Introduction
Breast cancer is the most prevalent malignancy among
women worldwide, with 2.26 million new cases reported in 2020
(accounting for 24.5% of all new cancer cases) and ~685,000
mortalities annually (1,2). Breast invasive carcinoma (BRCA) is the
most prevalent malignant type of breast cancer, characterized by
tumor cells penetrating the basement membrane of mammary ducts or
lobules and infiltrating surrounding tissues, with the potential to
metastasize to distant organs through the lymphatic system or
hematogenous circulation (3).
Although the widespread adoption of screening techniques and
advances in comprehensive treatment have improved the 5-year
survival rate of early-stage breast cancer to 90%, the 5-year
survival rate for patients with metastatic breast cancer remains
<30% (4,5). Notably, treatment modalities such as
intraoperative radiotherapy have shown a higher local recurrence
risk compared with whole-breast radiotherapy in patients with
early-stage breast cancer (6).
Current biomarkers, including estrogen receptor
(ER), progesterone receptor (PR), HER2, programmed death-ligand 1
(PD-L1) and tumor mutation burden, exhibit limitations in their
sensitivity and specificity (7).
However, emerging multi-gene signatures, including the nine-long
non-coding RNA prognostic model, require further validation for
clinical use (8). The tumor immune
microenvironment (TIME) influences BRCA progression, as M2
macrophages drive immunosuppression via C-X-C motif chemokine
ligand 1-mediated PI3K/AKT/NF-κB activation, elevating PD-L1
expression levels (9).
Additionally, in Traditional Chinese Medicine, it is believed that
decoction therapies suppress metastasis by modulating the
epithelial-to-mesenchymal transition (EMT) and immune cells such as
CD4+ T, T-helper 1 and monocytic myeloid-derived suppressor cells
(10).
Ras-GTPase-activating protein SH3 domain-binding
protein 1 (G3BP1) is an RNA-binding protein (RBP) that serves a key
role in regulating mRNA stability, stress granule (SG) formation
and signal transduction (7).
Emerging evidence has demonstrated that G3BP1 is frequently
upregulated in various malignancies and contributes to tumor
progression through multiple mechanisms: i) Serving as a core
scaffold protein of SGs to promote tumor cell survival under
chemotherapeutic or hypoxic stress conditions; ii) activating
oncogenic signaling pathways such as PI3K/AKT/mTOR to enhance tumor
cell proliferation and metastasis; and iii) modulating the
infiltration and function of immune cells within the tumor
microenvironment to establish an immunosuppressive niche (7,11,12).
Therefore, the present study aimed to investigate
the association between G3BP1 expression and the
clinicopathological characteristics and prognosis of BRCA, as well
as elucidate the biological role of G3BP1 in BRCA. Utilizing
multiple online databases, the differential expression of G3BP1
between BRCA tumor and normal tissues was compared, and the
association between G3BP1 expression levels and clinicopathological
features was assessed. Immunohistochemical (IHC) analysis was then
performed at the tissue level to validate the expression patterns
of G3BP1, while clinical follow-up data were analyzed to verify its
prognostic value. Gene set enrichment analysis was performed to
identify biological pathways associated with G3BP1, while immune
infiltration analysis was performed to uncover the biological
impact and potential mechanisms of G3BP1 in BRCA.
Materials and methods
Analysis of G3BP1 expression profiles
across human tissues
To comprehensively characterize the expression
profile of G3BP1 across transcriptional and proteomic hierarchies
in human tissues, multi-omics data from publicly accessible
repositories was leveraged. Pan-cancer mRNA expression of G3BP1 was
interrogated using the Tumor Immune Estimation Resource (TIMER)
database (https://cistrome.shinyapps.io/timer/) (13). Transcriptomic data from The Cancer
Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) BRCA project were
analyzed to evaluate differential G3BP1 expression between breast
tumor tissue and normal tissue. Proteomic quantification of G3BP1
in BRCA was performed using the Confirmatory Breast Cancer (CBC)
cohort from Clinical Proteomic Tumor Analysis Consortium (CPTAC)
dataset (https://cptac-data-portal.georgeton.edu/), with
Z-scores representing standardized deviations relative to the
median expression. IHC validation was conducted through the Human
Protein Atlas (HPA) platform, for comparison of G3BP1 protein
localization and intensity between malignant and non-malignant
breast tissue. The specific image data for G3BP1 in breast cancer
tissue can be accessed at: https://www.proteinatlas.org/ENSG00000145907-G3BP1/cancer/breast+cancer,
and for normal breast tissue at: https://www.proteinatlas.org/ENSG00000145907-G3BP1/tissue/breast.
All data and images regarding G3BP1 from the HPA are based upon
work from the HPA consortium and are available under a CC BY-SA 4.0
license (14).
Differential expression analysis of
G3BP1 and association between G3BP1 and clinicopathological
variables
Differential expression of G3BP1 between tumor and
adjacent normal tissues in TCGA-Breast Invasive Carcinoma
collection (TCGA-BRCA) was evaluated using non-parametric
statistical analyses. A Mann-Whitney U test was used for unpaired
samples (tumor compared with normal tissue), while the Wilcoxon
signed-rank test was used for paired samples. Associations between
G3BP1 expression levels and clinicopathological variables,
including age, sex, tumor (T) stage, lymph node (N) stage,
metastasis (M) stage and pathological stage, were assessed using
the Kruskal-Wallis test (for multi-group comparisons), followed by
Dunn's post-hoc test with Benjamini-Hochberg correction for
pairwise comparisons when a significant overall difference was
observed (P<0.05), and Mann-Whitney U test (for two-group
comparisons). P<0.05 was considered to indicate a statistically
significant difference (Benjamini-Hochberg correction).
Survival analysis based on the
expression of G3BP1 in BRCA
Kaplan-Meier survival analysis was performed to
assess the prognostic value of G3BP1 expression in TCGA-BRCA
collection, integrating RNA-sequencing data and clinical survival
metrics from the Kaplan-Meier plotter platform (http://kmplot.com). The analysis was conducted using
the default settings for the ‘Breast Cancer’ module, with the
following key parameters: The ‘autoselect best cut-off’ option was
applied to dichotomize patients into high and low G3BP1 expression
groups, and the probe set ‘201503_at’ was selected for G3BP1
quantification. Subtype-specific stratification was implemented for
luminal A (ER+/PR+, HER2- and low Ki-67), luminal B (ER+/PR+, HER2-
and high Ki-67 or HER2+), HER2-positive (ER-, PR- and HER2+) and
basal-like (largely triple-negative: ER-, PR- and HER2-) using
established classifier algorithms within the platform. Survival
disparities across subgroups were statistically evaluated using the
log-rank test, with P<0.05 being considered to indicate a
statistically significant difference. To comprehensively evaluate
the clinical impact of G3BP1 dysregulation in BRCA using multiple
survival endpoints: Overall survival (OS), disease-free survival
(DFS), distant metastasis-free survival (DMFS) and post-progression
survival (PPS).
Differentially expressed genes (DEGs)
analysis, volcano plot and heatmap
To identify the DEGs between the high G3BP1 and low
G3BP1 expression groups, differential analysis was performed using
the ‘DESeq2’ R package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
with a threshold set at log2 fold change (FC) >0.585
and false discovery rate (FDR) <0.05 (corresponding to a linear
scale of >1.5-fold change). Similar methods have been used in
previous bioinformatics research on breast cancers (15). DEGs were visualized through a
volcano plot generated using the ‘ggplot2’ R package (https://cran.r-project.org/web/packages/ggplot2/).
The x-axis of the volcano plot is labelled as ‘log2 FoldChange’ and
the red and green dots represent the upregulated and downregulated
genes that met the threshold, while the gray dots represent the
genes that did not meet the threshold. Additionally, a heatmap was
generated using the ‘pheatmap’ R package (https://cran.r-project.org/web/packages/pheatmap/) to
further illustrate the expression patterns of these DEGs across all
samples.
GSEA
Analysis was performed using the Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway database, Gene Ontology (GO)
gene sets, including cellular components (CC), biological processes
(BP) and molecular functions (MF), hallmark gene sets and
Immunologic Signature Database (ImmuneSigDB), obtained from the
Molecular Signatures Database (MsigDB, version 7.5.1) through its
official website (http://software.broadinstitute.org/gsea/downloads.jsp).
The ‘clusterProfiler’ package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
in R software was used for GSEA. The analysis was performed with
1,000 permutations to ensure statistical robustness. Gene sets
demonstrating a normalized enrichment score (NES) >1.0 and
achieving a nominal P<0.05 (after FDR correction) were
considered to be statistically significant. To validate
GSEA-predicted pathway associations, co-expression patterns between
G3BP1 and core PI3K/AKT/mTOR signaling molecules were analyzed
using transcriptomic data from the TIMER2.0 resource (http://timer.cistrome.org/).
Immunological correlation
analysis
To investigate the relationship between G3BP1
expression and TIME composition in BRCA, comprehensive immune cell
infiltration analyses were performed. Firstly, Spearman's rank
correlation was used to evaluate associations between G3BP1 mRNA
levels and the infiltration abundance of 22 immune cell types
derived using CIBERSORT deconvolution algorithms. Secondly,
correlations between G3BP1 and six major immune cell subsets (B
cells, CD4+ T cells, CD8+ T cells,
neutrophils, macrophages and dendritic cells) were further
validated using the TIMER database. To explore potential mechanisms
of G3BP1-mediated immune evasion, its co-expression with the immune
checkpoint molecule PD-L1, also known as cluster of differentiation
274 (CD274) was assessed via scatter plot analysis and Spearman's
rank correlation in the TIMER database.
Clinical data collection and IHC
staining
A total of 38 paired breast cancer specimens and
matching adjacent non-neoplastic tissues were retrospectively
collected from female patients (median age, 52 years; age range,
34–73 years) undergoing surgical resection at The Second Affiliated
Hospital of Anhui Medical University (Hefei, China) between January
2013 and October 2014.
Inclusion criteria comprised: i) Histologically
confirmed diagnosis of primary breast invasive carcinoma; ii)
surgically resected tumor tissue and adjacent normal tissue pairs
meeting predefined quality standards (tumor cellularity ≥30%;
intact protein preservation); and iii) complete clinicopathological
records.
Exclusion criteria comprised: i) Diagnosis of
non-breast primary malignancies or synchronous dual primaries
(except bilateral breast cancer); ii) incomplete clinical or
outcome documentation; and iii) specimens not meeting the G3BP1
detection standards. The study protocol was approved by the
Institutional Ethics Committee of The Second Affiliated Hospital of
Anhui Medical University (approval no. YX2019-055) and written
informed consent was obtained from all participants prior to tissue
procurement.
The samples were fixed in 4% formaldehyde at room
temperature for 24 h and subsequently embedded in paraffin. Serial
sections of 4 µm thickness were prepared, followed by baking at
60°C for 2 h, xylene dewaxing and gradient ethanol dehydration.
Following dehydration, antigen retrieval was performed. Sections
were then permeabilized with 0.1% Triton X-100 for 15 min and
blocked with 5% bovine serum albumin (cat. no. A7906; Sigma-Adrich;
Merck KGaA) at room temperature for 1 h to prevent non-specific
binding. IHC staining was performed using a monoclonal antibody
against G3BP1 (1:200; cat. no. GB115527; Wuhan Servicebio
Technology Co., Ltd.) with incubation at 37°C for 60 min, followed
by application of the secondary antibody (1:200; cat. no. PV6000;
Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) at 37°C for 30
min. Diaminobenzidine was used for chromogenic development for 3
min, followed by hematoxylin counterstaining (performed at room
temperature for 3 min) and gradient dehydration. The staining
results were evaluated under an Olympus light BX53 microscope
(Olympus Corporation; magnification, ×200).
Two senior pathologists independently assessed the
staining of breast cancer cells under high-power fields using a
double-blind method. To enhance objectivity and reduce observer
variability, digitized whole-slide images (WSIs) of stained
sections were additionally analyzed using QuPath (https://qupath.github.io/) digital pathology software
(version 0.6.0). G3BP1 exhibited cytoplasmic staining, with the
staining intensity being scored as follows: 0, no staining; 1,
faint yellow granules visible in cytoplasm; 2, brown-yellow
granules visible in cytoplasm; and 3, dark brown granules visible
in cytoplasm. A total of five representative high-power fields were
selected, and the percentage of positive cells in each field was
quantified: 0, negative; 1, <25%; 2, 26–50%; 3, 51–75%; and 4,
>75%. The staining area score was divided by the staining
intensity score. A score <4 points indicated low G3BP1
expression, while a score ≥4 points indicated high G3BP1
expression.
Using QuPath, automated cell detection and
classification algorithms were applied to the WSIs to quantify the
percentage of G3BP1-positive tumor cells and measure the average
optical density (OD) of staining intensity across the entire tumor
region or within annotated representative areas. The QuPath-derived
quantitative data (positive cell percentage and mean OD) were used
to corroborate the semi-quantitative manual H-scores. Consistency
between manual H-scores and QuPath quantification was assessed
using Spearman's rank correlation and Bland-Altman analysis. The
pre-defined thresholds for acceptable agreement were set as a
Spearman's correlation coefficient (ρ) >0.8, indicating a very
strong monotonic relationship, and a Bland-Altman bias <5%,
indicating a negligible mean difference between the two methods.
These thresholds are commonly adopted in digital pathology
validation studies to ensure that automated quantification is both
highly correlated with and metrically similar to expert manual
assessment (16,17).
Furthermore, differential expression of G3BP1
between breast cancer tissue and matching adjacent non-tumor tissue
was analyzed using IHC data curated from the HPA database
(https://www.proteinatlas.org). Protein
localization and staining intensity were assessed comparatively to
validate tissue-specific G3BP1 dysregulation in BRCA.
Statistical analysis
Multi-omics data and clinical validation were
integrated using a robust statistical framework. Differential
expression of G3BP1 in BRCA compared with normal tissues was
assessed through the aforementioned non-parametric tests
(Mann-Whitney U and Wilcoxon paired tests) using TCGA and CPTAC
data. Associations with clinicopathological features were evaluated
using Kruskal-Wallis and Mann-Whitney U tests. For the analysis of
associations between G3BP1 expression levels (high vs. low) and
categorical clinicopathological variables, Fisher's exact test was
employed. This test was chosen due to its appropriateness for small
sample sizes and when the assumptions of the χ2 test
(such as expected count in >20% of cells being ≤5) are violated.
Survival outcomes (OS, DFS, DMFS and PPS) were analyzed using
Kaplan-Meier curves with log-rank tests and Cox regression to
identify G3BP1 as an independent prognostic marker. DEGs were
screened (log2FC >0.585; FDR <0.05) and visualized
using volcano plots (‘ggplot2’) and heatmaps (‘pheatmap’). GSEA
with ‘clusterProfiler’-identified pathways was carried out using
KEGG, GO and hallmark gene sets (1,000 permutations; FDR <0.05;
NES >1.0). Immune infiltration correlations were analyzed using
Spearman's rank (CIBERSORT/TIMER). For G3BP1 protein expression in
BRCA compared with normal tissues, statistical analysis was
performed using the Wilcoxon signed-rank test (paired
non-parametric test). For recurrence and metastasis risk
assessment, multivariate Cox proportional hazards models were
constructed to adjust for clinically relevant covariates including
age and comorbidities, with hazard ratios (HR) and 95% confidence
intervals (CI) calculated to quantify prognostic effects. All
analyses were performed in R software with a two-tailed P<0.05,
ensuring methodological rigor and reproducibility.
Results
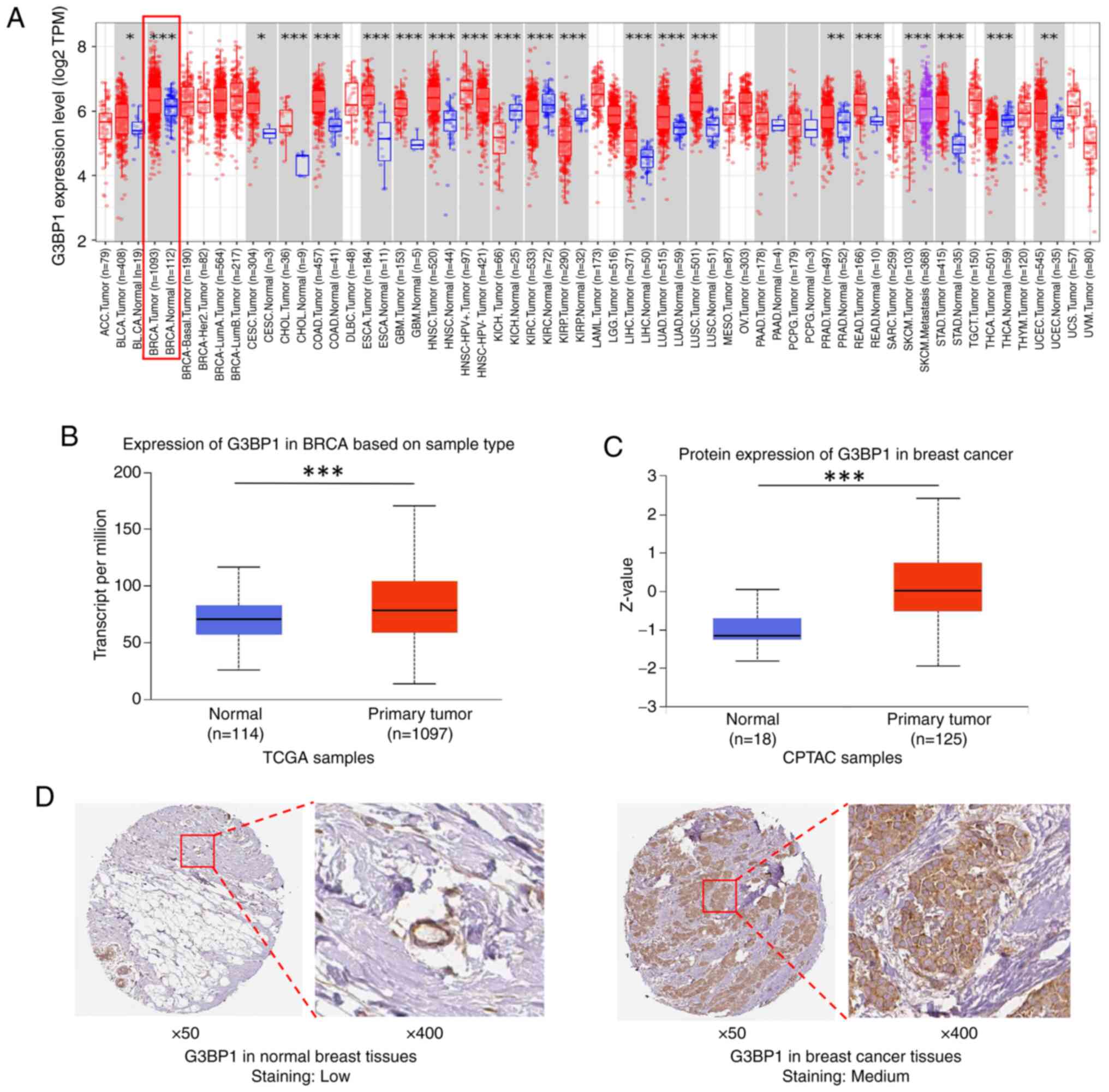
G3BP1 is upregulated in BRCA
Integrated multi-omics analyses revealed significant
upregulation of G3BP1 in BRCA tissue compared with normal tissue.
Transcriptomic data from the TIMER database demonstrated elevated
G3BP1 mRNA levels across multiple cancer types, including BRCA
(Fig. 1A). Analysis of TCGA
datasets confirmed that G3BP1 mRNA expression was significantly
higher in primary tumor tissues compared with normal breast tissue
controls (P=3.82×10−12 vs. normal; Fig. 1B). Subtype stratification of TCGA
samples showed significant upregulation in the luminal subtype
(luminal A and B combined; P=4.33×10−15 vs. normal) and
triple-negative breast cancer (P=0.012 vs. Normal; Fig. S1A) subtypes, while HER2-positive
tumors exhibited no significant difference (P=0.109 vs. normal;
Fig. S1A). At the protein level,
the CPTAC database showed increased G3BP1 abundance in BRCA tumors
(P=1.04×10−8 vs. normal; Fig. 1C), with significant elevation
observed across all molecular subtypes (luminal,
P=1.30×10−8; HER2-positive, P=0.019; TNBC,
P=0.010 vs. normal; Fig.
S1B). These proteomic findings were consistent with IHC
validation from HPA, which revealed moderate cytoplasmic G3BP1
expression in tumor tissue (‘medium staining’), whereas normal
breast tissue exhibited minimal (‘low’) staining (Fig. 1D). Collectively, these results
demonstrate G3BP1 is consistently upregulated in BRCA at both
transcriptional and translational levels, highlighting its
potential role in tumorigenesis.
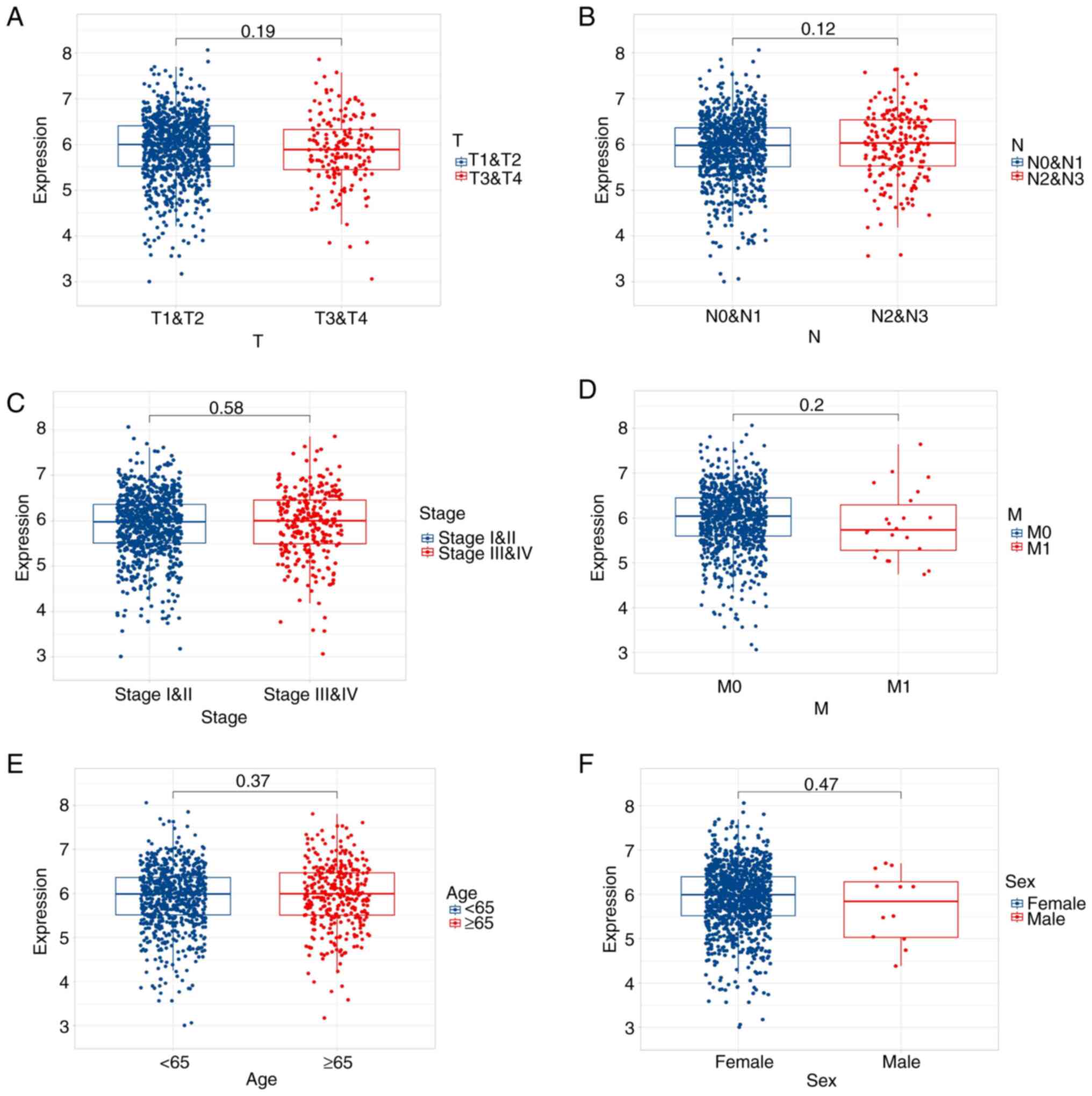
Differential expression of G3BP1
across clinicopathological characteristics of BRCA
To elucidate the clinical relevance of G3BP1 in
BRCA, G3BP1 expression was analyzed across key clinicopathological
parameters using transcriptomic data from TCGA. G3BP1 mRNA levels
exhibited stage-specific heterogeneity as T4 stage tumors
demonstrated significantly elevated G3BP1 expression compared with
T3 stage tumors (P=0.014), although no differences were observed
between T4 and T1 (P=0.180) or T2 (P=0.071) stages (Fig. 2A). Notably, N2 stage tumors
(characterized by extensive regional lymph node metastasis) showed
markedly higher G3BP1 expression compared with N0 and N1 stages
(P=0.005; Fig. 2B). This
underscored its association with lymphatic progression. By
contrast, G3BP1 expression was independent of pathological stage
(I&II vs. III&IV; P=0.580; Fig.
2C) and distant metastasis (M0 vs. M1; P=0.200; Fig. 2D). Demographically, no significant
associations were detected between G3BP1 expression and patient age
(≤65 vs. >65 years; P=0.370; Fig.
2E) or sex (male vs. female; P=0.470; Fig. 2F). These findings suggest that G3BP1
overexpression is selectively associated with local tumor
aggressiveness (stage T4) and advanced lymph node involvement
(stage N2). The significant association with advanced T stage
(local invasion) and N stage (lymphatic spread), coupled with the
lack of association with distant metastasis (M stage) or overall
pathological stage (which incorporates metastatic status),
collectively implicated its role primarily in regional invasion
rather than systemic dissemination.
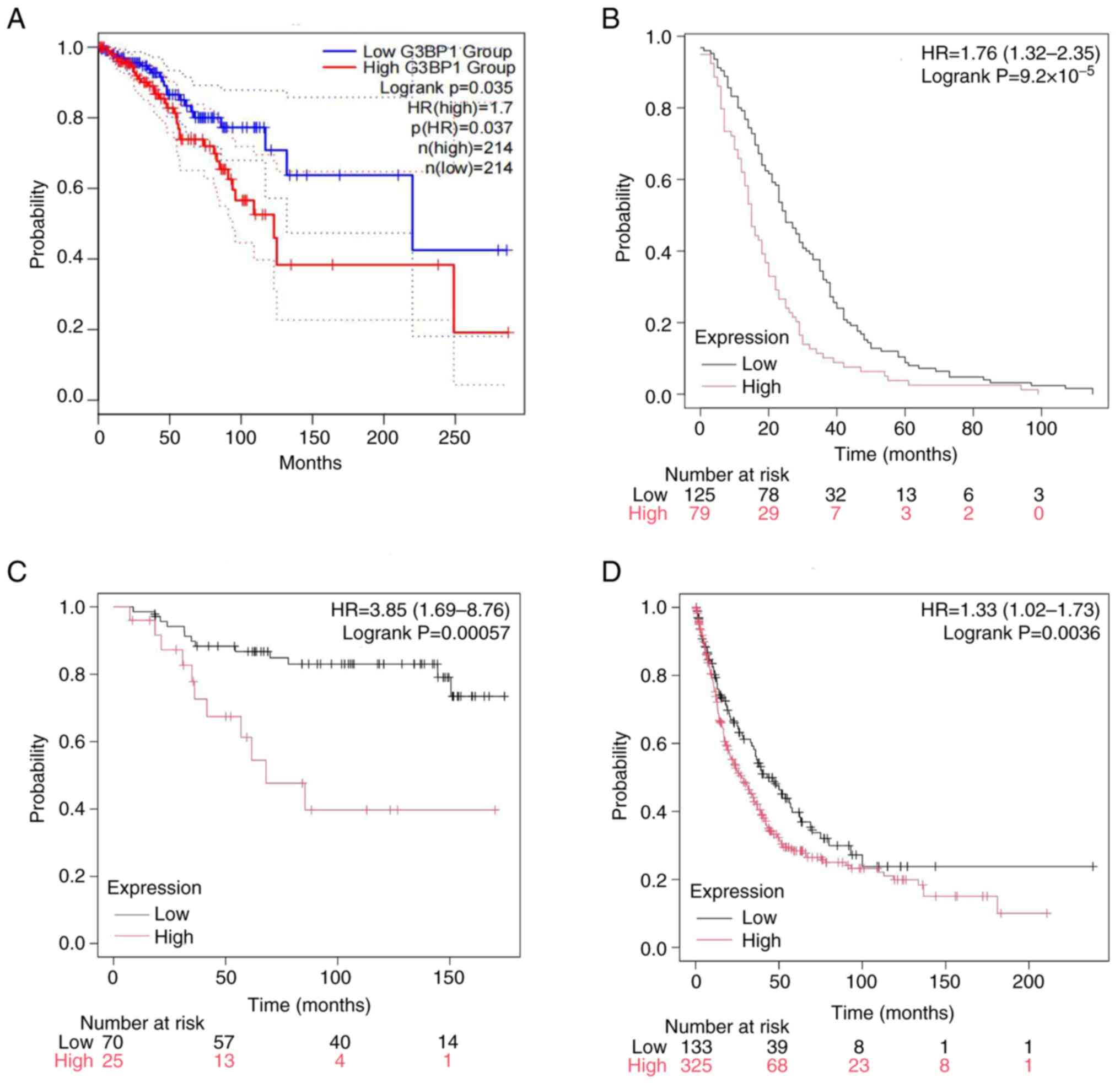
High expression of G3BP1 is associated
with poor prognosis in patients with BRCA
Survival analyses integrating multi-platform data
demonstrated a notable association between elevated G3BP1
expression and unfavorable clinical outcomes in BRCA. Kaplan-Meier
curves derived from TCGA cohorts revealed significantly shorter OS
in patients with high G3BP1 expression compared with the
low-expression group (P=0.037; Fig.
3A). External validation using the Kaplan-Meier plotter
platform further corroborated these findings, showing that high
G3BP1 levels were associated with reduced DFS
(P=9.20×10−5), DMFS (P=5.70×10−4) and PPS
(P=0.036; Fig. 3B-D).
In the Kaplan-Meier Plotter platform, survival
analysis stratified by G3BP1 expression levels revealed distinct
prognostic patterns across breast cancer subtypes (Fig. S2A-D). In the Luminal A cohort
(Fig. S2A), high G3BP1 expression
was significantly associated with worse OS (HR=1.36; 95% CI,
1.15–1.61; P=4.20×10−4). A similar adverse prognostic
trend was observed in luminal B tumors (Fig. S2B), where those with high G3BP1
expression exhibited reduced survival (HR=1.26; 95% CI, 1.04–1.53;
P=0.010). By contrast, HER2-positive tumors (Fig. S2C) showed no statistically
significant difference in survival between expression groups
(HR=1.37; 95% CI, 0.94–2.00; P=0.110). In patients with basal/TNBC
(Fig. S2D), high G3BP1
expression was significantly associated with a worse DFS (HR=1.59;
95% CI, 1.25–2.01; P=1.10×10−4).
G3BP1-associated molecular signatures
and pathway enrichment in BRCA
To uncover the molecular mechanisms underlying
G3BP1-driven progression of BRCA, DEGs between G3BP1-high and
G3BP1-low expression groups were systematically identified using
TCGA datasets. Under strict thresholds (log2FC
>0.585; FDR <0.05), 67 genes were identified as markedly
upregulated, while nine genes were downregulated. A volcano plot
was used to visualize the distribution of DEGs, with red and green
dots representing upregulated and downregulated genes, respectively
(Fig. 4A). A heatmap further
outlined the expression patterns of DEGs across samples, revealing
coordinated activation of pro-tumorigenic genes [such as leptin
(LEP), vascular endothelial growth factor D (VEGFD) and fatty acid
binding protein 4 (FABP4)] in the high G3BP1 expression group
(Fig. 4B).
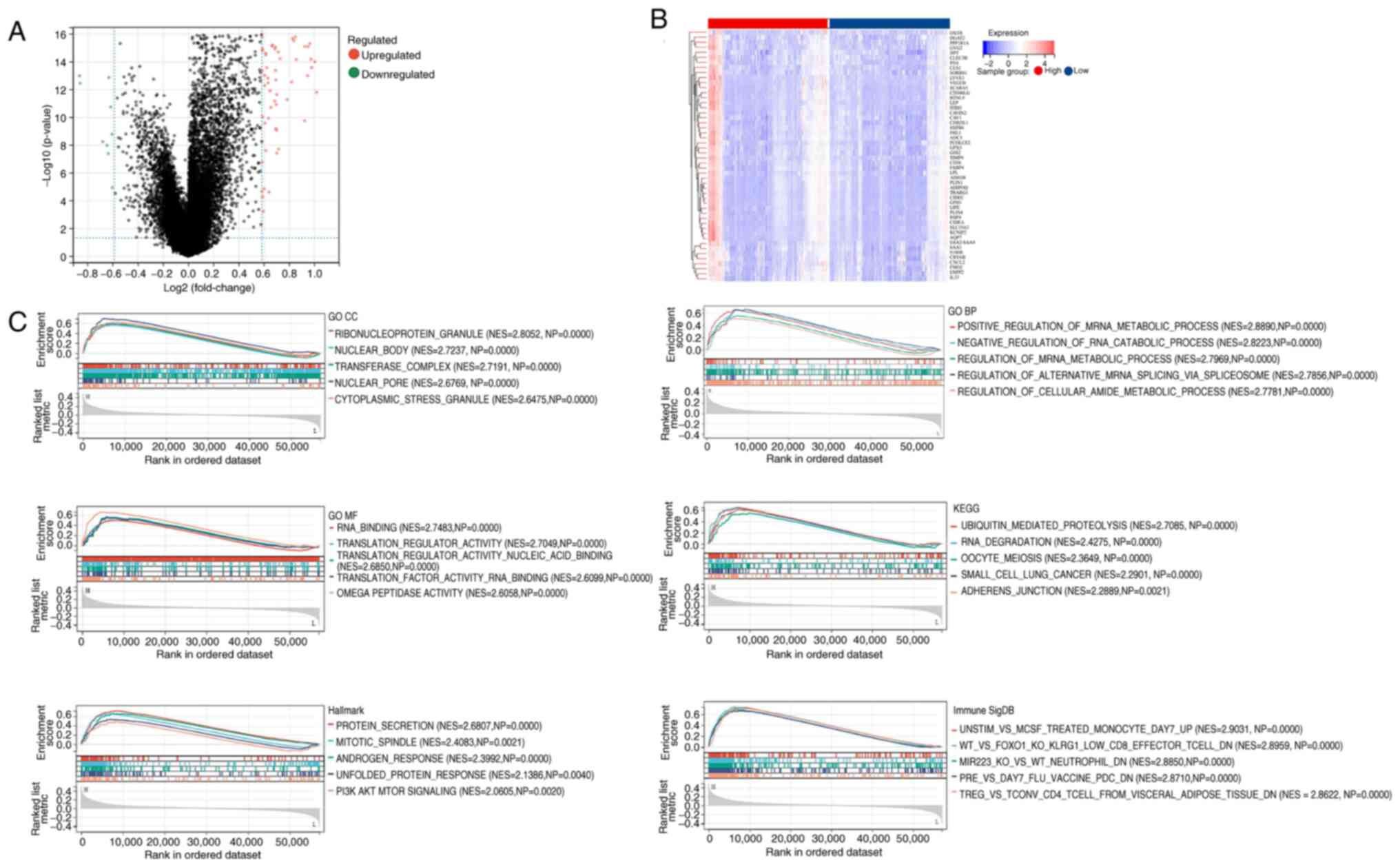 | Figure 4.Multi-dimensional profiling of
G3BP1-associated transcriptional dysregulation in breast invasive
carcinoma. (A) Differential gene screening via volcano plot. (B)
Stratified gene expression patterns by heatmap. (C) Multi-pathway
gene set enrichment analysis including GO CC, GO BP, GO MF, KEGG,
Hallmark and ImmuneSigDB. GO, Gene Ontology; CC, cellular
components; BP, biological processes; MF, molecular functions;
KEGG, Kyoto Encyclopedia of Genes and Genomes, G3BP1;
Ras-GTPase-activating protein SH3 domain-binding protein 1; NES,
normalized enrichment score; NP, nominal P-value. |
The results of the GSEA are presented, focusing only
on pathways with a NES >1 and an FDR of <0.05. The findings
indicate significant enrichment in pathways such as PI3K/AKT/mTOR
(NES=2.0605; P=0.0020) and ubiquitin-mediated protein hydrolysis
(NES=2.7085; P<0.0001), as shown in Fig. 4C. Additional key enriched pathways
in the high G3BP1 expression group are also presented in Fig. 4C. Furthermore, the top five pathways
with NES >1 along with their statistical metrics have been
listed in Table SI. These findings
suggest that G3BP1 promotes BRCA progression by activating
oncogenic pathways (such as PI3K/AKT/mTOR), disrupting proteostasis
and modulating nuclear complex dynamics. The convergence of DEGs
and enriched pathways underscores the therapeutic potential of
targeting G3BP1 in BRCA.
GSEA suggested a significant association between
G3BP1 and the PI3K/AKT/mTOR pathway (FDR <0.05). To validate
this finding, further analysis of the co-expression patterns of
G3BP1 and core signaling molecules in the TIMER2.0 database was
conducted (Fig. S3). G3BP1
exhibited a strong positive correlation with PIK3CA (encoding the
PI3K α catalytic subunit; ρ=0.583; P=3.98×10−101) and a
moderate correlation with PIK3CB (encoding PI3K β; ρ=0.488;
P=6.31×10−67), indicating its differential regulation of
PI3K subtypes. There was no significant correlation between AKT1
expression and G3BP1 (ρ=−0.014; P=0.638), but a significant
positive association was observed between AKT2 (ρ=0.264,
P=4.69×10−19) and AKT3 (ρ=0.230;
P=1.13×10−14), revealing the selective regulation of
G3BP1 on AKT subtypes. It was also observed that G3BP1
significantly correlated with the expression of mTOR (ρ=0.475;
P=6.9×10−63) and regulatory associated proteins of mTOR
complex 1 (ρ=0.536; P=7.83×10−83). Overall, G3BP1 may
drive the activation of PI3K/mTOR signaling by preferentially
coordinating the PIK3CA-AKT2/AKT3 axis.
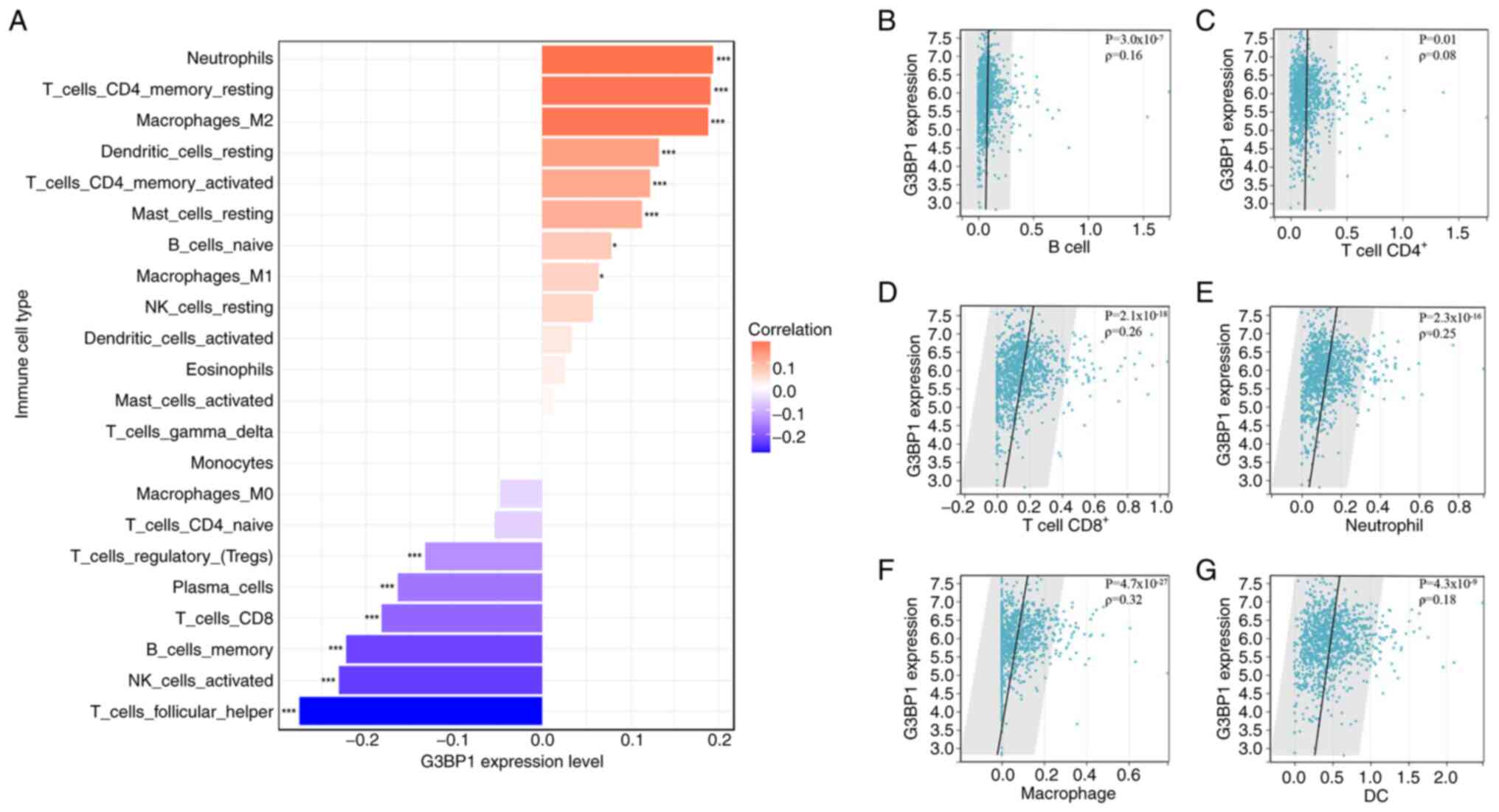
Relationship between G3BP1 expression,
immune microenvironment and PD-L1 expression in BRCA
Survival of patients with breast cancer is
associated with immune cell infiltration (16). The present study used Spearman's
rank correlation analysis to examine the relationship between G3BP1
expression levels in BRCA and the infiltration of 22 immune cell
types from CIBERSORT (Fig. 5A). The
results showed that G3BP1 expression was positively correlated with
‘neutrophils’, ‘T cells CD4 memory resting’, ‘macrophage M2’,
‘dendritic cells resting’, ‘T cells CD4 memory activated’, ‘masT
cells resting’, ‘B cells naïve’ and ‘macrophages M1’ (P<0.05),
while it was negatively correlated with ‘T cells regulatory’,
‘plasma cells’, ‘T cells CD8’, ‘B cells memory’, ‘natural killer
(NK) cells activated’ and ‘T cells follicular helper’ (P<0.001).
Additionally, the present study evaluated the correlation between
G3BP1 expression levels and the infiltration of six immune cell
types in TIMER (Fig. 5B-E). The
results demonstrated that G3BP1 expression was positively
correlated with the infiltration of B cells, T cell
CD4+, T cell CD8+, neutrophil, macrophage and
dendritic cells (all P<0.05).
To identify the mechanistic link between G3BP1 and
immune evasion, its relationship with PD-L1, also known as CD274, a
critical immune checkpoint molecule, was evaluated. Based on the
TIMER2.0 database, scatter plot analysis (Fig. S4) revealed a statistically
significant positive correlation between G3BP1 and CD274 mRNA
expression levels across BRCA samples (ρ=0.352;
P=2.44×10−33). The regression line with 95% CI
demonstrated a progressive increase in G3BP1 expression with
elevated CD274 levels. Data point density indicated clustering in
the mid-range expression zone, with outliers extending to higher
expression values. This significant correlation (P<0.001)
indicates G3BP1-driven PD-L1 expression as a key mechanism for
immune evasion in BRCA.
G3BP1 expression is upregulated in
BRCA and associated with clinicopathology and adverse
prognosis
IHC validation using 38 paired BRCA and adjacent
normal tissues confirmed notable upregulation of G3BP1 in tumor
tissues compared with normal tissues (P<0.001; Fig. 6A and B). The manual IHC scores and
QuPath quantification of each patient are shown in Table SII, with consistency analysis
indicating excellent consistency. Positive cell percentage showed
near-perfect correlation (ρ=0.892; P<0.001) with minimal bias
[-2.1%; 95% limits of agreement (LoA): −8.3 to +4.1%], while
average OD exhibited stronger correlation (ρ=0.924; P<0.001) and
negligible bias (+0.03; 95% LoA: −0.11 to +0.17). Both metrics
exceeded predefined validation thresholds (ρ>0.8; bias <5%),
confirming QuPath's reliability for standardized G3BP1
quantification (Table SIII).
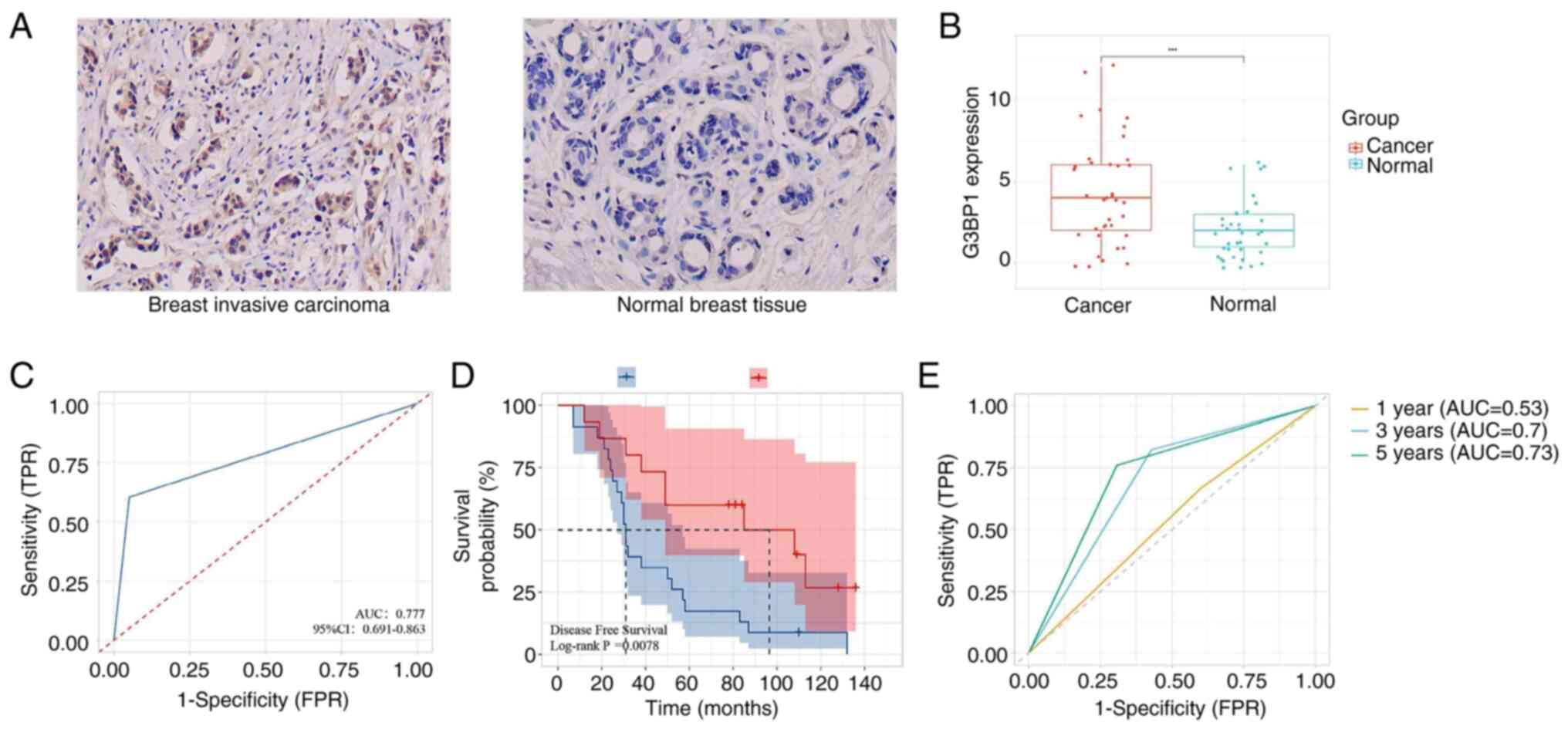 | Figure 6.G3BP1 expression, prognosis and
time-dependent ROC for BRCA. (A and B) G3BP1 protein was more
expressed in BRCA compared with normal tissues, analyzed using
immunohistochemistry. (C) The diagnostic value of G3BP1 expression
in BRCA. (D) Kaplan-Meier survival analysis showed that
upregulation G3BP1 had adverse DFS. (E) Time-dependent ROC curve of
G3BP1 expression in predicting 1-, 3- and 5-year DFS.
***P<0.001. BRCA, breast invasive carcinoma; DFS, disease free
survival; AUC, area under curve; G3BP1, Ras-GTPase-activating
protein SH3 domain-binding protein 1; ROC, receiver operating
characteristic; TPR, true positive rate; FPR, false positive
rate. |
Receiver operating characteristic (ROC) curve
analysis further demonstrated the diagnostic potential of G3BP1,
yielding an area under the curve (AUC) of 0.777 for distinguishing
BRCA from normal tissues (Fig. 6C),
indicating moderate diagnostic accuracy. Clinicopathological
analysis revealed that elevated G3BP1 expression was significantly
associated with advanced N stages (N2 and N3 vs. N0 and N1;
P=0.003), higher pathological stages (stage III and IV vs. I and
II; P=0.046), and tumor recurrence or metastasis (yes vs. no;
P=0.010) (Table I). No significant
associations were observed with age (P=0.285), menopausal status
(P=0.280), T stage (P=0.999), cardiovascular disease (P=0.999) or
diabetes (P=0.509), collectively indicating the specific role of
G3BP1 in driving lymphatic metastasis, local progression and
recurrence in breast cancer.
 | Table I.Association of G3BP1 expression and
clinicopathologic characteristics. |
Table I.
Association of G3BP1 expression and
clinicopathologic characteristics.
| Pathological
characteristic | n | Low G3BP1
expression | High G3BP1
expression | P-value for
Fisher's test |
|---|
| Age, years |
|
|
| 0.285 |
|
<45 | 27 | 9 | 18 |
|
|
≥45 | 11 | 6 | 5 |
|
| Menopausal
status |
|
|
| 0.280 |
|
Pre-menopause | 34 | 12 | 22 |
|
|
Menopause | 4 | 3 | 1 |
|
| T stage |
|
|
| 0.999 |
|
T1+T2 | 33 | 13 | 20 |
|
|
T3+T4 | 5 | 2 | 3 |
|
| N stage |
|
|
| 0.003 |
|
N0+N1 | 16 | 11 | 5 |
|
|
N2+N3 | 22 | 4 | 18 |
|
| Pathological
stage |
|
|
| 0.046 |
|
I+II | 17 | 10 | 7 |
|
|
III | 21 | 5 | 16 |
|
| Cardiovascular
disease |
|
|
| 0.999 |
| No | 34 | 12 | 22 |
|
|
Yes | 4 | 3 | 1 |
|
| Diabetes |
|
|
| 0.509 |
| No | 35 | 15 | 20 |
|
|
Yes | 3 | 0 | 3 |
|
|
Recurrence/metastasis |
|
|
| 0.010 |
| No | 7 | 6 | 1 |
|
|
Yes | 31 | 9 | 22 |
|
Kaplan-Meier survival analysis underscored the
prognostic relevance of G3BP1, with high expression levels
predicting significantly shorter DFS (P=0.007; Fig. 6D). Time-dependent ROC analysis
further validated the prognostic usefulness of G3BP1, achieving AUC
values of 0.530, 0.700 and 0.730 for predicting DFS at 1, 3 and
5-year intervals, respectively (Fig.
6E). After adjusting for age and complications (cardiovascular
disease and diabetes), multivariate Cox regression analysis showed
that the risk of recurrence or metastasis in the G3BP1
overexpression group significantly increased (HR=2.51; 95% CI,
1.07–5.90; P=0.034), supporting its independent value in the
prognosis assessment of breast cancer (Table SIV). Collectively, the integration
of multi-omics data and clinical validation solidifies G3BP1 as a
critical molecular driver linked to tumor aggressiveness and
unfavorable prognosis in BRCA.
Discussion
The present study comprehensively investigated the
clinical significance and biological implications of G3BP1 in BRCA
through integrated bioinformatics analysis and IHC validation. The
present findings demonstrated that G3BP1 is markedly overexpressed
in BRCA tissues at both mRNA and protein levels, and its high
expression is associated with advanced T stage, lymph node
metastasis and a poor prognosis. These results aligned with
emerging evidence that G3BP1, as a critical RBP serves an oncogenic
role in multiple cancers by regulating SG formation, mRNA
metabolism and signaling pathways such as PI3K/AKT/mTOR and
ubiquitin-mediated proteolysis (7,18).
Survival analyses from TCGA and Kaplan-Meier plotter
databases consistently revealed that high G3BP1 expression
predicted worse OS, DFS, DMFS and PPS in patients with BRCA. These
findings were supported by IHC validation in the present clinical
cohort, whereby elevated G3BP1 expression was markedly associated
with aggressive clinicopathological features, including advanced N
stage and pathological stage. ROC curve analysis further emphasized
the diagnostic potential of G3BP1, with an AUC of 0.777, suggesting
its usefulness in distinguishing BRCA from normal breast tissue,
consistent with previous studies on its diagnostic role in other
malignancies (19,20). Furthermore, time-dependent ROC
analysis demonstrated that the predictive efficacy of G3BP1 for
5-year DFS (AUC=0.700) was markedly superior to that of 1-year
prognosis (AUC=0.530), indicating its greater suitability for
medium-to-long-term prognostic evaluation.
GESA provided critical insights into the biological
pathways modulated by G3BP1 in BRCA. The enrichment of
ubiquitin-mediated proteolysis, PI3K/AKT/mTOR signaling and protein
secretion suggested that G3BP1 may drive tumor progression by
dysregulating protein homeostasis and oncogenic signaling networks
(7,21). The PI3K/AKT/mTOR pathway in
particular, is an established driver of breast cancer cell
proliferation, therapy resistance and metastasis (22), further supporting the role of G3BP1
in promoting aggressive tumor behavior. Additionally, immune
infiltration analysis revealed a complex relationship between G3BP1
expression and the TIME. G3BP1 exhibited a positive correlation
with immunosuppressive cell types (for example, macrophage M2 and
neutrophils) and a negative association with cytotoxic immune cells
(for example, CD8+ T and NK cells). This suggested that
G3BP1 may contribute to an immune-evasive phenotype, potentially
explaining its association with a poor prognosis. Recent studies
implicated RBPs in modulating immune checkpoint molecules (such as
PD-L1) and cytokine signaling (23,24),
suggesting G3BP1 could influence immunotherapy responses, a
hypothesis warranting further investigation.
The present data demonstrated that high G3BP1
expression was significantly associated with advanced N stage in
BRCA, implicating its role in promoting lymphatic metastasis. A
recent study reported that G3BP1 expression was notably upregulated
in breast cancer and that knockdown of G3BP1 suppressed the
proliferation and metastasis of breast cancer cells (25). In addition, G3BP1 has been shown to
promote the progression of nasopharyngeal carcinoma (NPC) by
activating the Janus kinase 2/STAT3 signaling pathway, which is
associated with lymph node metastasis and a poor prognosis in
patients with NPC (19).
Additionally, in non-small cell lung cancer, G3BP1 was shown to
interact with DEAH-Box Helicase 38 to activate the MAPK pathway and
EMT, promoting tumor cell migration and invasion, thereby
increasing the risk of lymph node metastasis (26). Subsequently, TIMER pan-cancer
analysis revealed G3BP1 upregulation in 18 out of 33 cancer types,
including cervical squamous cell carcinoma and endocervical
adenocarcinoma, stomach adenocarcinoma and lung adenocarcinoma,
which frequently exhibit lymphatic spread. Targeting G3BP1 may thus
represent a pan-cancer therapeutic strategy against lymphatic
metastasis.
To the best of our knowledge, the present study is
among the first to systematically link G3BP1 to BRCA progression
using multi-omics approaches. The consistency between TCGA data and
the present IHC results strengthened the reliability of the
conclusions drawn. Notably, the role of G3BP1 in SG formation may
also explain its upregulation in BRCA. Cancer cells frequently
encounter metabolic and oxidative stress, and G3BP1-mediated SG
assembly could enhance tumor cell survival under such conditions
(27,28). This adaptive mechanism may
contribute to chemotherapy resistance, a hypothesis supported by
the observation that high G3BP1 levels were associated with worse
PPS. The translational potential of G3BP1 lies in its dual role as
a diagnostic and therapeutic target. Due to its overexpression in
tumor tissues, G3BP1 could be used for liquid biopsy-based
detection of early-stage BRCA. Moreover, small-molecule inhibitors
targeting the RNA-binding domains of G3BP1 or its interaction with
SG components (for example, ubiquitin-specific peptidase 10 and
caprin 1) may offer novel therapeutic avenues (29,30).
Preclinical studies in other cancer types showed that G3BP1
knockdown suppresses tumor growth (17,19,24),
supporting its candidacy for targeted therapy in BRCA.
To directly resolve whether the pathway associations
of G3BP1 are primarily mediated through SGs or via direct molecular
interactions, future studies should employ SG perturbation assays
using CRISPR-mediated knockout of core SG components [such as T
cell intracellular antigen 1 (TIA-1)] and
pharmacological-integrated stress response inhibitors in BRCA cell
lines, followed by a comprehensive assessment of PI3K/AKT pathway
activity. With this, spatiotemporal mapping through live-cell
Förster resonance energy transfer imaging should distinguish direct
cytosolic binding from SG-dependent interactions between G3BP1 and
PI3K regulators. Functional validation using SG-defective G3BP1
mutants in isogenic knockout models should establish causal
relationships, while clinical correlation of SG markers (such as
TIA-1 and eukaryotic initiation factor 3 subunit η) with PI3K
activation signatures in BRCA samples using multiplex IHC should
confirm translational relevance (31–33).
The present study showed several limitations that
warrant discussion. Firstly, the IHC validation cohort (n=38)
derived from a single-center retrospective cohort may have limited
the statistical power for subgroup analyses. The present study
acknowledges that expanding sample size through multi-center
collaborations (for example, >100 paired samples) is essential
to validate the association between G3BP1 expression and breast
cancer. Secondly, the retrospective design carried inherent risks
of selection bias. Future prospective studies incorporating
longitudinal treatment response data could further strengthen the
clinical utility of G3BP1 as a prognostic biomarker. Thirdly, the
lack of clinically annotated cohorts receiving targeted therapies
(for example, PI3K inhibitors) or immunotherapy limited the direct
exploration of the predictive value of G3BP1. Fourthly,
experimental validation using small-interfering RNA/CRISPR models
and in vivo systems is essential to confirm the functional
mechanisms of G3BP1 (for example, the PI3K/AKT/mTOR pathway).
Investigating the interaction between G3BP1 and immune cells via
co-culture experiments, including macrophage polarization and T
cell inhibition assays, is equally important. Addressing these gaps
could substantially strengthen the scientific foundation for
translating G3BP1 into clinical applications, potentially
facilitating its development as both a diagnostic biomarker and a
therapeutic target in precision oncology.
In conclusion, the present study identified G3BP1 as
a critical prognostic biomarker in BRCA, with elevated expression
strongly associated with advanced tumor progression and diminished
survival outcomes. However, the small IHC validation cohort (n=38)
constrained subtype-specific generalization and
bioinformatics-predicted mechanisms (for example, PI3K/AKT
activation and immune evasion) require further functional
validation. Future multicenter cohorts and experimental models
should address these gaps and evaluate the translational potential
in precision oncology frameworks.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research Fund of Anhui
Institute of Translational Medicine (grant no. 2023zhyx-C65), Anhui
Provincial Health Scientific Research Project (grant nos.
AHWJ2024BAb30020 and AHWJ2023BAb20009) and Postgraduate Innovation
Research and Practice Program of Anhui Medical University (grant
no. YJS20240102).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JJL conceptualized the present study and was
involved in data curation (processing and annotating clinical and
pathological data from the internal patient cohort), formal
analysis, methodology, writing, reviewing and editing. ZQZ was also
involved in conceptualization, data curation (collecting and
organizing raw multi-omics data from public databases), formal
analysis and writing the original draft. JS was involved in formal
analysis, validation and writing the original draft. JHW was
involved in data curation (standardizing and validating IHC image
data and associated clinical metadata), methodology, validation and
writing the original draft. XZ was involved in data analysis,
interpretation and writing the original draft. WS was involved in
the methodology and validation. JL was involved in methodology, and
validation. HZ was involved in funding acquisition and methodology.
YWY was involved in funding acquisition, methodology, supervision
and visualization. FFL was involved in conceptualization, funding
acquisition, methodology, supervision, visualization, writing and
editing. JJL and FFL confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All procedures followed the ethical guidelines of
the Declaration of Helsinki in 1964 and its subsequent amendments.
The study received approval from the Ethics Committee of the Second
Affiliated Hospital of Anhui Medical University (approval no.
YX2019-055). This study declares that written informed consent was
obtained from all participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Newman L: Oncologic anthropology: Global
variations in breast cancer risk, biology, and outcome. J Surg
Oncol. 128:959–966. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdulla RA, Kareem NA, Assadi RA,
Sanaullah AAR, Nandagopal S, Wazil SM and Muttappallymyalil J:
Impact of breast cancer awareness program on breast screening
utilization among women in the United Arab Emirates: A
cross-sectional study. BMC Public Health. 25:5782025. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carlson RW, Allred DC, Anderson BO,
Burstein HJ, Carter WB, Edge SB, Erban JK, Farrar WB, Forero A,
Giordano SH, et al: Invasive breast cancer. J Natl Compr Canc Netw.
9:136–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ihle CL, Wright-Hobart SJ and Owens P:
Therapeutics targeting the metastatic breast cancer bone
microenvironment. Pharmacol Ther. 239:1082802022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baumann Z, Auf der Maur P and Bentires-Alj
M: Feed-forward loops between metastatic cancer cells and their
microenvironment-the stage of escalation. EMBO Mol Med.
14:e142832022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang L, Sun M, Yang S, Chen Y and Li T:
Intraoperative radiotherapy is not a better alternative to whole
breast radiotherapy as a therapeutic option for early-stage breast
cancer. Front Oncol. 11:7379822021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo J, Huang R, Mei Y, Lu S, Gong J, Wang
L, Ding L, Wu H, Pan D and Liu W: Application of stress granule
core element G3BP1 in various diseases: A review. Int J Biol
Macromol. 282:1372542024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun M, Liu X, Xia L, Chen Y, Kuang L, Gu X
and Li T: A nine-lncRNA signature predicts distant relapse-free
survival of HER2-negative breast cancer patients receiving taxane
and anthracycline-based neoadjuvant chemotherapy. Biochem
Pharmacol. 189:1142852021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Gu S, Wang L, Zhao L, Li T, Zhao
X and Zhang L: M2 macrophages promote PD-L1 expression in
triple-negative breast cancer via secreting CXCL1. Pathol Res
Pract. 260:1554582024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Ye CX, Chen HT, Li T, Ma LT and
Guo Y: Jianpi-Tiaoqi decoction inhibits tumour proliferation and
lung metastasis in tumour-bearing mice with triple-negative breast
cancer. Clin Exp Pharmacol Physiol. 51:e139002024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hashemi M, Taheriazam A, Daneii P,
Hassanpour A, Kakavand A, Rezaei S, Hejazi ES, Aboutalebi M,
Gholamrezaie H, Saebfar H, et al: Targeting PI3K/Akt signaling in
prostate cancer therapy. J Cell Commun Signal. 17:423–443. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tong W, Qin N, Lu T, Liu L, Liu R, Chen J
and Luo N: Integrating bulk and single-cell RNA sequencing reveals
SH3D21 promotes hepatocellular carcinoma progression by activating
the PI3K/AKT/mTOR pathway. PLoS One. 20:e03027662025. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng C, Guo H, Mo Y and Liu G:
Integrating bioinformatics and drug sensitivity analyses to
identify molecular characteristics associated with targeting
necroptosis in breast cancer and their clinical prognostic
significance. Recent Pat Anticancer Drug Discov. 19:681–694. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aeffner F, Adissu HA, Boyle MC, Cardiff
RD, Hagendorn E, Hoenerhoff MJ, Klopfleisch R, Newbigging S,
Schaudien D, Turner O and Wilson K: Digital microscopy, image
analysis, and virtual slide repository. ILAR J. 59:66–79. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bankhead P, Loughrey MB, Fernández JA,
Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ,
Coleman HG, et al: QuPath: Open source software for digital
pathology image analysis. Sci Rep. 7:168782017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ali HR, Chlon L, Pharoah PD, Markowetz F
and Caldas C: Patterns of immune infiltration in breast cancer and
their clinical implications: A gene-expression-based retrospective
study. PLoS Med. 13:e10021942016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ge Y, Jin J, Chen G, Li J, Ye M and Jin X:
Endometrial cancer (EC) derived G3BP1 overexpression and mutant
promote EC tumorigenesis and metastasis via SPOP/ERα axis. Cell
Commun Signal. 21:3032023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhan Y, Wang W, Wang H, Xu Y, Zhang Y,
Ning Y, Zheng H, Luo J, Yang Y, Zang H, et al: G3BP1 interact with
JAK2 mRNA to promote the malignant progression of nasopharyngeal
carcinoma via activating JAK2/STAT3 signaling pathway. Int J Biol
Sci. 20:94–112. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xing FL, Li BR, Fang YJ, Liang C, Liu J,
Wang W, Xu J, Yu XJ, Qin Y and Zhang B: G3BP2 promotes tumor
progression and gemcitabine resistance in PDAC via regulating
PDIA3-DKC1-hENT in a stress granules-dependent manner. Acta
Pharmacol Sin. 46:474–488. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo J, Zhao Y, Sui H, Liu L, Liu F, Yang
L, Gao F, Wang J, Zhu Y, Li L, et al: USP21-mediated G3BP1
stabilization accelerates proliferation and metastasis of
esophageal squamous cell carcinoma via activating Wnt/β-Catenin
signaling. Oncogenesis. 13:232024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu K, Wu Y, He P, Fan Y, Zhong X, Zheng H
and Luo T: PI3K/AKT/mTOR-targeted therapy for breast cancer. Cells.
11:25082022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Q, Yang Z, Hao X, Dandreo LJ, He L,
Zhang Y, Wang F, Wu X and Xu L: Niclosamide improves cancer
immunotherapy by modulating RNA-binding protein HuR-mediated PD-L1
signaling. Cell Biosci. 13:1922023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cook ME, Bradstreet TR, Webber AM, Kim J,
Santeford A, Harris KM, Murphy MK, Tran J, Abdalla NM, Schwarzkopf
EA, et al: The ZFP36 family of RNA binding proteins regulates
homeostatic and autoreactive T cell responses. Sci Immunol.
7:eabo09812022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu S, Tian S, Lin T, He X, Eze Ideozu J,
Wang R, Wang Y, Yue D and Geng H: G3BP1 regulates breast cancer
cell proliferation and metastasis by modulating PKCζ. Front Genet.
13:10348892022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mi K, Zeng L, Chen Y, Ning J, Zhang S,
Zhao P and Yang S: DHX38 enhances proliferation, metastasis, and
EMT progression in NSCLC through the G3BP1-mediated MAPK pathway.
Cell Signal. 113:1109622024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yao Z, Liu Y, Chen Q, Chen X, Zhu Z, Song
S, Ma X and Yang P: The divergent effects of G3BP orthologs on
human stress granule assembly imply a centric role for the core
protein interaction network. Cell Rep. 43:1146172024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Redding A and Grabocka E: Stress granules
and hormetic adaptation of cancer. Trends Cancer. 9:995–1005. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schulte T, Panas MD, Han X, Williams L,
Kedersha N, Fleck JS, Tan TJC, Dopico XC, Olsson A, Morro AM, et
al: Caprin-1 binding to the critical stress granule protein G3BP1
is influenced by pH. Open Biol. 13:2203692023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sheehan CT, Hampton TH and Madden DR:
Tryptophan mutations in G3BP1 tune the stability of a cellular
signaling hub by weakening transient interactions with Caprin1 and
USP10. J Biol Chem. 298:1025522022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Xu C, Qian X, Wang G, Han C, Hua H,
Dong M, Chen J, Yu H, Zhang R, et al: Myeloid PTEN loss affects the
therapeutic response by promoting stress granule assembly and
impairing phagocytosis by macrophages in breast cancer. Cell Death
Discov. 10:3442024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yeong J, Tan T, Chow ZL, Cheng Q, Lee B,
Seet A, Lim JX, Lim JCT, Ong CCH, Thike AA, et al: Multiplex
immunohistochemistry/immunofluorescence (mIHC/IF) for PD-L1 testing
in triple-negative breast cancer: A translational assay compared
with conventional IHC. J Clin Pathol. 73:557–562. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen CH, Chen IC, Hsu CL, Lu TP, Wang MY,
Tsai LW, Huang CS, Lu YS and Lin CH: Characterization of the tumor
immune microenvironment in pregnancy-associated breast cancer
through multiplex immunohistochemistry and transcriptome analyses.
Breast Cancer Res. 27:1542025. View Article : Google Scholar : PubMed/NCBI
|