Introduction
MZL is an indolent B-cell neoplasm thought to
originate from post-germinal center marginal zone B cells, commonly
presenting in the spleen, lymph nodes and mucosa-associated
lymphoid tissues (MALT) (1).
Histologically, the neoplastic cells typically display a
monomorphic population of small-to-medium lymphocytes with slightly
irregular nuclei, inconspicuous nucleoli and moderate amounts of
pale cytoplasm (2,3). The pathogenesis of MZL is closely
linked to chronic antigenic stimulation, frequently driven by
infectious agents such as Helicobacter pylori in gastric
MALT lymphoma. Additionally, recurrent molecular aberrations,
including somatic hypermutation of immunoglobulin genes, specific
chromosomal abnormalities [e.g., t(11;18), trisomies 3 and 18, and
deletion 6q23], and mutations in genes such as Notch receptor 2
(NOTCH2), Kruppel-like factor 2 (KLF2), TNF alpha-induced protein 3
and TBL1X/Y related 1, are critical contributors to lymphomagenesis
and disease progression (4,5). Current frontline therapies primarily
employ anti-CD20 monoclonal antibodies (e.g., rituximab), often
combined with chemotherapy regimens (e.g., bendamustine or CHOP
variants), which substantially improve response rates and survival
outcomes (6). Nevertheless,
inherent and acquired chemoresistance and frequent disease relapse
persist as significant clinical challenges, limiting long-term
curability and highlighting the need for innovative therapeutic
approaches (7). Consequently,
targeting molecular pathways beyond conventional chemotherapy
represents a pivotal research direction. In this context,
dysregulated autophagy, a conserved lysosomal degradation pathway
essential for maintaining cellular homeostasis, and its therapeutic
potential have emerged as significant research areas in oncology.
Aberrant expression of key autophagy-related proteins has been
documented across multiple malignancies, including lymphoma
(8), ovarian (9), lung (10), breast (11) and colorectal cancers (12), suggesting that this pathway may
represent a therapeutic target. MZL constitutes roughly 5–10% of
non-Hodgkin lymphoma cases globally, with an annual incidence
estimated at 2–3 per 100,000 population (13,14).
Although considered indolent, MZL typically follows a chronic,
relapsing clinical course, characterized by median overall survival
exceeding 10 years but frequent recurrence following conventional
immunochemotherapy. Relapsed or refractory disease remains
therapeutically challenging, emphasizing the urgent need for novel
molecular targets and more effective treatments (5,15).
Autophagy dysregulation has been implicated in
various lymphoma subtypes, including diffuse large B-cell lymphoma
(DLBCL), mantle cell lymphoma (MCL) and follicular lymphoma (FL),
where reduced Beclin-1 or microtubule-associated protein
1A/1B-light chain 3 (LC3) expression and increased accumulation of
p62 correlate with adverse prognosis and chemoresistance (16,17).
These findings indicate that autophagy dysfunction may be a shared
oncogenic mechanism across B-cell malignancies. Clinically, several
agents targeting autophagy and apoptosis pathways, such as
chloroquine, hydroxychloroquine and the Bcl-2 inhibitor venetoclax,
have demonstrated encouraging efficacy in preclinical studies and
early-phase clinical trials (18),
further highlighting the therapeutic relevance of targeting this
pathway. Although abnormal expression of primary
autophagy-associated proteins such as Beclin-1, LC3 and p62 has
been well-characterized in lymphomas like DLBCL, MCL and FL
(19–21), their expression patterns in MZL have
not yet been extensively studied. To bridge this knowledge gap and
evaluate the therapeutic implications of autophagy modulation in
MZL, this research examined the expression levels of crucial
autophagy indicators, including Beclin-1, LC3, sequestosome
(SQSTM)1/p62 and Bcl-2, in clinical tissue samples obtained from
patients diagnosed with MZL.
Patients and methods
Study and control cohorts
The demographic and clinicopathological data of the
patients are summarized in Table I.
MZL cohort: Formalin-fixed, paraffin-embedded (FFPE) tissue blocks
were analyzed from 16 patients with newly diagnosed,
treatment-naïve MZL (11 males, 5 females; median age, 64.5 years;
range, 45–78 years). All specimens were collected at initial
diagnosis between January 2014 and December 2024 from Shandong
Provincial Hospital Affiliated to Shandong First Medical University
(Jinan, Shandong). Diagnoses were strictly confirmed according to
current World Health Organization lymphoma classification criteria
(1). Patients with previous
radiotherapy or chemotherapy exposure were excluded. Subtypes
included 10 nodal MZL, 4 MALT lymphomas and 2 splenic MZL. MZL
cases were retrospectively identified from the pathology archives,
and all newly diagnosed, treatment-naïve patients with adequate
FFPE tissue and complete baseline data during the study period were
included. The control cohort consisted of FFPE tissue blocks from
16 patients with RLH (12 males, 4 females; median age, 62 years;
range, 50–75 years). RLH cases were consecutively identified from
the pathology archives of the same center during the same period
based on the availability of treatment-naïve FFPE tissue blocks,
rather than being artificially matched to the MZL group. By
including all eligible MZL and RLH cases from the same institution
and time window, rather than arbitrarily selecting or artificially
matching controls, the risk of selection bias was minimized.
Baseline demographic and clinicopathological characteristics,
including age, sex distribution and involved sites, did not differ
significantly between the MZL and RLH groups.
 | Table I.Demographic and clinicopathological
characteristics of the study cohort. |
Table I.
Demographic and clinicopathological
characteristics of the study cohort.
| Characteristic | MZL (n=16) | RLH (n=16) |
|---|
| Sex |
|
|
|
Male | 11 | 12 |
|
Female | 5 | 4 |
| Median age, years
(range) | 64.5 (45–78) | 62 (50–75) |
| Pathological
diagnosis |
|
|
|
MALT | 4 | - |
|
NMZL | 10 | - |
|
SMZL | 2 | - |
Immunohistochemistry (IHC)
Antibodies and reagents
Anti-Beclin-1 antibody (cat. no. ab207612; Abcam)
was used at a dilution of 1:100. Anti-SQSTM1/p62 antibody (cat. no.
ab109012; Abcam) was used at a dilution of 1:2,000. LC3A/LC3B
polyclonal antibody (cat. no. ab109012; Abcam) was used at a
dilution of 1:200. Anti-Bcl-2 antibody (cat. no. ab2137583; Abcam)
was used at a dilution of 1:500. HRP-conjugated goat anti-rabbit
IgG (H+L) secondary antibody (cat. no. ab205718; Abcam) was applied
at a dilution of 1:1,000. A diaminobenzidine (DAB) chromogen kit
(cat. no. DA1010; Beijing Solarbio Science & Technology Co.,
Ltd.), 5% bovine serum albumin (BSA; Wuhan Boster Biological
Technology, Ltd.) and 3% H2O2 (Wuhan Boster
Biological Technology, Ltd.) were also utilized.
Experimental procedures
Tissue sections were first baked, deparaffinized,
rehydrated and washed with PBS. Antigen retrieval was then
performed via high-pressure treatment using EDTA buffer, after
which slides were cooled to ambient temperature. Blocking of
endogenous peroxidase activity was achieved by treating sections
with 3% H2O2, and non-specific
antigen-antibody interactions were minimized by incubating slides
in BSA. Primary antibodies were applied overnight at 4°C, and
subsequently, slides underwent washing in PBS before incubation at
37°C for 40 min with secondary antibodies conjugated to HRP.
Following PBS washes, staining was visualized using DAB chromogen,
counterstained with hematoxylin, differentiated and blued. Finally,
slides were mounted with coverslips and neutral gum prior to
imaging.
Quantitative analysis
Pathological assessments were independently
performed by two blinded, experienced pathologists using optical
microscopy (magnification, ×400). For each specimen, five
non-overlapping, randomly selected high-power fields were used for
semiquantitative analysis. Protein expression levels were
quantified using ImageJ software (1.54p; National Institutes of
Health) to calculate the average optical density (AOD): AOD=Σ
integrated optical density/Σ positive pixel area.
Statistical analysis
All statistical analyses were carried out using
GraphPad Prism version 9.0 (Dotmatics). Expression differences of
autophagy-related proteins between groups were assessed by applying
unpaired two-tailed Student's t-tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
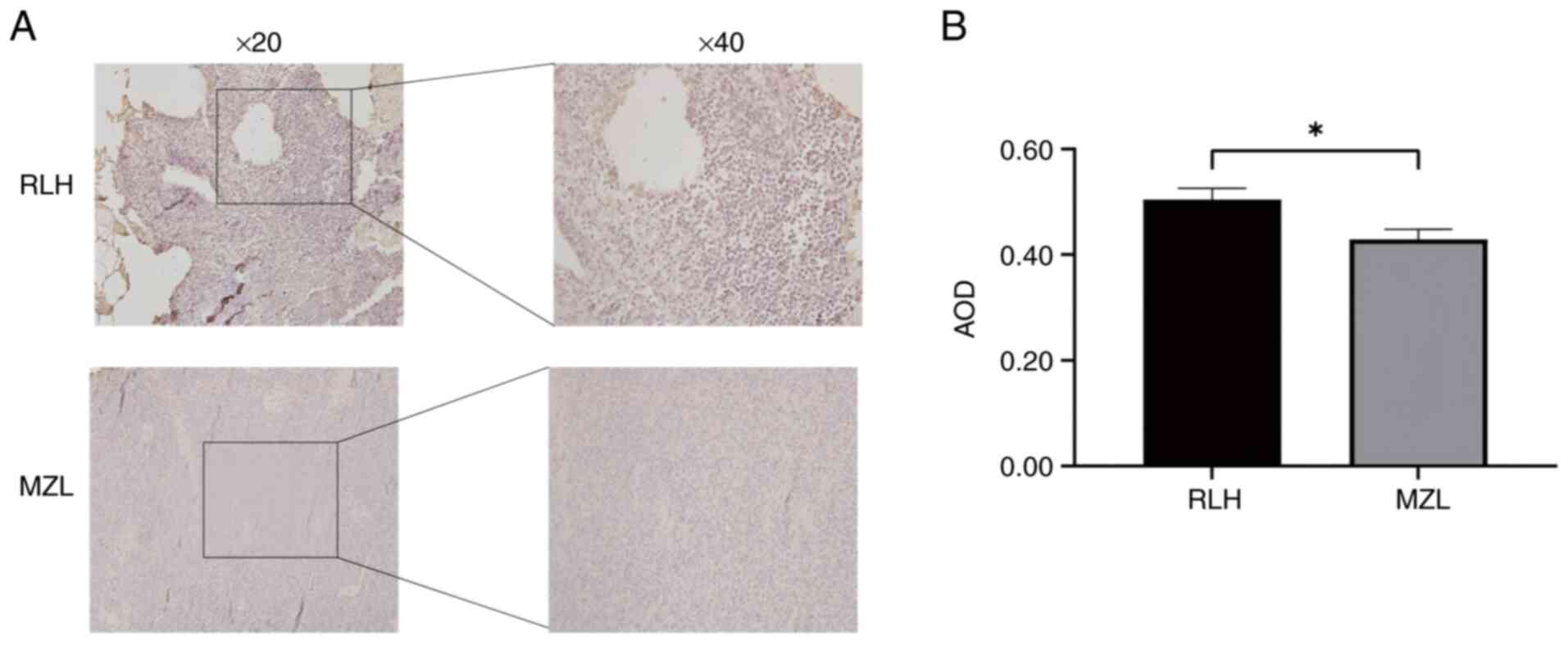
Suppressed Beclin-1 expression in MZL
tissues
Beclin-1 exhibited cytoplasmic granular
(yellowish-brown) staining in both RLH and MZL tissues by IHC
analysis. Quantitative evaluation demonstrated a significant
reduction in Beclin-1 expression in MZL tissues (AOD=0.43±0.19)
compared to RLH controls (AOD=0.50±0.21; P=0.0139) (Fig. 1).
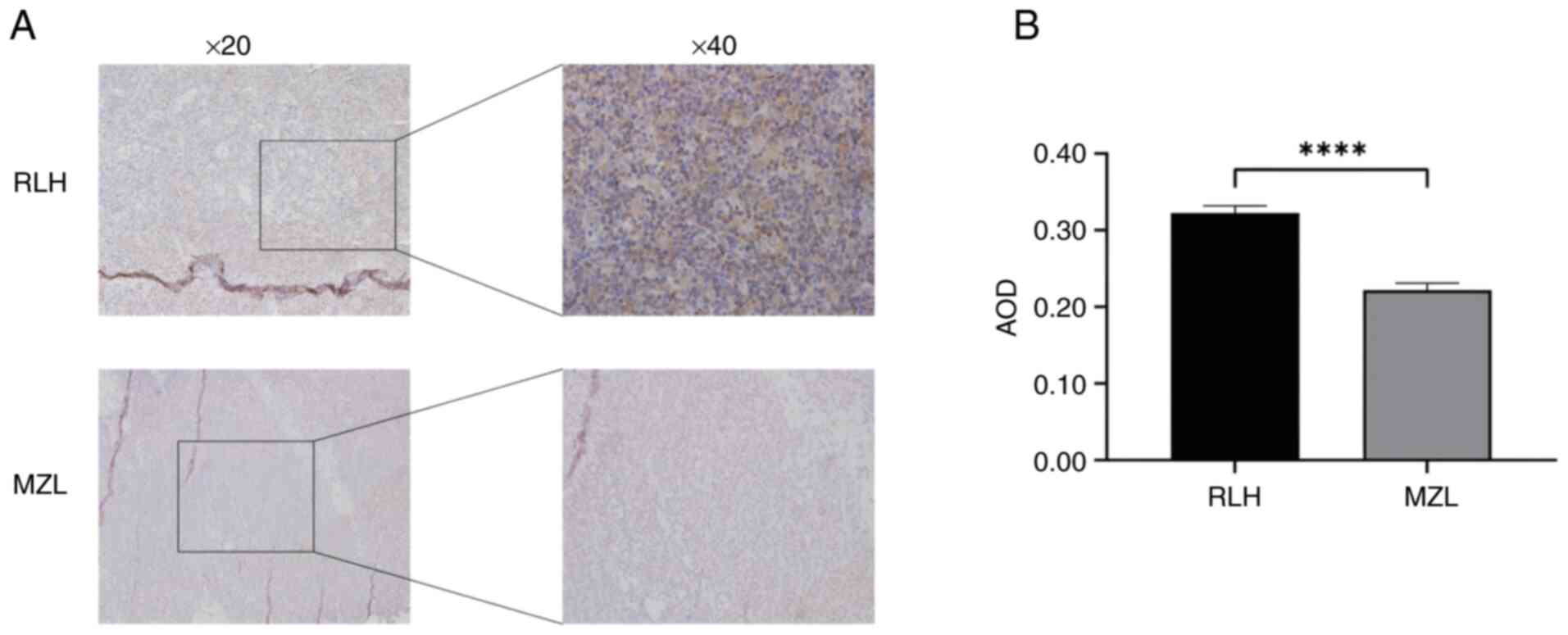
Impaired autophagic flux in MZL via
reduced LC3 expression
LC3 displayed characteristic cytoplasmic granular
staining in all specimens (Fig. 2).
Compared to RLH controls (AOD=0.32±0.09), MZL tissues showed
significantly reduced LC3 expression (AOD=0.22±0.09; P<0.0001),
indicating defective autophagosome formation.
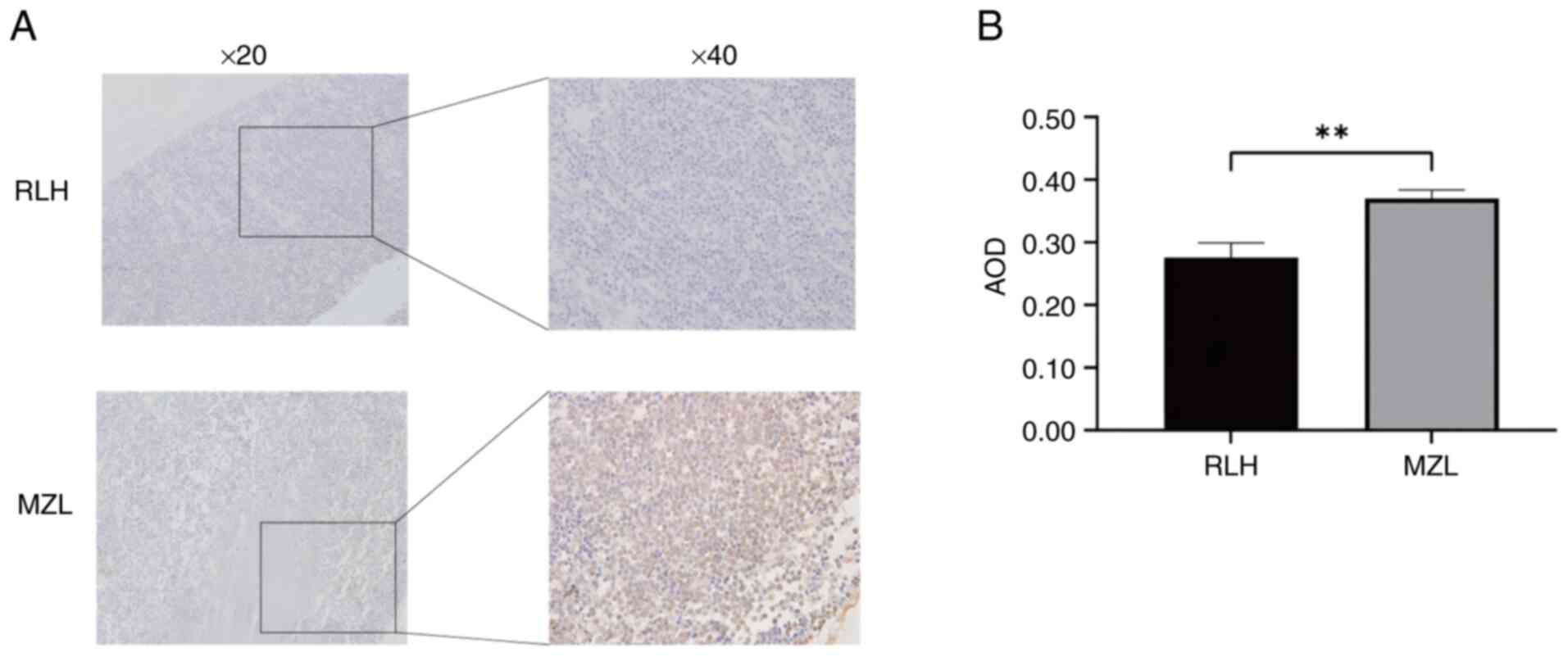
Accumulation of p62 indicates
autophagic dysfunction in MZL
Consistent with impaired autophagic flux, p62
expression was significantly elevated in MZL tissues
(AOD=0.37±0.13) compared with RLH controls (AOD=0.28±0.23;
P=0.0017) (Fig. 3). Cytoplasmic
granular accumulation (yellowish-brown staining) of this selective
autophagy receptor was confirmed by IHC, suggesting defective
substrate clearance as a potential mechanism underlying MZL
pathology.
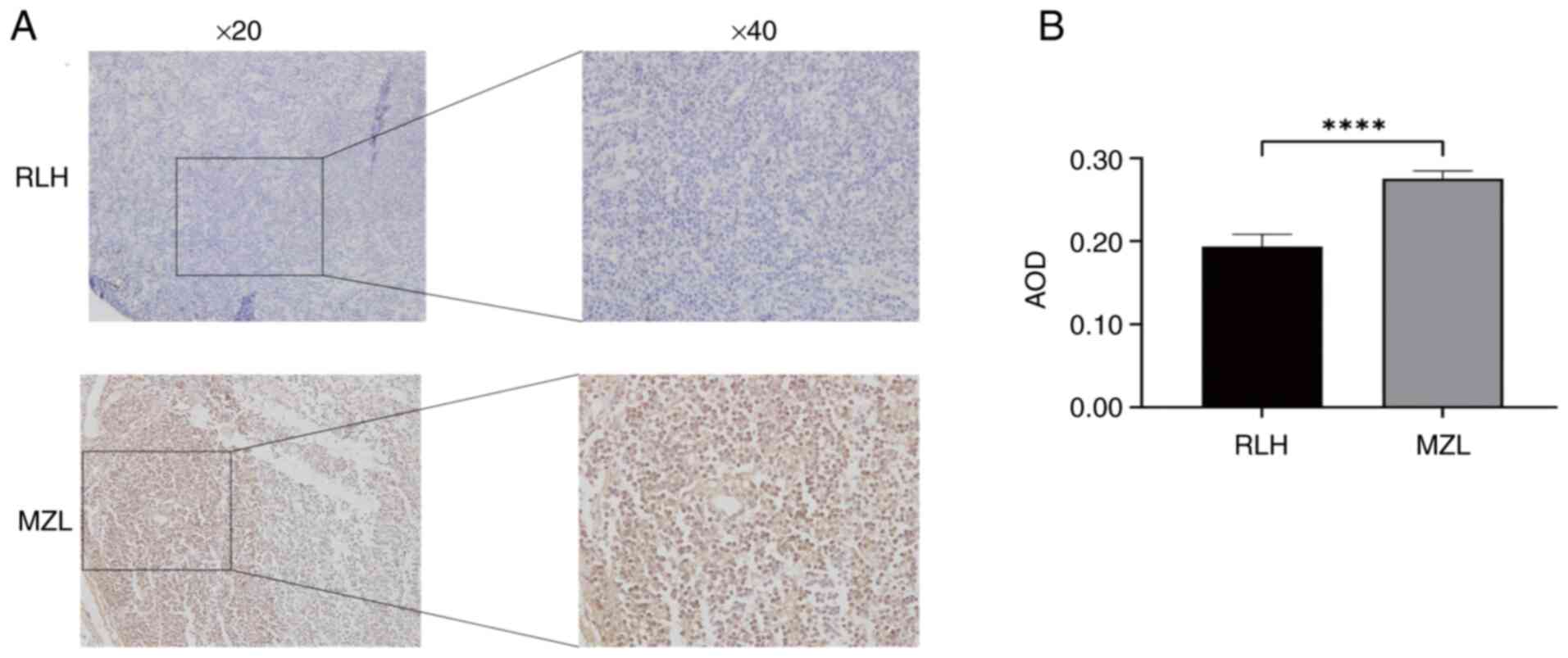
Elevated anti-apoptotic Bcl-2
expression in MZL
IHC demonstrated cytoplasmic Bcl-2 accumulation
(granular yellowish-brown staining) in all tissues. MZL tissues
displayed significantly increased expression (AOD=0.28±0.09)
compared to RLH controls (AOD=0.19±0.15; P<0.0001) (Fig. 4), indicating enhanced survival
signaling typical of lymphomagenesis. This molecular alteration
represents a potential therapeutic target.
Discussion
Autophagy is a key catabolic process that maintains
balance in cells via the lysosomal breakdown of damaged organelles
and protein aggregates, as well as redundant components in the body
(22). Autophagy is a dynamic
process that occurs in five sequential steps (22–24),
which can result in dysfunctions in various key steps. At the
initiation of autophagy, signaling pathways that involve mTOR
complex 1 (mTORC1) lead to the inhibition of unc-51 like autophagy
activating kinase 1/2 complexes that inhibit the initiation of
autophagy (25). At the vesicle
nucleation step, Bcl-2:Beclin-1 complexes inhibit autophagosome
formation via the inhibition of Beclin-1/VPS34 retromer complex
component complexes (26–28). Furthermore, in pathologic scenarios,
there is a possibility of defective autophagy, leading to a
reduction in the usual physiological roles of autophagy, which in
turn contributes to cancer development. Dysautophagy in cancer
allows cancer cells to adapt in a host that is under various
pathologic stresses, including hypoxia as well as starvation
(29,30).
Autophagy's role remains particular in the context
of tumorigenesis, having a dual role that seems contradictory in
nature (31–34). Autophagy is a tumor suppressor
mechanism that eliminates dysfunctional organelles, misfolded
proteins, as well as reactive oxygen species, thereby maintaining
genomic stability and preventing tumorigenesis. However, in already
present malignancies, dysfunctional/mutated autophagy pathways are
known to promote tumor survival during metabolic, as well as
hypoxic stresses, thereby leading to increased tumor progression as
well as resistance (35,36). These data observed in MZL cases in
the current study, showing dysfunctional Beclin-1 and reduced LC3
expression levels, as well as increased p62 accumulation, are
possibly suggestive of a transition from a protective to a
dysfunctional form of autophagy, leading to survival in the tumor
microenvironment (37).
Furthermore, certain known mutations in MZL will possibly interact
with the autophagy pathways. NOTCH2 mutations leading to
increased B-cell survival as well as B-cell activation possibly
lead to a reduction in autophagy as a result of increased mTOR, as
well as NF-κB pathways in those downstream pathways. Furthermore,
mutations in KLF2 leading to a reduction in KLF2 activity possibly
result in increased NF-κB pathways in a possibly perpetually
increased phase of activity, leading to alterations in metabolism
of cells, as well as functioning as known modifiers of the process
of autophagy (38,39). Therefore, dysfunctional autophagy
observed in MZL is possibly a result of a multitude of these
mutations affecting the pathways.
As increasing studies have focused on MZL, the
intricate relationship between autophagy and MZL pathogenesis has
slowly been revealed. Autophagy plays a role in MZL development and
progression through various pathways, such as B-cell receptor
signaling modulation, tumor microenvironment modulation, increased
drug resistance, as well as crosstalk between tumor suppressor
pathways (40,41). Goldsmith et al (42) revealed that autophagy supports
metabolic rerouting in RAS-mutant cancer cells. Poillet-Perez et
al (43) also found that
autophagy suppresses T-cell immunity functions by attenuating
stimulator of interferon response cGAMP interactor 1 pathway
activity, whereas hepatocyte-specific autophagy deficiency
moderately increased T-cell immunity against tumors. Furthermore,
in 2024, Choi et al (44)
found that myeloid-specific deficiency of autophagy reduced
tumor-associated macrophages, increasing the accumulation of
myeloid-derived suppressor cells. However, the mechanism of
disrupted autophagy in MZL pathogenesis remains to be fully
elucidated.
Beclin-1 inhibits tumor initiation under
physiological circumstances. Conversely, in a tumor
microenvironment, Beclin-1 expression is reduced. This leads to a
failure in the formation of autophagosomes, thereby increasing the
susceptibility of cells to transformation and tumorigenesis. Jiang
et al (10) confirmed that
Beclin-1 expression is significantly reduced in non-small cell lung
cancer tissues compared with normal lung tissues. Pattingre and
Levine (45) observed that Beclin-1
haplo deletion resulted in reduced autophagy in B lymphocytes. This
further increased the susceptibility of mice to spontaneous B-cell
lymphoma. It was revealed that B-cell lymphomas often display high
Bcl-2 expression. This not only prevents apoptosis but also has a
role in increasing B-cell lymphoma by functioning as a negative
regulator of Beclin-1 expression-initiated autophagy (46). Furthermore, in the present analysis,
it was discovered that Beclin-1 was downregulated, whereas Bcl-2
was upregulated in MZL patients' tissue, suggesting that there is a
problem in the formation of vesicles in MZL-related autophagy. In
addition, it may be speculated that the abnormal expression of
Beclin-1 and Bcl-2, regulated by the tumor microenvironment, leads
to phagophores in MZL, making it impossible to remove metabolic
waste in the form of autophagosomes, paving a way towards the
transformation of cancerous cells in MZL. However, the current
results indicated that this autophagy-related, Bcl-2-targeted
hypothesis needs further clarification due to insufficient data
supporting the use of Bcl-2 inhibitors in MZL. Nevertheless, some
small molecules of Bcl-2 inhibitor drugs like ABT-199 (47) and ABT-263 (48) are already in the clinical trials
phase. Furthermore, the present results demonstrate that Bcl-2
inhibitors have promising potential utility in patients MZL as a
treatment drug. Future mechanistic and preclinical studies are
warranted to determine whether modulating phagophore formation via
Bcl-2 inhibition can influence MZL progression.
Dysregulation of autophagy is often found in the
elongation process of the vesicle, in which LC3 is a crucial
component. In this process, LC3 is targeted to the autophagosomal
membrane, where it monitors autophagy activity, as well as
targeting the components of the core autophagy process to the
phagophore membrane. It has been irrefutably shown that lack of LC3
leads to a marked reduction in the efficiency of autophagosome
formation and fusion with lysosomes (49–51).
In addition, the process of bringing key components of autophagy to
the phagophore membrane is regulated by LC3 in a
p62/SQSTM1-mediated way. p62 binds to ubiquitinated target
proteins, directing them towards the autophagosomal membrane via a
specific interaction with LC3. Importantly, this is possible as p62
itself is a target of autophagy. As such, p62 accumulation is
expected in most cases of dysfunctional autophagy activity
(52,53). This results in the activation of
mTOR and NF-κB pathways that initiate tumorigenic processes. This
suggested that in autophagy-deficient animal models, p62
overexpression is observed, increasing oxidative stress in
conjunction with increased tumorigenic activity (54,55).
This is also observed in the present analysis: LC3 is
downregulated, while p62 is increased in MZL tumor tissues.
Together, these alterations support the present hypothesis that
excessive p62 accumulation, combined with LC3 repression, reflects
a block in phagophore/autophagosome formation and membrane growth,
thereby contributing to tumor-promoting autophagy dysfunction in
MZL. Future studies will involve transgenic strategies that aim to
increase LC3 expression in conjunction with p62 inhibition
strategies via small molecule drugs that inhibit p62 accumulation
to provoke a positive response in MZL by overcoming phagophore
membrane maturation.
The present findings provide compelling evidence
that MZL exhibits coordinated dysregulation of core autophagy
machinery. Significant downregulation of Beclin-1 (P<0.05) and
LC3 (P<0.0001), key regulators of autophagosome initiation and
maturation, alongside elevated expression of the autophagy receptor
p62 (P<0.001) and anti-apoptotic Bcl-2 (P<0.0001), highlights
impaired autophagic flux as a potential hallmark of MZL
pathobiology. Differential expression of LC3, p62, Bcl-2 and
Beclin-1 between MZL tissues and RLH tissues could help in the
precise identification of MZL. Future studies could aim at the
development of a multiplexed IHC antibody kit that will help in the
precise identification of MZL, as there is a lack of a specific
biomarker in the field that will prompt the clinician to initiate
appropriate treatment strategies. Besides Bcl-2 inhibition, other
strategies targeting the phenomenon of autophagy in hematologic
malignancies are under investigation (56,57).
These include mTOR pathway inhibitors (everolimus, rapamycin),
which inhibit the initiation of autophagy through mTORC1 pathway
modulation, and agents that target autophagy, such as chloroquine
and hydroxychloroquine, which inhibit the fusion of the
autophagosome with the lysosome. Exploring these agents,
individually or in rational combinations, may provide further
therapeutic avenues for MZL.
The present study also has several limitations.
Firstly, the sample size is small, even if considering the subtypes
of MZL: Nodal, MALT and spleen involvement. Due to this small
sample size, certain parameters could not be studied in a subtype
analysis. Secondly, this analysis was mostly done by IHC. IHC is
appropriate to characterize protein localization in clinical
specimens, which, however, lack any molecular confirmation. It is
proposed that analysis involving western blotting analysis or
certain genetic studies in a large series of specimens could
provide new perspectives. Lastly, survival data are not available
in this analysis. This makes certain parameters, including survival
outcomes, impossible to analyze in this particular experiment.
Furthermore, since certain studies involve alterations in a
particular group of genes, as also observed in this experiment, in
a variety of lymphomas as well as certain solid cancers, this
pattern of alterations is more of a supporting factor than a
confirmatory one.
In conclusion, the present analysis identified a
distinct molecular signature of autophagic dysfunction across MZL
subtypes, characterized by significant suppression of Beclin-1
(P<0.05) and LC3-II (P<0.0001) expression, coupled with
pathological accumulation of p62 (P<0.001) and Bcl-2
(P<0.0001). Collectively, these results provide preliminary
evidence that coordinated dysregulation of key autophagy-associated
proteins, specifically reduced Beclin-1 and LC3 expression and
increased accumulation of p62 and Bcl-2, may represent a distinct
molecular hallmark of MZL pathobiology. Although these findings
offer valuable insights into the potential role of autophagy in
MZL, they should be interpreted as hypothesis-generating rather
than definitive. Validation through larger, independent cohorts and
complementary functional studies will be necessary to confirm these
observations and explore their therapeutic implications.
Acknowledgements
Not applicable.
Funding
The study was supported by a grant from the Taishan Youth
Scholar Foundation of Shandong Province (grant no.
tsqn201812140).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JSR designed the study. AL performed the analysis
and interpretation of images. NS performed the analysis and
interpretation of data. XZ performed the analysis and
interpretation of data, and contributed to manuscript drafting and
critical revisions of the intellectual content. XZ and NS confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All methods were carried out in accordance with
relevant guidelines and regulations. The study was approved by the
Ethics Committee of Shandong Provincial Hospital Affiliated to
Shandong First Medical University (approval no. NSFC:NO.2022-209;
approval date, March 1st, 2024). Written informed consent for
participation in this study, including the use of their tissue
samples for scientific research, was provided by the participants'
legal guardians/next of kin.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD and Jaffe ES: The 2016 revision of the World Health Organization
classification of lymphoid neoplasms. Blood. 127:2375–2390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Juárez-Salcedo LM and Castillo JJ:
Lymphoplasmacytic lymphoma and marginal zone lymphoma. Hematol
Oncol Clin North Am. 33:639–656. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zucca E, Arcaini L, Buske C, Johnson PW,
Ponzoni M, Raderer M, Ricardi U, Salar A, Stamatopoulos K,
Thieblemont C, et al: Marginal zone lymphomas: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 31:17–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rodríguez-Sevilla JJ and Salar A: Recent
advances in the genetic of MALT Lymphomas. Cancers (Basel).
14:1762021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rossi D, Bertoni F and Zucca E:
Marginal-zone lymphomas. N Engl J Med. 386:568–581. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coiffier B, Thieblemont C, Van Den Neste
E, Lepeu G, Plantier I, Castaigne S, Lefort S, Marit G, Macro M,
Sebban C, et al: Long-term outcome of patients in the LNH-98.5
trial, the first randomized study comparing rituximab-CHOP to
standard CHOP chemotherapy in DLBCL patients: a study by the Groupe
d'Etudes des Lymphomes de l'Adulte. Blood. 116:2040–2045. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo C, Wu G, Huang X, Ma Y, Zhang Y, Song
Q, Xie M, Sun Y, Huang Y, Huang Z, et al: Efficacy and safety of
new anti-CD20 monoclonal antibodies versus rituximab for induction
therapy of CD20+ B-cell non-Hodgkin lymphomas: a systematic review
and meta-analysis. Sci Rep. 11:32552021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kyriazopoulou L, Karpathiou G,
Hatzimichael E, Peoc'h M, Papoudou-Bai A and Kanavaros P: Autophagy
and cellular senescence in classical Hodgkin lymphoma. Pathol Res
Pract. 236:1539642022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ying H, Qu D, Liu C, Ying T, Lv J, Jin S
and Xu H: Chemoresistance is associated with Beclin-1 and PTEN
expression in epithelial ovarian cancers. Oncol Lett. 9:1759–1763.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang L, Liang X, Liu M, Wang W, Ma J, Guo
Q, Han L, Yang C and Nan K: Reduced expression of liver kinase B1
and Beclin1 is associated with the poor survival of patients with
non-small cell lung cancer. Oncol Rep. 32:1931–1938. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu T, Li Y, Gong L, Lu JG, Du XL, Zhang
WD, He XL and Wang JQ: Multi-step process of human breast
carcinogenesis: A role for BRCA1, BECN1, CCND1, PTEN and UVRAG. Mol
Med Rep. 5:305–312. 2012.PubMed/NCBI
|
|
12
|
Wang L, Zhang H, Sun M, Yin Z and Qian J:
High mobility group box 1-mediated autophagy promotes neuroblastoma
cell chemoresistance. Oncol Rep. 34:2969–2976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheah CY, Zucca E, Rossi D and Habermann
TM: Marginal zone lymphoma: Present status and future perspectives.
Haematologica. 107:35–43. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fiorentino V, Pizzimenti C, Pierconti F,
Lentini M, Ieni A, Caffo M, Angileri F, Tuccari G, Fadda G, Martini
M and Larocca LM: Unusual localization and clinical presentation of
primary central nervous system extranodal marginal zone B-cell
lymphoma: A case report. Oncol Lett. 26:4082023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zucca E, Rossi D and Bertoni F: Marginal
zone lymphomas. Hematol Oncol. 41 (Suppl 1):S88–S91. 2023.
View Article : Google Scholar
|
|
16
|
Ye J, Zhang J, Zhu Y, Wang L, Jiang X, Liu
B and He G: Targeting autophagy and beyond: Deconvoluting the
complexity of Beclin-1 from biological function to cancer therapy.
Acta Pharm Sin B. 13:4688–4714. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nicotra G, Mercalli F, Peracchio C,
Castino R, Follo C, Valente G and Isidoro C: Autophagy-active
beclin-1 correlates with favourable clinical outcome in non-Hodgkin
lymphomas. Mod Pathol. 23:937–950. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Avsec D, Djordjevič AT, Kandušer M,
Podgornik H, Škerget M and Mlinarič-Raščan I: Targeting autophagy
triggers apoptosis and complements the action of venetoclax in
chronic lymphocytic leukemia cells. Cancers (Basel). 13:45572021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Valente G, Morani F, Nicotra G, Fusco N,
Peracchio C, Titone R, Alabiso O, Arisio R, Katsaros D, Benedetto C
and Isidoro C: Expression and clinical significance of the
autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed Res
Int. 2014:4626582014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Huang F, Wang J, You R, Huang Q
and Chen Y: SQSTM1/p62 predicts prognosis and upregulates the
transcription of CCND1 to promote proliferation in mantle cell
lymphoma. Protoplasma. 262:635–647. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McCarthy A, Marzec J, Clear A, Petty RD,
Coutinho R, Matthews J, Wilson A, Iqbal S, Calaminici M, Gribben JG
and Jia L: Dysregulation of autophagy in human follicular lymphoma
is independent of overexpression of BCL-2. Oncotarget.
5:11653–11668. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li-Harms X, Milasta S, Lynch J, Wright C,
Joshi A, Iyengar R, Neale G, Wang X, Wang YD, Prolla TA, et al:
Mito-protective autophagy is impaired in erythroid cells of aged
mtDNA-mutator mice. Blood. 125:162–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu L, Chen Y and Tooze SA: Autophagy
pathway: Cellular and molecular mechanisms. Autophagy. 14:207–215.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alers S, Löffler AS, Wesselborg S and
Stork B: Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy:
Cross talk, shortcuts, and feedbacks. Mol Cell Biol. 32:2–11. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levine B, Sinha S and Kroemer G: Bcl-2
family members: Dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Usman RM, Razzaq F, Akbar A, Farooqui AA,
Iftikhar A, Latif A, Hassan H, Zhao J, Carew JS, Nawrocki ST, et
al: Role and mechanism of autophagy-regulating factors in
tumorigenesis and drug resistance. Asia Pac J Clin Oncol.
17:193–208. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vitto VAM, Bianchin S, Zolondick AA,
Pellielo G, Rimessi A, Chianese D, Yang H, Carbone M, Pinton P,
Giorgi C and Patergnani S: Molecular mechanisms of autophagy in
cancer development, progression, and therapy. Biomedicines.
10:15962022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rangel M, Kong J, Bhatt V, Khayati K and
Guo JY: Autophagy and tumorigenesis. FEBS J. 289:7177–7198. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Russell RC and Guan KL: The multifaceted
role of autophagy in cancer. EMBO J. 41:e1100312022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Levy JM and Thorburn A: Autophagy in
cancer: Moving from understanding mechanism to improving therapy
responses in patients. Cell Death Differ. 27:843–857. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chavez-Dominguez R, Perez-Medina M,
Lopez-Gonzalez JS, Galicia-Velasco M and Aguilar-Cazares D: The
double-edge sword of autophagy in cancer: From tumor suppression to
pro-tumor activity. Front Oncol. 10:5784182020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ahmadi-Dehlaghi F, Mohammadi P, Valipour
E, Pournaghi P, Kiani S and Mansouri K: Autophagy: A challengeable
paradox in cancer treatment. Cancer Med. 12:11542–11569. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pizzimenti C, Fiorentino V, Ruggeri C,
Franchina M, Ercoli A, Tuccari G and Ieni A: Autophagy involvement
in non-neoplastic and neoplastic endometrial pathology: The state
of the art with a focus on carcinoma. Int J Mol Sci. 25:121182024.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pandey A, Goswami A, Jithin B and Shukla
S: Autophagy: The convergence point of aging and cancer. Biochem
Biophys Rep. 42:1019862025.PubMed/NCBI
|
|
37
|
Debnath J, Gammoh N and Ryan KM: Autophagy
and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol.
24:560–575. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pizzimenti C, Fiorentino V, Franchina M,
Martini M, Giuffrè G, Lentini M, Silvestris N, Di Pietro M, Fadda
G, Tuccari G and Ieni A: Autophagic-related proteins in brain
gliomas: Role, mechanisms, and targeting agents. Cancers (Basel).
15:26222023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pizzimenti C, Curcio A, Fiorentino V,
Germanò A, Martini M, Ieni A and Tuccari G: Immunoexpression of
autophagy-related proteins in a single-center series of sporadic
adult conventional clival chordomas. Oncol Lett. 29:322025.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
García Ruiz O, Sánchez-Maldonado JM,
López-Nevot MÁ, García P, Macauda A, Hernández-Mohedo F, Veiga J,
Figuerola J, Videvall E and Martínez-de la Puente J: Autophagy in
Hematological Malignancies. Cancers (Basel). 14:50722022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Buchner M and Müschen M: Targeting the
B-cell receptor signaling pathway in B lymphoid malignancies. Curr
Opin Hematol. 21:341–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Goldsmith J, Levine B and Debnath J:
Autophagy and cancer metabolism. Methods Enzymol. 542:25–57. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Poillet-Perez L, Sharp DW, Yang Y, Laddha
SV, Ibrahim M, Bommareddy PK, Hu ZS, Vieth J, Haas M, Bosenberg MW,
et al: Autophagy promotes growth of tumors with high mutational
burden by inhibiting a T-cell immune response. Nat Cancer.
1:923–934. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Choi J, Park G, Lee SSY, Dominici E,
Becker L, Macleod KF, Kron SJ and Hwang S: Context-dependent roles
for autophagy in myeloid cells in tumor progression. bioRxiv.
16:2024.07.12.603292. 2024.
|
|
45
|
Pattingre S and Levine B: Bcl-2 inhibition
of autophagy: A new route to cancer? Cancer Res. 66:2885–2888.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu HD and Qin ZH: Beclin 1, Bcl-2 and
autophagy. Adv Exp Med Biol. 1206:109–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Minson A, Tam C, Dickinson M and Seymour
JF: Targeted agents in the treatment of indolent b-cell non-hodgkin
lymphomas. Cancers (Basel). 14:12762022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Warren CFA, Wong-Brown MW and Bowden NA:
BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis.
10:1772019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lamark T and Johansen T: Mechanisms of
selective autophagy. Annu Rev Cell Dev Biol. 37:143–169. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Schaaf MBE, Keulers TG, Vooijs MA and
Rouschop KMA: LC3/GABARAP family proteins: Autophagy-(un)related
functions. FASEB J. 30:3961–3978. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Galluzzi L and Green DR:
Autophagy-independent functions of the autophagy machinery. Cell.
177:1682–1699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shaid S, Brandts CH, Serve H and Dikic I:
Ubiquitination and selective autophagy. Cell Death Differ.
20:21–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Amaravadi R, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Moscat J and Diaz-Meco MT: p62: A
versatile multitasker takes on cancer. Trends Biochem Sci.
37:230–236. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wei H, Wang C, Croce CM and Guan JL:
p62/SQSTM1 synergizes with autophagy for tumor growth in vivo.
Genes Dev. 28:1204–1216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Feng Y, Chen X, Cassady K, Zou Z, Yang S,
Wang Z and Zhang X: The role of mTOR inhibitors in hematologic
disease: From bench to bedside. Front Oncol. 10:6116902020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ali ES, Mitra K, Akter S, Ramproshad S,
Mondal B, Khan IN, Islam MT, Sharifi-Rad J, Calina D and Cho WC:
Recent advances and limitations of mTOR inhibitors in the treatment
of cancer. Cancer Cell Int. 22:2842022. View Article : Google Scholar : PubMed/NCBI
|