Introduction
Lung cancer remains the leading cause of
cancer-related mortality worldwide, accounting for 18.7% of all
cancer deaths, with non-small cell lung cancer (NSCLC) comprising
~85% of cases (1). For patients
with early-stage NSCLC, surgical resection offers the best chance
for long-term survival (2).
However, recurrence after curative surgery remains a major
challenge (3). Emerging evidence
highlights the prognostic importance of a pathological feature
known as spread through air spaces (STAS), a pattern of invasion
where tumor cells disseminate into alveolar spaces beyond the tumor
margin, occurring in ~50% of resected NSCLC cases (4,5). The
presence of STAS has been associated with early recurrence and poor
survival, with 5-year recurrence-free and overall survival rates
nearly halved compared with STAS-negative cases, particularly in
patients undergoing limited resections (6). This suggests that lobectomy or wider
surgical margins may be more appropriate for patients with STAS to
reduce the risk of recurrence. However, its identification
currently relies entirely on postoperative pathological examination
of resected specimens, which remains the standard for STAS
diagnosis (7). This creates a key
gap in preoperative planning, as STAS status is unknown when
deciding surgical extent or resection margin.
Due to this limitation, accurately predicting STAS
preoperatively would aid thoracic surgeons in selecting surgical
strategies, planning lymph node dissection and counseling patients
about recurrence risks. Although several studies (8,9) have
developed prediction models for STAS, most of them rely on
radiomics features extracted from high-resolution CT images, which
necessitate advanced image segmentation, texture analysis and
specialized computational tools. These requirements limit the
feasibility of such models in routine clinical workflows,
particularly in community hospitals or resource-limited settings.
Therefore, there is a clear need for accessible and generalizable
models that use only routinely available clinical features.
Artificial intelligence, particularly in machine
learning (ML), has created novel opportunities to improve
predictive modeling in clinical research. Unlike traditional
statistical methods, ML algorithms can capture complex and
non-linear interactions among high-dimensional variables and have
demonstrated notably increased performance in a range of medical
applications, including pulmonary nodule characterization and lung
cancer risk prediction (10–12).
Despite these advantages, the limited interpretability of ML models
has hindered their widespread clinical adoption. To address this
concern, explainable artificial intelligence techniques such as
SHapley Additive exPlanations (SHAP) have been introduced to
improve model transparency. SHAP assigns interpretable contribution
scores to individual features, offering both global insights into
overall feature importance and local explanations for individual
predictions (13,14).
The present study aimed to develop a practical and
explainable ML model to predict STAS using only standard
preoperative data, without relying on radiomics. The present study
trained and compared five supervised ML algorithms based on
routinely available clinical, radiological and serological
variables to identify the optimal predictive model. Furthermore,
SHAP analysis was applied to interpret feature contributions and
visualize the decision-making process of the model. This
explainable, data-driven model is intended to support personalized
surgical planning and potentially improve preoperative risk
stratification in patients with NSCLC in the future.
Patients and methods
Study population
The present study was a retrospective, single-center
study that included patients aged ≥18 years who underwent
curative-intent pulmonary resection for NSCLC at Anqing Municipal
Hospital (Anqing, China) between January 2021 and December 2023.
Patients were identified using the electronic medical records.
Inclusion criteria were as follows: i)
Histopathologically confirmed NSCLC after surgical resection; and
ii) resectable NSCLC staged retrospectively according to the 9th
edition of the TNM classification (International Association for
the Study of Lung Cancer, 2024) (15), which was applied during data
analysis to ensure consistency with the most recent staging
standards. The re-staging process was performed based on available
pathological and imaging data by two thoracic surgeons to ensure
accuracy and reproducibility. Exclusion criteria were as follows:
i) Receipt of neoadjuvant chemotherapy, radiotherapy, immunotherapy
or targeted therapy prior to surgery; ii) history of other
malignancies; iii) multiple primary tumors in the same lobe of the
lung; and iv) incomplete clinical, radiological or pathological
data.
The present study was approved by the Ethics
Committee of Anqing Municipal Hospital (approval no. 2025-152). The
committee confirmed that the study design met ethical standards and
that written informed consent was obtained from all participants
prior to inclusion. No formal sample-size calculation was performed
due to the retrospective nature of the present study and all
eligible patients within the study period were included. Patients
with missing data on any included variables were excluded from the
analysis.
Data collection
Variables for STAS included: Age, sex, smoking
history, lesion location and pathological tumor (T) stage
(categorized as T0, T1 or T2 according to the 9th edition of the
TNM classification).
Radiological features based on preoperative chest CT
included nodule types [pure ground-glass opacity (pGGO), part-solid
or solid], presence of lobulation, spiculation and vacuole sign.
Due to the absence of universally standardized criteria for nodule
classification, nodule types were defined according to widely
accepted radiological principles to minimize inter-observer
variability and ensure reproducibility (16). The nodule types were as follows: i)
pGGO: A lesion indicating homogeneous increased attenuation of the
lung parenchyma without any solid component obscuring the
underlying lung architecture; ii) part-solid: A lesion containing
both ground-glass and solid components, with the solid portion
partially obscuring the lung parenchymal markings; and iii) solid:
A lesion in which the underlying lung architecture is completely
obscured by soft-tissue attenuation. All CT images were reviewed by
two experienced radiologists blinded to the outcome.
Serum tumor markers included squamous cell carcinoma
antigen (SCC), carcinoembryonic antigen (CEA), cytokeratin 19
fragment (CYFRA21-1), neuron-specific enolase (NSE) and ferritin.
Continuous variables such as tumor marker levels and nodule
diameter were transformed into categorical variables: CEA (≤5.0
ng/ml as normal), CYFRA21-1 (≤3.3 ng/ml), NSE (≤17.0 ng/ml), SCC
(≤1.5 ng/ml) and ferritin (21.8-274.6 ng/ml for men, 13.0-150.0
ng/ml for women). All variable categorizations were predefined and
applied consistently across training and test sets.
Outcome definition
The primary outcome of the present study was the
presence of STAS. STAS was defined in accordance with the 2021
World Health Organization Classification of Thoracic Tumors
(7). According to this definition,
STAS is identified when free tumor cells are identified within
alveolar spaces beyond the main tumor edge, with a minimum distance
of at least two alveolar septa from the tumor margin.
All specimens were obtained from surgical resections
and independently evaluated by experienced pulmonary pathologists
blinded to clinical and imaging information. STAS+
status was assessed on hematoxylin and eosin (H&E)-stained
sections, which were prepared according to routine standardized
histopathological protocols (17),
by two experienced pulmonary pathologists using light microscopy
and was assigned only when detached tumor clusters (micropapillary,
solid nests or single cells) were clearly visible beyond the tumor
edge.
Statistical analysis and model
development
Baseline characteristics were summarized as median
and interquartile range (IQR) for continuous variables and as
counts with percentages (n; %) for categorical variables. To
develop a predictive model for STAS, five supervised ML algorithms
were used: Logistic regression, random forest, eXtreme Gradient
Boosting (XGBoost), support vector machine (SVM) and naïve Bayes
(18–22).
The dataset was randomly divided into a training and
a test set in an 8:2 ratio. Model training and evaluation were
performed using 5-fold cross-validation on the training set to
reduce random variability and prevent overfitting. Model
performance was assessed using accuracy, sensitivity, specificity
and area under the receiver operating characteristic (ROC) curve
(AUC). Calibration performance was quantified using the Brier score
(23), which measures the mean
squared difference between predicted probabilities and actual
outcomes, with lower values indicating improved probability
calibration. Mean ROC curves were computed by averaging across
folds. Decision curve analysis was conducted to assess the net
clinical benefit across various threshold probabilities. To
comprehensively assess model consistency, a Taylor diagram was
generated to evaluate the correlation coefficient, standard
deviation and root mean square error between predicted
probabilities and observed outcomes across models.
To optimize model performance, grid search with
5-fold cross-validation was applied to tune hyperparameters for all
five models on the training dataset. The complete parameter search
spaces and optimal settings for each model are summarized in
Table SI.
The optimal model was selected based on the highest
AUC in the test set. No feature selection techniques were applied
prior to model training. All categorical variables, including sex,
smoking history and radiological features, were converted into
dummy variables using one-hot encoding. Continuous variables such
as tumor marker levels had already been transformed into
categorical variables and therefore no additional normalization or
scaling was required. The model outputs were treated as continuous
probability scores and no fixed cut-off thresholds or predefined
risk categories were used.
To enhance interpretability, global SHapley Additive
exPlanations (SHAP) analysis was performed for the best-performing
model, which was selected based on the highest AUC in the test set
and further validated against using DeLong's test. Following
established methodological practices reported in recent oncology
and radiology research (24,25),
the SHAP analysis quantified the overall contribution of each
feature across the entire dataset. Summary and dependence plots
were then generated to visualize feature importance, effect
direction and potential interaction patterns.
All analyses were performed using Python (version
3.9), with implementation via the scikit-learn (https://scikit-learn.org/stable/index.html) and
XGBoost libraries (https://xgboost.ai/).
Results
Baseline characteristics
A total of 584 patients with resected NSCLC were
included, among whom 259 (44.4%) were classified as
STAS+ and 325 (55.6%) as STAS−. The median
age was 62 years (IQR, 55–69). Of the patients, 318 (54.5%) were
women and 133 (22.8%) reported a history of smoking. Most patients
were classified as T1 stage (n=467, 80.0%), with fewer at T2
(n=112, 19.1%) and T3 stages (n=5, 0.9%). Common radiological signs
included lobulation (n=353, 60.5%), spicule sign (n=334, 57.2%) and
vacuolar sign (n=84, 14.4%). Nodule types were primarily subsolid
(n=326, 55.8%), followed by solid (n=199, 34.1%) and pure
ground-glass nodules (n=59, 10.1%). The most frequent tumor
locations were the right upper (n=207, 35.4%) and left upper lobes
(n=153, 26.2%). Abnormal levels of tumor markers were present in a
minority of patients: CYFRA21-1 (n=113, 19.4%), NSE (n=42, 7.2%),
CEA (n=53, 9.1%), SCC (n=20, 3.4%) and ferritin (n=93, 15.9%).
Detailed baseline characteristics are summarized in Table I.
 | Table I.Baseline characteristics of the
present study population (n=584). |
Table I.
Baseline characteristics of the
present study population (n=584).
| Variable | Patients |
|---|
| Age, years
(range) | 62 (55–69) |
| Sex |
|
|
Female | 318 (54.5) |
| Male | 266 (45.5) |
| Smoking history
(Yes) | 133 (22.8) |
| T staging |
|
| T1 | 467 (80.0) |
| T2 | 112 (19.1) |
| T3 | 5 (0.9) |
| Lobulation
(Yes) | 353 (60.5) |
| Spicule sign
(Yes) | 334 (57.2) |
| Vacuolar sign
(Yes) | 84 (14.4) |
| Tumor location |
|
| Right
lower lung | 103 (17.6) |
| Right
upper lung | 207 (35.5) |
| Right
middle lung | 32 (5.5) |
| Left
lower lung | 89 (15.2) |
| Left
upper lung | 153 (26.2) |
| Nodule type |
|
|
Subsolid | 326 (55.8) |
|
Solid | 199 (34.1) |
|
Ground-glass | 59 (10.1) |
| SCC |
|
|
Abnormal | 20 (3.4) |
|
Normal | 564 (96.6) |
| CEA |
|
|
Abnormal | 53 (9.1) |
|
Normal | 531 (90.9) |
| CYFRA21-1 |
|
|
Abnormal | 113 (19.4) |
|
Normal | 471 (80.6) |
| NSE |
|
|
Abnormal | 42 (7.2) |
|
Normal | 542 (92.8) |
| Ferritin |
|
|
Abnormal | 93 (15.9) |
|
Normal | 491 (84.1) |
| STAS |
|
|
Positive | 259 (44.4) |
|
Negative | 325 (55.6) |
Model performance evaluation
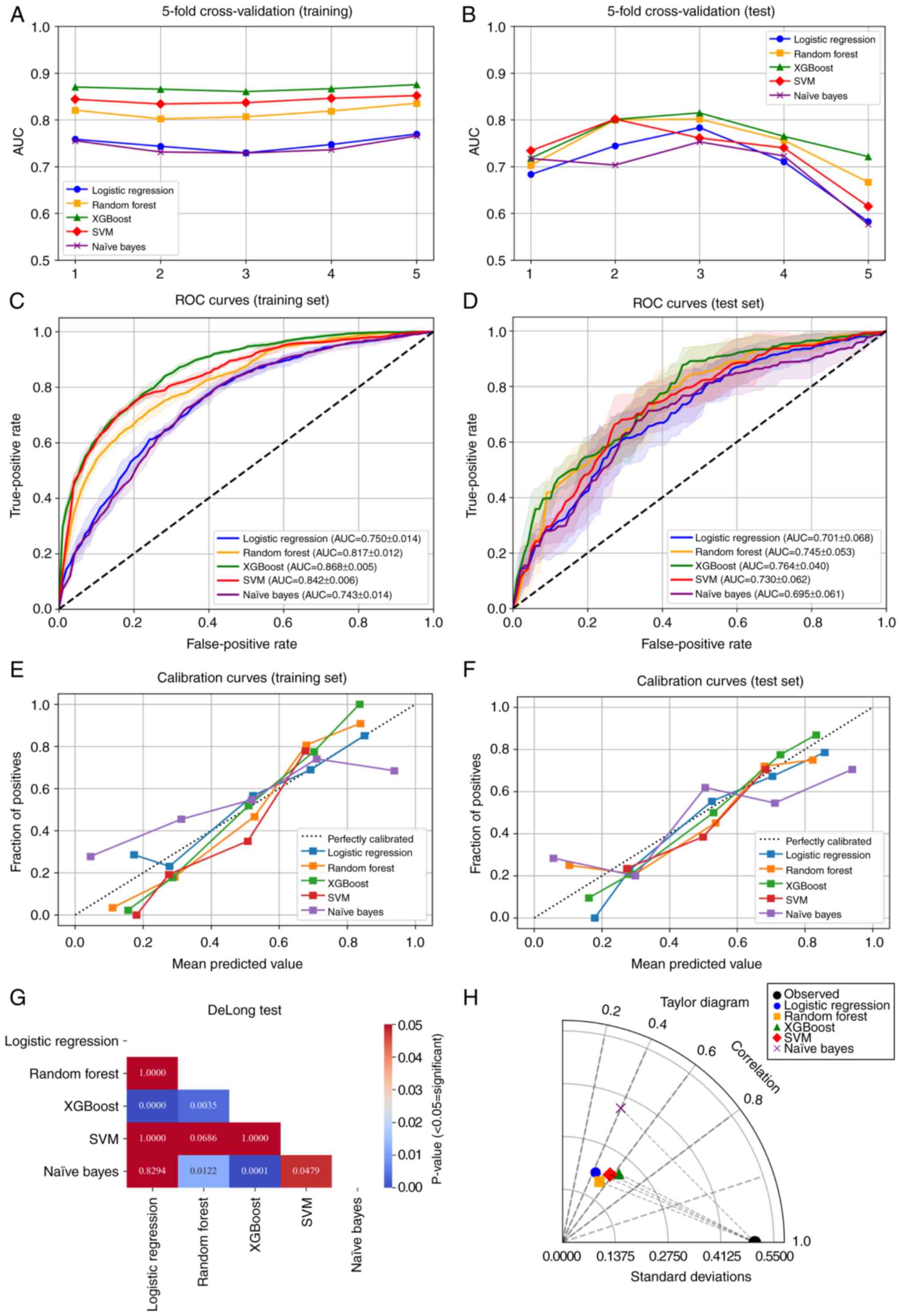
In the training set, 5-fold cross-validation
demonstrated stable performance (Fig.
1A) and the model generalized well to the test set (Fig. 1B). Among the five algorithms,
XGBoost demonstrated the best overall performance, achieving the
highest mean AUC across five cross-validation folds in the training
set (0.868; 95% CI, 0.860-0.875; Fig.
1C) and maintaining robust performance in the test set (mean
AUC =0.764; 95% CI, 0.718-0.815; Fig.
1D). In addition to the highest AUC, XGBoost exhibited balanced
accuracy, sensitivity and specificity and achieved the lowest Brier
scores (0.158 in the training set and 0.197 in the test set),
indicating notably increased discrimination and calibration among
the tested models (Table II).
 | Table II.Performance of machine learning
models. |
Table II.
Performance of machine learning
models.
| Model | Dataset | Accuracy | Sensitivity | Specificity | AUC (95% CI) | Brier score |
|---|
| Logistic
regression | Training | 0.685 | 0.724 | 0.647 | 0.750
(0.729-0.770) | 0.203 |
|
| Test | 0.638 | 0.677 | 0.600 | 0.701
(0.582-0.783) | 0.223 |
| Random forest | Training | 0.726 | 0.774 | 0.677 | 0.817
(0.802-0.835) | 0.182 |
|
| Test | 0.680 | 0.733 | 0.630 | 0.745
(0.667-0.802) | 0.206 |
| XGBoost | Training | 0.775 | 0.743 | 0.808 | 0.868
(0.860-0.875) | 0.157 |
|
| Test | 0.680 | 0.657 | 0.700 | 0.764
(0.718-0.815) | 0.197 |
| Support vector
machine | Training | 0.751 | 0.819 | 0.682 | 0.842
(0.834-0.852) | 0.173 |
|
| Test | 0.669 | 0.751 | 0.588 | 0.730
(0.615-0.801) | 0.212 |
| Naïve Bayes | Training | 0.677 | 0.636 | 0.718 | 0.743
(0.729-0.766) | 0.240 |
|
| Test | 0.660 | 0.621 | 0.700 | 0.695
(0.576-0.753) | 0.269 |
DeLong's tests demonstrated that XGBoost yielded
significantly higher AUCs compared with logistic regression, naïve
Bayes and random forest (all P<0.05), whereas its difference
with SVM did not reach statistical significance (P>0.05;
Fig. 1G). Calibration curves
(Fig. 1E and F) further
demonstrated closer agreement between predicted probabilities and
observed outcomes for XGBoost. The Taylor diagram demonstrated that
the XGBoost model was positioned closest to the reference point,
indicating the highest overall agreement with the observed data
among all algorithms (Fig. 1H).
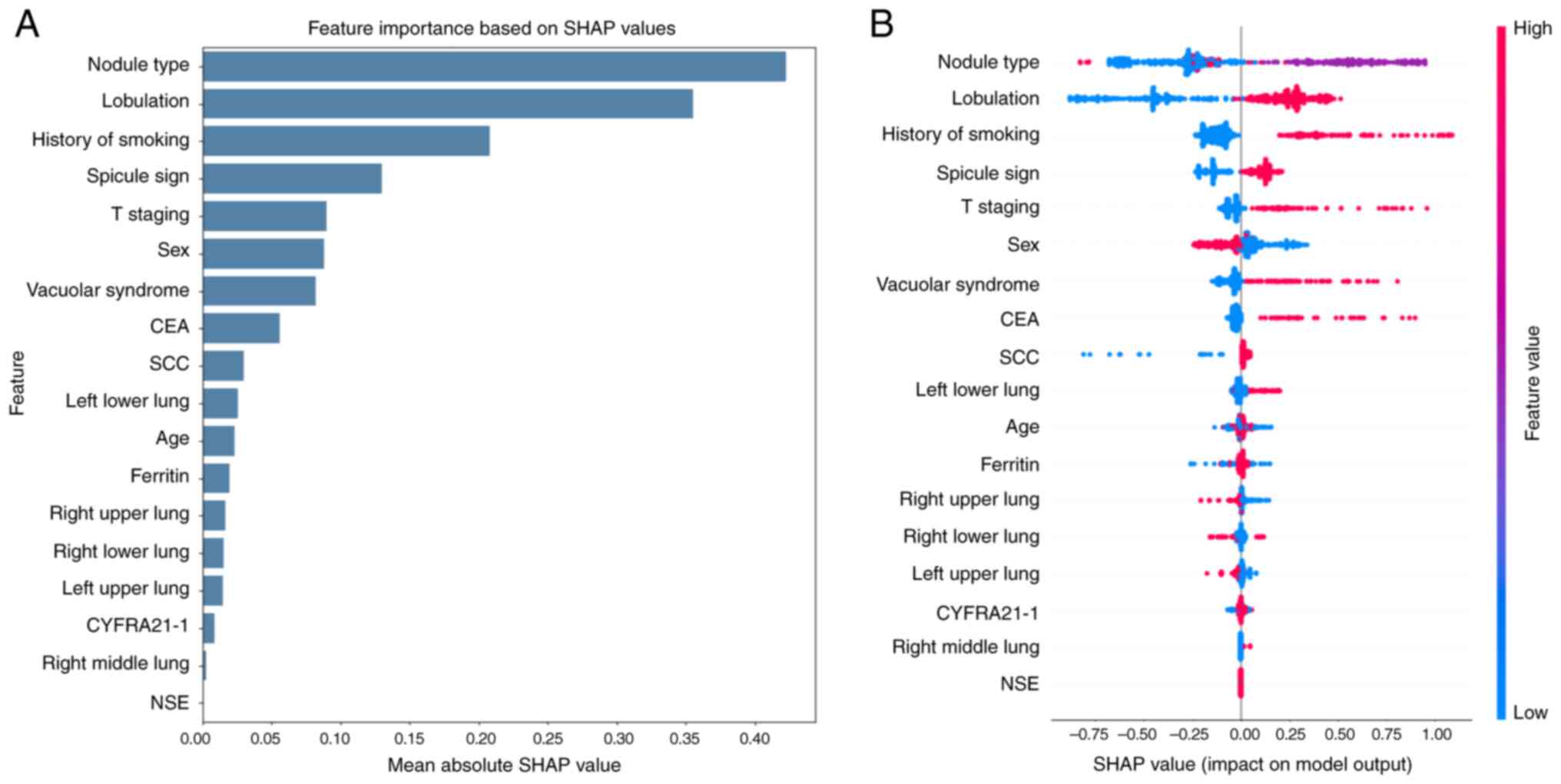
Feature importance analysis
To enhance model interpretability, the present study
employed SHAP to analyze the contribution of individual features in
the XGBoost model. The feature importance plot (Fig. 2A) demonstrated that the nodule type,
lobulation and smoking history were the top three predictors
contributing most to the decision-making of the model. The Beeswarm
plot (Fig. 2B) further illustrated
the direction and magnitude of the effect of each feature. For
example, solid nodule types and presence of lobulation positively
contributed to STAS prediction. The history of smoking was also
associated with higher predicted risk. By contrast, features such
as NSE and CYFRA21-1 exhibited minimal contribution.
Discussion
In the present study, an XGBoost-based model was
developed using real-world preoperative data to predict STAS in
surgically resectable NSCLC. Unlike previous approaches that rely
on postoperative pathology or advanced imaging techniques, the
present study model enables early, non-invasive prediction of STAS
and provides thoracic surgeons with a valuable decision-support
tool to optimize preoperative surgical planning.
Methodologically, to enhance model robustness,
reduce selection bias and determine the most appropriate algorithm
for STAS prediction under real-world clinical constraints, the
present study evaluated five distinct ML models. Among them,
XGBoost consistently outperformed the others on both the training
(AUC=0.868) and test (AUC=0.764) sets. As a gradient boosting
framework, XGBoost is particularly adept at capturing complex
non-linear relationships and feature interactions, and has
demonstrated robust performance across a wide range of clinical
diagnostic and prognostic tasks (26–28).
These prior findings were consistent with the present study
results, further supporting the suitability of XGBoost in the
context of oncology. Therefore, the present study ultimately
selected XGBoost as the final model for STAS prediction in this
study.
In terms of predictive findings, the present study
confirmed and extended existing knowledge regarding radiological
and clinical associations of STAS, while offering improved
generalizability and clinical practicality. Several prior studies
have investigated the association between STAS and radiological or
pathological features. For example, radiomics-based models have
achieved high AUCs by incorporating handcrafted features and
clinical data, but often suffer from limited generalizability due
to complex processing pipelines (29). Large retrospective analyses have
also associated STAS with programmed cell death-ligand 1
expression, spiculation and lobulated margins, yet these features
alone provide inconsistent predictive power (30). A recent study using an XGBoost and
SHAP framework in lung adenocarcinoma reported higher AUCs and
identified similar predictive features, such as nodule type and
lobulation, but relied heavily on radiomic features and was
restricted to a single histological subtype (31). By contrast, the present study model
offers broader applicability across NSCLC subtypes and requires
only routinely collected variables, making it more feasible for
real-world deployment.
Building on the performance of the model, the
contribution of individual features to STAS prediction was further
explored. The strong predictive value of nodule type and lobulation
aligns with prior studies that have consistently associated these
semantic imaging features with histological patterns associated
with STAS. Specifically, previous studies have reported that solid
and part-solid nodules, compared with pure ground-glass nodules,
are more frequently associated with aggressive histological
subtypes such as micropapillary and solid patterns (32,33).
These lesions typically reflect higher tumor cellularity and
invasiveness, making them more prone to tumor cell dissemination
into peripheral airspaces (34).
Lobulation, another key predictive feature, similarly corresponds
with biological plausibility. Radiologically, lobulated tumor
margins indicate an irregular and heterogeneous tumor-stromal
interface, which may reflect underlying extracellular matrix
remodeling, neoangiogenesis and disrupted cell adhesion at the
invasive front. These alterations facilitate tumor cell detachment
and alveolar spread, thereby elevating STAS risk (35–37).
This interpretation is supported by multiple histopathological
studies that have associated lobulated tumors with increased
invasiveness and worse clinical outcomes (38,39).
Smoking history was also identified as a key
predictor in the present study model. A recent predictive modeling
study involving 1,212 patients with clinical stage IA lung
adenocarcinoma (15) reported that
smoking history was a consistent and independent predictor of STAS
in both logistic regression models, with one achieving an AUC of
0.807 (40). Biologically, smoking
contributes to NSCLC pathogenesis through mechanisms such as
genomic instability (for example, TP53 mutations), chronic
inflammation and epithelial-mesenchymal transition (41–43),
all of which promote tumor invasiveness and may facilitate airspace
spread. These findings reinforce the interpretability and clinical
relevance of the present study model. Furthermore, smoking history
is easily obtainable in routine preoperative evaluations, further
supporting its utility as a practical feature in STAS risk
prediction.
In contrast to the strong predictive signals from
imaging and clinical factors, conventional serum tumor markers
including CEA, CYFRA21-1 and NSE, contributed minimally to STAS
prediction in the present study model. This finding aligns with
previous studies, some of which included CEA in their models but
reported non-significant associations with STAS status (P=0.359) or
low odds ratios, suggesting limited discriminatory power (44,45).
Although one study identified CEA as part of a combined
clinical-radiological nomogram, its individual predictive value was
relatively weak compared with radiological features such as density
type and the distal ribbon sign (35). From a biological perspective, these
serum markers primarily reflect systemic tumor burden rather than
specific patterns of local invasion such as STAS (46). Furthermore, their serum levels are
subject to variation due to non-tumoral factors and often lack
specificity in early-stage disease. The present study findings thus
reinforce the utility of readily accessible imaging features and
clinical variables over traditional tumor markers for the
preoperative risk assessment of STAS.
Despite these promising findings, several
limitations should be acknowledged. First, this was a retrospective
study based on a single-center cohort, which may limit the
generalizability of the present study model. External validation
using multicenter datasets with broader population diversity is
necessary to confirm its robustness. Second, although the present
study selected clinically accessible features, the interpretation
of certain CT signs (for example, lobulation) may be subject to
inter-observer variability. Future research incorporating automated
image analysis or radiomics-based augmentation may further enhance
reproducibility. Third, while SHAP plots improved model
transparency, the current model still lacks direct histological
validation of the features associated with STAS, which would be
valuable for biological interpretability. Lastly, the present study
dataset did not include genomic or molecular data, which might
contribute additional predictive value and deserves exploration in
future studies.
In conclusion, the present study developed an
interpretable XGBoost model using routinely available preoperative
clinical and semantic CT features to predict STAS in surgically
resectable NSCLC. The model demonstrated favorable predictive
performance and identified clinically notable features such as
nodule type, lobulation and smoking history. By enabling early,
non-invasive risk stratification, this approach may assist thoracic
surgeons in tailoring surgical strategies prior to resection.
Future prospective and multicenter studies are warranted to
externally validate the model and facilitate its potential
integration into clinical workflows.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CY and YY conceived and designed the study. CY
collected and analyzed the data. GD, BZ, LX and FC contributed to
data interpretation and key revision of the manuscript. CY drafted
the initial manuscript. YY supervised the study and revised the
manuscript for key intellectual content. CY and YY confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Anqing Municipal Hospital (approval no. 2025-152;
Anqing, China). Written informed consent was obtained from all
participants prior to data collection and all patient data were
anonymized before analysis to protect privacy.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hendriks LEL, Remon J, Faivre-Finn C,
Garassino MC, Heymach JV, Kerr KM, Tan DSW, Veronesi G and Reck M:
Non-small-cell lung cancer. Nat Rev Dis Primers. 10:712024.
View Article : Google Scholar
|
|
2
|
Rathore K, Weightman W, Palmer K, Hird K
and Joshi P: Survival analysis of early-stage NSCLC patients
following lobectomy: Impact of surgical techniques and other
variables on long-term outcomes. Heart Lung Circ. 34:639–646. 2025.
View Article : Google Scholar
|
|
3
|
Lou F, Sima CS, Rusch VW, Jones DR and
Huang J: Differences in patterns of recurrence in early-stage
versus locally advanced non-small cell lung cancer. Ann Thorac
Surg. 98:1755–1761. 2014. View Article : Google Scholar
|
|
4
|
Xie H, Su H, Zhu E, Gu C, Zhao S, She Y,
Ren Y, Xie D, Zheng H, Wu C, et al: Morphological subtypes of tumor
spread through air spaces in non-small cell lung cancer: Prognostic
heterogeneity and its underlying mechanism. Front Oncol.
11:6083532021. View Article : Google Scholar
|
|
5
|
Han YB, Kim H, Mino-Kenudson M, Cho S,
Kwon HJ, Lee KR, Kwon S, Lee J, Kim K, Jheon S, et al: Tumor spread
through air spaces (STAS): Prognostic significance of grading in
non-small cell lung cancer. Mod Pathol. 34:549–561. 2021.
View Article : Google Scholar
|
|
6
|
Kutlay C, Gülhan SŞE, Acar LN, Aslan M and
Tanrıkulu FB: Impact of spread through air spaces (STAS) and
lymphovascular invasion (LVI) on prognosis in NSCLC: A
comprehensive pathological evaluation. Updates Surg. 77:1205–1213.
2025. View Article : Google Scholar
|
|
7
|
Nicholson AG, Tsao MS, Beasley MB, Borczuk
AC, Brambilla E, Cooper WA, Dacic S, Jain D, Kerr KM, Lantuejoul S,
et al: The 2021 WHO classification of lung tumors: Impact of
advances since 2015. J Thorac Oncol. 17:362–387. 2022. View Article : Google Scholar
|
|
8
|
Liu C, Meng A, Xue XQ, Wang YF, Jia C, Yao
DP, Wu YJ, Huang Q, Gong P and Li XF: Prediction of early lung
adenocarcinoma spread through air spaces by machine learning
radiomics: A cross-center cohort study. Transl Lung Cancer Res.
13:3443–3459. 2024. View Article : Google Scholar
|
|
9
|
Wang X, Ma C, Jiang Q, Zheng X, Xie J, He
C, Gu P, Wu Y, Xiao Y and Liu S: Performance of deep learning model
and radiomics model for preoperative prediction of spread through
air spaces in the surgically resected lung adenocarcinoma: A
two-center comparative study. Transl Lung Cancer Res. 13:3486–3499.
2024. View Article : Google Scholar
|
|
10
|
Swanson K, Wu E, Zhang A, Alizadeh AA and
Zou J: From patterns to patients: Advances in clinical machine
learning for cancer diagnosis, prognosis, and treatment. Cell.
186:1772–1791. 2023. View Article : Google Scholar
|
|
11
|
Zhang B, Shi H and Wang H: Machine
learning and AI in cancer prognosis, prediction, and treatment
selection: A critical approach. J Multidiscip Healthc.
16:1779–1791. 2023. View Article : Google Scholar
|
|
12
|
Zou Y, Mao Q, Zhao Z, Zhou X, Pan Y, Zuo Z
and Zhang W: Intratumoural and peritumoural CT-based radiomics for
diagnosing lepidic-predominant adenocarcinoma in patients with pure
ground-glass nodules: A machine learning approach. Clin Radiol.
79:e211–e218. 2024. View Article : Google Scholar
|
|
13
|
Parisineni SRA and Pal M: Enhancing trust
and interpretability of complex machine learning models using local
interpretable model agnostic shap explanations. Int J Data Sci
Anal. 18:457–466. 2023. View Article : Google Scholar
|
|
14
|
Li Y, Ding J, Wu K, Qi W, Lin S, Chen G
and Zuo Z: Ensemble machine learning classifiers combining CT
radiomics and clinical-radiological features for preoperative
prediction of pathological invasiveness in lung adenocarcinoma
presenting as part-solid nodules: A multicenter retrospective
study. Technol Cancer Res Treat. 24:153303382513513652025.
View Article : Google Scholar
|
|
15
|
Fong KM, Rosenthal A, Giroux DJ, Nishimura
KK, Erasmus J, Lievens Y, Marino M, Marom EM, Putora PM, Singh N,
et al: The international association for the study of lung cancer
staging project for lung cancer: Proposals for the revision of the
M descriptors in the forthcoming ninth edition of the TNM
classification for lung cancer. J Thorac Oncol. 19:786–802. 2024.
View Article : Google Scholar
|
|
16
|
Zuo Z, Wang P, Zeng W, Qi W and Zhang W:
Measuring pure ground-glass nodules on computed tomography:
Assessing agreement between a commercially available deep learning
algorithm and radiologists' readings. Acta Radiol. 64:1422–1430.
2023. View Article : Google Scholar
|
|
17
|
Dunn C, Brettle D, Cockroft M, Keating E,
Revie C and Treanor D: Quantitative assessment of H&E staining
for pathology: Development and clinical evaluation of a novel
system. Diagn Pathol. 19:422024. View Article : Google Scholar
|
|
18
|
Christodoulou E, Ma J, Collins GS,
Steyerberg EW, Verbakel JY and van Calster B: A systematic review
shows no performance benefit of machine learning over logistic
regression for clinical prediction models. J Clin Epidemiol.
110:12–22. 2019. View Article : Google Scholar
|
|
19
|
Venkat V, Clark K, Jeng XJ, Yao TC, Tsai
HJ, Lu TP, Hsiao TH, Lin CH, Holloway S, Hoyo C, et al: Exploring
random forest in genetic risk score construction. Genet Epidemiol.
49:e700222025. View Article : Google Scholar
|
|
20
|
Kim KS, Yoon TJ, Ahn J and Ryu JA:
Development and validation of a machine learning model for early
prediction of acute kidney injury in neurocritical care: A
comparative analysis of XGBoost, GBM, and random forest algorithms.
Diagnostics (Basel). 15:20612025. View Article : Google Scholar
|
|
21
|
Byvatov E and Schneider G: Support vector
machine applications in bioinformatics. Appl Bioinformatics.
2:67–77. 2003.
|
|
22
|
Zhang J, Hao L, Xu Q and Gao F: Radiomics
and clinical characters based gaussian naive bayes (GNB) model for
preoperative differentiation of pulmonary pure invasive mucinous
adenocarcinoma from mixed mucinous adenocarcinoma. Technol Cancer
Res Treat. 23:153303382412584152024. View Article : Google Scholar
|
|
23
|
Yang W, Jiang J, Schnellinger EM, Kimmel
SE and Guo W: Modified Brier score for evaluating prediction
accuracy for binary outcomes. Stat Methods Med Res. 31:2287–2296.
2022. View Article : Google Scholar
|
|
24
|
Ling T, Zuo Z, Huang M, Wu L, Ma J, Huang
X and Tang W: Prediction of mucinous adenocarcinoma in colorectal
cancer with mucinous components detected in preoperative biopsy
diagnosis. Abdom Radiol (NY). 50:2794–2805. 2025. View Article : Google Scholar
|
|
25
|
Ling T, Zuo Z, Huang M, Ma J and Wu L:
Stacking classifiers based on integrated machine learning model:
Fusion of CT radiomics and clinical biomarkers to predict lymph
node metastasis in locally advanced gastric cancer patients after
neoadjuvant chemotherapy. BMC Cancer. 25:8342025. View Article : Google Scholar
|
|
26
|
Zhou Y, Zhao J, Zou F, Tan Y, Zeng W,
Jiang J, Hu J, Zeng Q, Gong L, Liu L and Zhong L: Interpretable
machine learning models based on body composition and inflammatory
nutritional index (BCINI) to predict early postoperative recurrence
of colorectal cancer: Multi-center study. Comput Methods Programs
Biomed. 269:1088742025. View Article : Google Scholar
|
|
27
|
Yang M, Chen Y, Zhou X, Yu R, Huang N and
Chen J: Machine learning models for prediction of NPVR ≥80% with
HIFU ablation for uterine fibroids. Int J Hyperthermia.
42:24737542025. View Article : Google Scholar
|
|
28
|
Lü X, Wang C, Tang M, Li J, Xia Z, Fan S,
Jin Y and Yang Z: Pinpointing potent hits for cancer immunotherapy
targeting the TIGIT/PVR pathway using the XGBoost model,
centroid-based virtual screening, and MD simulation. Comput Biol
Chem. 118:1084502025. View Article : Google Scholar
|
|
29
|
Chen S, Wang X, Lin X, Li Q, Xu S, Sun H,
Xiao Y, Fan L and Liu S: CT-based radiomics predictive model for
spread through air space of IA stage lung adenocarcinoma. Acta
Radiol. 66:477–486. 2025. View Article : Google Scholar
|
|
30
|
Wang S, Xu M, Liu Y, Hou X, Gao Z, Sun J
and Shen L: PD-L1 expression and its association with
clinicopathological and computed tomography features in surgically
resected non-small cell lung cancer: A retrospective cohort study.
Sci Rep. 15:243232025. View Article : Google Scholar
|
|
31
|
Wang P, Cui J, Du H, Qian Z, Zhan H, Zhang
H, Ye W, Meng W and Bai R: Preoperative prediction of STAS risk in
primary lung adenocarcinoma using machine learning: An
interpretable model with SHAP analysis. Acad Radiol. 32:4266–4277.
2025. View Article : Google Scholar
|
|
32
|
Su Y, Tao J, Lan X, Liang C, Huang X,
Zhang J, Li K and Chen L: CT-based intratumoral and peritumoral
radiomics nomogram to predict spread through air spaces in lung
adenocarcinoma with diameter ≤ 3 cm: A multicenter study. Eur J
Radiol Open. 14:1006302025. View Article : Google Scholar
|
|
33
|
Xie M, Gao J, Ma X, Wu C, Zang X, Wang Y,
Deng H, Yao J, Sun T, Yu Z, et al: Consolidation radiographic
morphology can be an indicator of the pathological basis and
prognosis of partially solid nodules. BMC Pulm Med. 22:3692022.
View Article : Google Scholar
|
|
34
|
Zhang X, Qiao W, Shen J, Jiang Q, Pan C,
Wang Y, Bidzińska J, Dai F and Zhang L: Clinical, pathological, and
computed tomography morphological features of lung cancer with
spread through air spaces. Transl Lung Cancer Res. 13:2802–2812.
2024. View Article : Google Scholar
|
|
35
|
Wang Y, Lyu D, Zhang D, Hu L, Wu J, Tu W,
Xiao Y, Fan L and Liu S: Nomogram based on clinical characteristics
and radiological features for the preoperative prediction of spread
through air spaces in patients with clinical stage IA non-small
cell lung cancer: A multicenter study. Diagn Interv Radiol.
29:771–785. 2023. View Article : Google Scholar
|
|
36
|
Liu X, Ding Y, Ren J, Li J, Wang K, Sun S,
Zhang W, Xu M, Jing Y, Gao G, et al: Analysis of factors affecting
the diagnostic efficacy of frozen sections for tumor spread through
air spaces in lung adenocarcinoma. Cancers (Basel). 17:21682025.
View Article : Google Scholar
|
|
37
|
Qi L, Li X, He L, Cheng G, Cai Y, Xue K
and Li M: Comparison of diagnostic performance of spread through
airspaces of lung adenocarcinoma based on morphological analysis
and perinodular and intranodular radiomic features on chest CT
images. Front Oncol. 11:6544132021. View Article : Google Scholar
|
|
38
|
Warth A, Muley T, Kossakowski CA, Goeppert
B, Schirmacher P, Dienemann H and Weichert W: Prognostic impact of
intra-alveolar tumor spread in pulmonary adenocarcinoma. Am J Surg
Pathol. 39:793–801. 2015. View Article : Google Scholar
|
|
39
|
Shi J, Xu K, Liu X, Shi M, Ji C and Ye B:
Anaplastic lymphoma kinase rearrangement and tumor spread through
air spaces is associated with worse clinical outcomes for resected
stage IA lung adenocarcinoma. Clin Lung Cancer.
S1525-7304(25)00222-0. 2025.(Epub ahead of print). View Article : Google Scholar
|
|
40
|
Huang G, Wang L, Zhao Z, Wang Y, Li B,
Huang Z, Yu X, Liang N and Li S: Development and internal
validation of predictive models for spread through air spaces in
clinical stage IA lung adenocarcinoma. Gen Thorac Cardiovasc Surg.
Apr 28–2025.(Epub ahead of print). View Article : Google Scholar
|
|
41
|
Fu H, Liu K, Zheng Y, Zhao J, Xie T and
Ding Y: Upregulation of ARHGAP18 by miR-613 inhibits cigarette
smoke extract-induced apoptosis and epithelial-mesenchymal
transition in bronchial epithelial cells. Int J Chron Obstruct
Pulmon Dis. 20:2525–2537. 2025. View Article : Google Scholar
|
|
42
|
Díaz-Gay M, Zhang T, Hoang PH, Leduc C,
Baine MK, Travis WD, Sholl LM, Joubert P, Khandekar A, Zhao W, et
al: The mutagenic forces shaping the genomes of lung cancer in
never smokers. Nature. 644:133–144. 2025. View Article : Google Scholar
|
|
43
|
Zhang W, Chen B, Zhao C, Yang D, Shima M,
Fan W, Yoda Y, Li S, Guo C, Chen Y, et al: Personal exposure to
PM2.5 and O3 induced heterogeneous
inflammatory responses and modifying effects of smoking: A
prospective panel study in COPD patients. J Hazard Mater.
494:1384712025. View Article : Google Scholar
|
|
44
|
Wang J, Yao Y, Tang D and Gao W: An
individualized nomogram for predicting and validating spread
through air space (STAS) in surgically resected lung
adenocarcinoma: A single center retrospective analysis. J
Cardiothorac Surg. 18:3372023. View Article : Google Scholar
|
|
45
|
Yang Y, Li L, Hu H, Zhou C, Huang Q, Zhao
J, Duan Y, Li W, Luo J, Jiang J, et al: A nomogram integrating the
clinical and CT imaging characteristics for assessing spread
through air spaces in clinical stage IA lung adenocarcinoma. Front
Immunol. 16:15197662025. View Article : Google Scholar
|
|
46
|
van den Heuvel M, Holdenrieder S,
Schuurbiers M, Cigoianu D, Trulson I, van Rossum H and Lang D:
Serum tumor markers for response prediction and monitoring of
advanced lung cancer: A review focusing on immunotherapy and
targeted therapies. Tumour Biol. 46((S1)): S233–S268. 2023.
|