plasma | 47/798 | 5.89 |
|
|
| B, LBC vs.
urine |
|
|
Specimen | Count | Proportion,
% | P-value |
|
| LBC | 170/798 | 21.3 | <0.01 |
| Urine | 23/798 | 2.88 |
|
|
| C, Plasma vs.
urine |
|
|
Specimen | Count | Proportion,
% | P-value |
|
| Plasma | 47/798 | 5.89 | <0.01 |
| Urine | 23/798 | 2.88 |
|
Discussion
The early detection of CC precursor lesions is
crucial for improving patient prognosis, as timely intervention can
markedly reduce progression to invasive cancer (5). LB provides a unique opportunity to
identify molecular changes at an early stage, even before
morphological alterations become evident in conventional cytology
or histopathology (6). In this
context, circulating miRNAs have emerged as promising biomarkers,
allowing the detection of early oncogenic alterations and enhancing
the sensitivity of current screening strategies (30,31).
However, selecting the most appropriate specimen for LB-based
diagnostics remains a critical challenge (32). Therefore, the present study
performed a comparative analysis of plasma, urine and LBC as
potential specimens for CC precursor lesions detection, using raw
and normalized counts of detected miRNAs.
The initial analysis revealed substantial
differences in the levels of detected miRNAs between plasma, urine
and LBC. LBC exhibited higher levels of detected miRNAs than plasma
and urine, suggesting its potential superiority in capturing miRNA
biomarkers for the detection of CC precursor lesions. This
observation is in-line with previous studies highlighting LBC as a
potential specimen for LB-based identification of CC precursor
lesions (10,33). By contrast, plasma and urine
demonstrated comparable levels of detected miRNAs, indicative of a
relative similarity between these two specimens. Although plasma
and urine demonstrated comparable levels of detected miRNAs, plasma
remains the preferred specimen for LB because it directly reflects
systemic processes and captures miRNAs released from circulating
cells (34).
Although there was no primary aim to compare
diagnostic groups in the present study, an exploratory analysis of
raw miRNA counts was performed to assess differences in detection
between the case and control groups. However, no differences were
observed, suggesting that interindividual variability, rather than
biological factors, may underlie the similar detection rates. This
is consistent with previous studies reporting that circulating
miRNA profiles are affected by factors such as cellular origin,
sample handling and inherent physiological variation between
individuals (35,36). Given the small sample size and early
stage of the disease, such variability is expected and has been
cited as a challenge in LB interpretation (37,38).
These findings are therefore complementary and do not support
differential expression analysis.
Data normalization provided more information about
the diagnostic utility of plasma and urine. Scatterplots
demonstrated uniformity and similarity in detected miRNA levels
between plasma and LBC, highlighting the potential of plasma as a
promising specimen for miRNA detection. By contrast, urine revealed
greater variability in miRNA levels, posing challenges to
diagnostic accuracy. Density plots accentuated the disparities
between plasma and urine, with plasma demonstrating a distribution
profile more similar to LBC. This observation underlines the
diagnostic superiority of plasma over urine, as the latter
exhibited notable differences with the standard specimen. Moreover,
the findings of the present study highlight the importance of
selecting the most appropriate specimen for LB-based diagnostics in
CC precursor lesions. Although LBC has emerged as a pioneer in
capturing miRNA biomarkers, plasma exhibited a comparable
performance, offering advantages in diagnostic accuracy and
clinical utility (39,40). The aforementioned observations align
with previous studies that emphasize the diagnostic potential of
plasma-derived miRNAs in CC and its precursor lesions (21,41).
The robustness of plasma as a specimen for LB can be attributed to
its direct reflection of systemic processes and release of miRNA
from circulating cells (4,42), making it a valuable resource for
early cancer detection and monitoring.
By contrast, urine, despite its non-invasive
collection method and potential usefulness in certain scenarios,
presents a challenge due to its inherent variability in miRNA
levels. This variability can be attributed to contamination and
compositional differences (43),
which need to be addressed when utilizing urine for LB-based
diagnosis. However, despite the aforementioned challenges of using
the NanoString technology in the present study, other studies have
reported the use of miRNAs as biomarkers in urine using
quantitative PCR. Aftab et al (11) identified a combination of
miR-145-5p, miR-218-5p and miR-34a-5p in urine that achieved
notable results. This combination showed 100% sensitivity and 92.8%
specificity in distinguishing between patients with pre-cancer and
cancer from healthy controls. Moreover, the levels of these miRNAs
in urine were associated with those observed in serum and tumor
tissues. Notably, the expression of miR-34a-5p and miR-218-5p
emerged as independent prognostic factors for the overall survival
of patients with CC (11).
Furthermore, the NanoString nCounter system served a
key role in improving the reliability of the findings in the
present study by delivering consistent and reproducible miRNA
quantification across all specimens. Its ability to handle multiple
samples simultaneously facilitated a comprehensive comparison of
miRNA profiles between LBC, plasma and urine. Additionally, the
sensitivity of the platform in detecting miRNAs, even from degraded
samples, allowed the generation of robust expression profiles,
supporting the exploration of miRNAs as potential biomarkers for CC
screening (44).
The present has several notable strengths,
particularly its comprehensive approach to evaluating multiple
specimens (LBC, plasma and urine) for miRNA detection in CC
precursor lesions. It provides valuable insight into identifying
the optimal medium for non-invasive CC screening. Moreover, the use
of NanoString technology to analyze a broad panel of 798 miRNAs
strengthens the reliability of the data by providing a
comprehensive assessment of miRNA expression levels across
specimens. However, despite the valuable insights gained from the
comparative analysis of specimens for CC precursor lesions
detection, several limitations restrict the interpretation and
generalization of the findings. For example, the present study did
not include groups with low-grade lesions (CIN 1) and CC. Their
inclusion would have enabled a more comprehensive analysis of miRNA
detection across disease stages. Without these groups, conclusions
on the effectiveness of LBC, plasma and urine as specimens are
limited. The modest sample size and inherent heterogeneity of the
study population may also limit the statistical power and
robustness of the conclusions. In addition, due to the limited
number of samples, no differential expression analysis was
performed to identify specific miRNAs between groups. This decision
was made to avoid generating potentially unreliable results without
adequate statistical power. Considering this, the present study
focused on evaluating the overall detection performance of
NanoString technology across different specimen types. Future
studies with larger cohorts will allow for robust differential
expression analyses to explore biomarker candidates with greater
statistical confidence. Furthermore, the cross-sectional design of
the study precludes the establishment of causal relationships, and
the biological variability inherent in circulating miRNA levels may
have confounded the results. Addressing these limitations through
large-scale studies with standardized protocols and comprehensive
biomarker panels will be critical to advancing the field of
LB-based diagnostics in CC and its precursor lesions.
In conclusion, the results of the present study
demonstrate the superiority of LBC, followed by plasma, over urine
when using NanoString technology to detect miRNA biomarkers in CC
precursor lesions and control groups (Fig. 5). The comparative analysis
highlights the diagnostic superiority of plasma over urine in
capturing miRNA biomarkers for CC precursor lesions detection,
whilst LBC stands out as the most effective specimen overall. These
findings provide valuable information for clinicians and
researchers in selecting the most appropriate specimen for LB-based
diagnostics, paving the way for greater diagnostic accuracy and
improved patient outcomes in treating CC and its precursor
lesions.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was funded by National Support Program for
Oncological Care (PRONON), Ministry of Health (grant no.
NUP-25000.023997.2018/34) entitled: ‘Identificação de biomarcadores
para screening e detecção precoce de tumores no contexto do Sistema
Único de Saúde (SUS)’, Barretos Cancer Hospital Research Incentive
Program (PAIP) (grant no. not applicable) and a scholarship from
the FAPESP process (grant no. 2022/12283-6).
Availability of data and materials
The raw and normalized NanoString data generated in
the present study may be found in the National Centre for
Biotechnology Information Gene Expression Omnibus under accession
number GSE302097 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE302097.
All other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
SC and AJAF developed and led the study, performed
data reviews and analyses, and prepared the manuscript. JCPR helped
with the study design. RLC assisted with the NanoString
experiments. WH participated in the data analysis. RDR helped with
the study design and critically reviewed the manuscript. RMR helped
with the study design, provided advice during the study development
and critically reviewed the manuscript. MMCM conceived and guided
the development of the study and critically reviewed the
manuscript. SC and AJAF confirm the authenticity of all the raw
data. All authors have substantially revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Barretos Cancer Hospital (approval no.
3.926.525). Each research participant provided written informed
consent for their samples to be used in the research. All
information that could be used to identify the study participants
was kept confidential and encrypted in a secure database to ensure
full confidentiality of clinical information, laboratory findings
and the anonymity of each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
|
|
2
|
Albulescu A, Plesa A, Fudulu A, Iancu IV,
Anton G and Botezatu A: Epigenetic approaches for cervical
neoplasia screening (review). Exp Ther Med. 22:14812021. View Article : Google Scholar
|
|
3
|
Poulet G, Massias J and Taly V: Liquid
biopsy: General concepts. Acta Cytologica. 63:449–455. 2019.
View Article : Google Scholar
|
|
4
|
De Rubis G, Rajeev Krishnan S and Bebawy
M: Liquid biopsies in cancer diagnosis, monitoring, and prognosis.
Trends Pharmacol Sci. 40:172–186. 2019. View Article : Google Scholar
|
|
5
|
Dasari S, Wudayagiri R and Valluru L:
Cervical cancer: Biomarkers for diagnosis and treatment. Clin Chim
Acta. 445:7–11. 2015. View Article : Google Scholar
|
|
6
|
Ma L, Guo H, Zhao Y, Liu Z, Wang C, Bu J,
Sun T and Wei J: Liquid biopsy in cancer current: Status,
challenges and future prospects. Sig Transduct Target Ther.
9:3362024. View Article : Google Scholar
|
|
7
|
Strander B, Andersson-Ellström A, Milsom
I, Rådberg T and Ryd W: Liquid-based cytology versus conventional
Papanicolaou Smear in an organized screening program: A prospective
randomized study. Cancer. 111:285–291. 2007. View Article : Google Scholar
|
|
8
|
Gibson J, Young S, Leng B, Zreik R and Rao
A: Molecular diagnostic testing of cytology specimens: Current
applications and future considerations. J Am Soc Cytopathol.
3:280–294. 2014. View Article : Google Scholar
|
|
9
|
Marrugo-Ramírez J, Mir M and Samitier J:
Blood-based cancer biomarkers in liquid biopsy: A promising
Non-invasive alternative to tissue biopsy. Int J Mol Sci.
19:28772018. View Article : Google Scholar
|
|
10
|
Causin RL, da Silva LS, Evangelista AF,
Leal LF, Souza KCB, Pessôa-Pereira D, Matsushita GM, Reis RM,
Fregnani JHTG and Marques MMC: MicroRNA biomarkers of High-grade
cervical intraepithelial neoplasia in liquid biopsy. Biomed Res
Int. 2021:66509662021. View Article : Google Scholar
|
|
11
|
Aftab M, Poojary SS, Seshan V, Kumar S,
Agarwal P, Tandon S, Zutshi V and Das BC: Urine miRNA signature as
a potential Non-invasive diagnostic and prognostic biomarker in
cervical cancer. Sci Rep. 11:103232021. View Article : Google Scholar
|
|
12
|
van den Helder R, Steenbergen RDM, van
Splunter AP, Mom CH, Tjiong MY, Martin I, Rosier-van Dunné FMF, van
der Avoort IAM, Bleeker MCG and van Trommel NE: HPV and DNA
methylation testing in urine for cervical intraepithelial neoplasia
and cervical cancer detection. Clin Cancer Res. 28:2061–2068. 2022.
View Article : Google Scholar
|
|
13
|
Poljak M, Cuschieri K, Alemany L and
Vorsters A: Testing for human papillomaviruses in urine, blood, and
oral specimens: An update for the laboratory. J Clin Microbiol.
61:e0140322013.
|
|
14
|
Jeannot E, Latouche A, Bonneau C,
Calméjane MA, Beaufort C, Ruigrok-Ritstier K, Bataillon G, Larbi
Chérif L, Dupain C, Lecerf C, et al: Circulating HPV DNA as a
marker for early detection of relapse in patients with cervical
cancer. Clin Cancer Res. 27:5869–5877. 2021. View Article : Google Scholar
|
|
15
|
Gothwal M, Nalwa A, Singh P, Yadav G,
Bhati M and Samriya N: Role of cervical cancer biomarkers p16 and
Ki67 in abnormal cervical cytological smear. J Obstet Gynaecol
India. 71:72–77. 2021. View Article : Google Scholar
|
|
16
|
Pan L, Yan G, Chen W, Sun L, Wang J and
Yang J: Distribution of circulating tumor cell phenotype in early
cervical cancer. Cancer Manag Res. 11:5531–5536. 2019. View Article : Google Scholar
|
|
17
|
Vishnoi A and Rani S: MiRNA Biogenesis and
Regulation of Diseases: An Overview. Rani S: MicroRNA Profiling.
Volume 1509. Springer New York, New York, NY, USA: pp. 1–10. 2017,
https://link.springer.com/protocol/10.1007/978-1-4939-6524-3_1March
28–2022 View Article : Google Scholar
|
|
18
|
He Y, Lin J, Ding Y, Liu G, Luo Y, Huang
M, Xu C, Kim TK, Etheridge A, Lin M, et al: A systematic study on
dysregulated microRNAs in cervical cancer development. Int J
Cancer. 138:1312–1327. 2016. View Article : Google Scholar
|
|
19
|
Causin RL, de Freitas AJA, Trovo Hidalgo
Filho CM, dos Reis R, Reis RM and Marques MMC: A systematic review
of MicroRNAs involved in cervical cancer progression. Cells.
10:6682021. View Article : Google Scholar
|
|
20
|
Wei H, Wen-Ming C and Jun-Bo J: Plasma
miR-145 as a novel biomarker for the diagnosis and radiosensitivity
prediction of human cervical cancer. J Int Med Res. 45:1054–1060.
2017. View Article : Google Scholar
|
|
21
|
Ma G, Song G, Zou X, Shan X, Liu Q, Xia T,
Zhou X and Zhu W: Circulating plasma microRNA signature for the
diagnosis of cervical cancer. Cancer Biomark. 26:491–500. 2019.
View Article : Google Scholar
|
|
22
|
Metcalf GAD: MicroRNAs: Circulating
biomarkers for the early detection of imperceptible cancers via
biosensor and Machine-learning advances. Oncogene. 43:2135–2142.
2024. View Article : Google Scholar
|
|
23
|
Khieu M and Butler SL: High-Grade Squamous
Intraepithelial Lesion of the Cervix. StatPearls Treasure Island
(FL): StatPearls Publishing; 2025, http://www.ncbi.nlm.nih.gov/books/NBK430728/August
28–2025
|
|
24
|
Rao A, Young S, Erlich H, Boyle S,
Krevolin M, Sun R, Apple R and Behrens C: Development and
characterization of the cobas human papillomavirus test. J Clin
Microbiol. 51:1478–1484. 2013. View Article : Google Scholar
|
|
25
|
Neuber AC, Tostes CH, Ribeiro AG,
Marczynski GT, Komoto TT, Rogeri CD, da Silva VD, Mauad EC, Reis RM
and Marques MMC: The biobank of barretos cancer hospital: 14 years
of experience in cancer research. Cell Tissue Bank. 23:271–284.
2022. View Article : Google Scholar
|
|
26
|
R: The R Project for Statistical
Computing. https://www.r-project.org/October
2–2024
|
|
27
|
Wickham H: ggplot2. Springer Cham; Cham,
Switzerland: pp. 24–129. 2016, https://link.springer.com/book/10.1007/978-3-319-24277-4October
2–2024
|
|
28
|
Waggott D, Chu K, Yin S, Wouters BG, Liu
FF and Boutros PC: NanoStringNorm: An extensible R package for the
pre-processing of NanoString mRNA and miRNA data. Bioinformatics.
28:1546–1548. 2012. View Article : Google Scholar
|
|
29
|
Deo A, Carlsson J and Lindlöf A: How to
choose a normalization strategy for miRNA quantitative real-time
(qPCR) arrays. J Bioinform Comput Biol. 9:795–812. 2011. View Article : Google Scholar
|
|
30
|
Laengsri V, Kerdpin U, Plabplueng C,
Treeratanapiboon L and Nuchnoi P: Cervical cancer markers:
Epigenetics and microRNAs. Lab Med. 49:97–111. 2018. View Article : Google Scholar
|
|
31
|
Freitas AJA, Causin RL, Varuzza MB, Calfa
S, Hidalgo Filho CMT, Komoto TT, Souza CP and Marques MMC: Liquid
biopsy as a tool for the diagnosis, treatment, and monitoring of
breast cancer. Int J Mol Sci. 23:99522022. View Article : Google Scholar
|
|
32
|
Adhit KK, Wanjari A, Menon S and K S:
Liquid biopsy: An evolving paradigm for Non-invasive disease
diagnosis and monitoring in medicine. Cureus. 15:e501762023.
|
|
33
|
Xie H, Norman I, Hjerpe A, Vladic T,
Larsson C, Lui WO, Östensson E and Andersson S: Evaluation of
microRNA-205 expression as a potential triage marker for patients
with low-grade squamous intraepithelial lesions. Oncol Lett.
13:35862017. View Article : Google Scholar
|
|
34
|
Martins I, Ribeiro IP, Jorge J, Gonçalves
AC, Sarmento-Ribeiro AB, Melo JB and Carreira IM: Liquid biopsies:
Applications for cancer diagnosis and monitoring. Genes.
12:3492021. View Article : Google Scholar
|
|
35
|
McDonald JS, Milosevic D, Reddi HV, Grebe
SK and Algeciras-Schimnich A: Analysis of circulating microRNA:
Preanalytical and analytical challenges. Clin Chem. 57:833–840.
2011. View Article : Google Scholar
|
|
36
|
Pritchard CC, Kroh E, Wood B, Arroyo JD,
Dougherty KJ, Miyaji MM, Tait JF and Tewari M: Blood cell origin of
circulating microRNAs: A cautionary note for cancer biomarker
studies. Cancer Prev Res (Phila). 5:492–497. 2012. View Article : Google Scholar
|
|
37
|
Witwer KW: Circulating microRNA biomarker
studies: Pitfalls and potential solutions. Clin Chem. 61:56–63.
2015. View Article : Google Scholar
|
|
38
|
Moldovan L, Batte KE, Trgovcich J, Wisler
J, Marsh CB and Piper M: Methodological challenges in utilizing
miRNAs as circulating biomarkers. J Cell Mol Med. 18:371–390. 2014.
View Article : Google Scholar
|
|
39
|
You W, Wang Y and Zheng J: Plasma miR-127
and miR-218 might serve as potential biomarkers for cervical
cancer. Reprod Sci. 22:1037–1041. 2015. View Article : Google Scholar
|
|
40
|
Wang H, Zhang D, Chen Q and Hong Y: Plasma
expression of miRNA-21, −214, −34a, and −200a in patients with
persistent HPV infection and cervical lesions. BMC Cancer.
19:9862019. View Article : Google Scholar
|
|
41
|
Hoelzle CR, Arnoult S, Borém CRM, Ottone
M, Magalhães KCSF, da Silva IL and Simões RT: MicroRNA levels in
cervical cancer samples and relationship with lesion grade and HPV
Infection. Microrna. 10:139–145. 2021. View Article : Google Scholar
|
|
42
|
Cafforio P, Palmirotta R, Lovero D,
Cicinelli E, Cormio G, Silvestris E, Porta C and D'Oronzo S: Liquid
biopsy in cervical cancer: Hopes and pitfalls. Cancers (Basel).
13:39682021. View Article : Google Scholar
|
|
43
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The MicroRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar
|
|
44
|
Buus R, Szijgyarto Z, Schuster EF, Xiao H,
Haynes BP, Sestak I, Cuzick J, Paré L, Seguí E, Chic N, et al:
Development and validation for research assessment of Oncotype
DX® Breast Recurrence Score, EndoPredict® and
Prosigna®. NPJ Breast Cancer. 7:152021. View Article : Google Scholar
|