Introduction
Cervical cancer is the fourth most prevalent
malignancy diagnosed in women worldwide, following breast,
colorectal and lung cancer (1).
Numerous risk factors have been associated with cervical cancer,
including human papillomavirus (HPV) infection, early onset of
sexual activity, multiple sexual partners, the use of oral
contraceptives and smoking. In this context, HPV vaccination has
demonstrated safety and effectiveness as a primary prevention
strategy for cervical intraepithelial neoplasia (CIN) and cervical
cancer (2). CIN is a premalignant
stage in the development of cervical cancer and provides an
opportunity for early detection and intervention. Although
persistent infection with HPV is considered the major cause of the
disease, only a small proportion of HPV-infected women develop
cervical malignancy, which limits the utility of HPV testing due to
its low positive predictive value for high-grade lesions (3,4). Other
main screening techniques for cervical cancer include the Pap
smear, visual inspection with acetic acid and Lugol's iodine and
liquid-based cytology. Several countries, including Türkiye, United
Kingdom, Australia, Canada and The Netherlands, have implemented
cytology-based testing and HPV testing as part of their cancer
screening programs (5). There is
increasing interest in the identification of novel molecular
markers to improve the identification of individuals at high risk
of cervical cancer, with the aim of expanding the reach of
screening programs in the community, improving accessibility and
increasing their diagnostic accuracy.
In almost every type of human cell, there are
hundreds to thousands of mitochondria, each containing its own
genome, a feature that distinguishes mitochondria from other
organelles. Human mitochondrial DNA (mtDNA) is a circular,
double-stranded DNA of ~16,569 nucleotides that is involved in a
variety of cellular functions (6).
It contains a total of 37 genes encoding 13 proteins essential for
the oxidative phosphorylation system, as well as 22 transfer RNAs
and 2 ribosomal RNAs (7). Several
characteristics distinguish mtDNA from nuclear DNA (nDNA). The most
well-known of these is the maternal inheritance of mtDNA, which
differs from the Mendelian pattern of inheritance. Another feature
is that the mitochondrial genome is polyploid, meaning that each
cell has multiple copies of mtDNA depending on the cell type
(8). mtDNA is present in multiple
copies per cell, with the copy number typically ranging from 1,000
to 10,000 and varying according to the type of cell (9). Notably, mtDNA is particularly
vulnerable to damage and copy number alterations, due to exposure
to reactive oxygen species (ROS) and other metabolites generated
within mitochondria, the lack of protective histones, and
inadequate repair and protection mechanisms (10). Alterations in mtDNA copy number
(mtCN) can potentially result in impaired mitochondrial function
and increased ROS production. Variations in mtCN have been observed
in numerous pathological conditions, including neuropsychiatric
disorders, autoimmune diseases, chronic inflammation and various
types of cancer (11–13). In this context, alongside
well-characterized alterations in nDNA, mtDNA has also been
extensively studied in a range of cancer types (9).
Changes in mtCN, reflecting the balance between
mitochondrial biogenesis and degradation, have emerged as potential
indicators of cellular dysfunction and tumorigenesis. It has been
suggested that mtCN increases during the early stages of cancer to
compensate for metabolic defects in damaged mitochondria, However,
in advanced stages, cumulative damage may result in mtDNA
degradation and a subsequent reduction in mtCN (14,15).
In light of this, previous studies have elucidated the involvement
of mtCN alterations in the pathogenesis of various cancer types in
different patient cohorts. mtCN changes reported in the literature
vary according to cancer stage, the characteristics of the study
cohort and additional patient-related factors (16). This variability indicates that, when
mtDNA is considered as a biomarker in cancer, expectations of
increased or decreased copy numbers in tumor samples cannot be
generalized and may not be independent of diagnosis, stage and
patient characteristics. Therefore, it is essential to
quantitatively determine the alterations of mtDNA within the
specific population for which a predictive model is being
developed.
Considering that high-risk HPV oncoproteins E6 and
E7 may induce DNA damage by affecting ROS production by different
mechanisms (17), it can be
hypothesized that alterations in mtCN associated with mtDNA damage
in cervical cancer cells may contribute to cervical carcinogenesis.
Supporting this hypothesis, Sun et al (18) demonstrated that the mtCN in cells
from cervical smear samples from patients with cervical cancer was
significantly higher than that in corresponding samples from
cancer-free controls. Furthermore, Warowicka et al (19) reported an association between
increased mtCN, as well as mtDNA mutations, with the pathogenesis
of cervical cancer. A recent study has demonstrated a significant
association between altered mtCN and cervical cancer, even in
HPV-negative cases. Al-Awadhi et al (20) demonstrated elevated mtCN levels in
both high-risk HPV-positive and HPV-negative cervical samples,
suggesting that this increase may represent an adaptive response to
mitochondrial oxidative stress and energy deficiency.
Despite accumulating evidence that high-risk HPV
oncoproteins (E6/E7) can increase oxidative stress and perturb
mitochondrial homeostasis, the direction and magnitude of mtCN
changes appear to vary across settings. In cervical pathology,
previous studies have primarily compared cancer or high-grade
squamous intraepithelial lesions (HSILs) with samples from control
subjects without evaluating mtCN alterations across the full
spectrum of CIN. In addition, they have rarely included HPV status
in the same analysis. Consequently, it remains unclear whether mtCN
exhibits a stepwise pattern with increasing histological severity
and whether such a pattern is influenced by HPV positivity. To
address this gap, the present study quantified mtCN across normal,
CIN1 [low-grade squamous intraepithelial lesion (LSIL)], CIN2/3
(HSIL) and invasive cervical cancer tissues. In addition, the
combined contribution of mtCN, HPV status and smoking status was
evaluated in relation to the classification of high-risk
disease.
Materials and methods
Study group
A total of 100 participants who presented at the
Gynecology and Obstetrics Clinic of Ankara Etlik City Hospital
(Ankara, Türkiye) between July 2023 and September 2023 and provided
cervical samples were included in the present study. All
participants were female, and their median age was 53 years, with
an age range of 45–65 years. Prior to intervention, written
informed consent was obtained from each patient. The patients were
selected based on histopathological diagnosis, and included 32
healthy individuals and 52 patients with cervical pathology. The
latter comprised 21 cases of CIN1 (LSIL), 23 cases of CIN2/3 (HSIL)
and 8 cases of cervical cancer. The remaining 16 samples were
excluded due to insufficient DNA yield or suboptimal quality.
Personal and clinical data of the participants,
including age, medical history, histopathological features and HPV
genotype were collected from the digital medical archive. The
samples were collected from patients aged 45–65 years, with efforts
made to minimize age-related variations in mtDNA by ensuring a
comparable age distribution across groups. To standardize mtCN,
individuals with a history of chronic exposure to environmental or
occupational agents, narcotic substances or drugs, as well as those
with chronic inflammation of the internal genital organs or
adjacent tissues, were excluded from the study. Smoking status was
recorded and evaluated as a variable in the analysis. The control
group included both HPV-negative and HPV-positive cases; HPV16 and
HPV18 were classified as high-risk. The present study was approved
by the Ethics Committee of Ankara Etlik City Hospital
(AEŞH-EK1-2023-015).
DNA extraction
Genomic DNA was extracted from formalin-fixed,
paraffin-embedded (FFPE) cervical tissue sections using the
AmoyDx® FFPE DNA Kit (Amoy Diagnostics Co., Ltd.),
following the manufacturer's protocol. The FFPE tissue blocks had
been archived for ≤12 months. Serial 5-µm FFPE sections were cut
for nucleic acid extraction. Briefly, deparaffinization was
performed using the standard xylene/ethanol method and DNA was
isolated via the silica column-based procedure of the kit. The
purified DNA was eluted and stored at −20°C until further analysis.
DNA concentration and purity were evaluated using a NanoDrop
ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.) and samples with an optical density (OD)260/OD280
ratio between 1.8 and 2.0 were considered suitable for downstream
sequencing.
HPV status assessment
The HPV status of the cervical samples was
determined. This was performed by analysis of the extracted DNA
using the cobas® HPV Test on the cobas® 4800
testing system according to the manufacturer's instructions (Roche
Diagnostics for both). This qPCR-based assay detects 14 high-risk
HPV genotypes, providing individual results for HPV16 and HPV18 and
pooled detection for 12 additional types (HPV31, 33, 35, 39, 45,
51, 52, 56, 58, 59, 66 and 68). The specific primer sequences used
in this commercial assay are proprietary to the manufacturer and
not disclosed. The analytical sensitivity and specificity of the
cobas® HPV Test for the detection of CIN2+ lesions have
been reported in clinical validation studies to be 90–99 and ~87%,
respectively (21). All analyses
were performed in the institutional laboratory following standard
quality control procedures.
Measurement of mtCN
The mtCN normalized to nDNA in cervical cells was
determined using quantitative PCR (qPCR). The difference in
quantification cycle (ΔCq) values of the NADH dehydrogenase subunit
1 (ND1) and hemoglobin subunit β (HBB) genes were used to evaluate
the mtCN and nDNA copy number, respectively. Amplification was
performed using QuantiNova SYBR Green PCR Master Mix (Qiagen, Inc.)
and a commercial primer assay (RT2 qPCR Primer Assay;
Qiagen, Inc.) on the CFX96 Touch Real-Time PCR Detection System
(Bio-Rad Laboratories, Inc.). The primer sequences are presented in
Table SI. The reference gene,
hemoglobin subunit beta (HBB), was selected as a single-copy
nuclear locus with stable representation in cervical epithelial DNA
and no known regulation by mitochondrial pathways, consistent with
previous validations (11–13).
qPCR was performed for 40 cycles using a two-step
protocol, comprising denaturation at 95°C for 15 sec, followed by
combined annealing and extension at 60°C for 60 sec, in accordance
with the manufacturer's recommendations for SYBR Green chemistry.
The assay results were accepted when the following criteria were
met: i) Cq values <35 for both the mtDNA target ND1 and the
single-copy nuclear reference (HBB); ii) no amplification in
no-template controls (Cq undetermined or >40); iii) melting
curve analysis demonstrated a single, specific peak with a
consistent melting temperature across replicates (±0.5°C); and iv)
technical replicates differed by ≤0.3 Cq.
The relative mtCN differences between the study
groups and controls were calculated using the 2−ΔΔCq
method and expressed as fold change (22).
Power analysis
An a priori sample size calculation was
performed using G*Power software (version 3.1.9.7; Heinrich Heine
University Düsseldorf; http://www.gpower.hhu.de) based on a two-tailed
independent t-test with an α value of 0.05 and statistical power of
0.80. The analysis was designed to compare a high-risk group,
comprising patients with HSIL or invasive cancer, with a low-risk
group, comprising patients with LSIL or normal cervical cytology
based on mtCN fold-change (2−ΔΔCq) as the continuous
outcome measure. A conservative effect size corresponding to a
large Cohen's d of 0.80 was assumed to prevent the overestimation
of results, based on previous studies reporting significant
mitochondrial alterations, including elevated mtCN, in cervical
cell exfoliates, and major mitochondrial genomic alterations in
dysplastic and cancerous tissues (20,22,23).
Under the assumption of an equal group allocation, the required
sample size was 25 per group. A 20% attrition/mismeasurement buffer
was established; therefore, the prespecified target sample size was
increased to 30 per group (total, n=60). These assumptions were
informed by published evidence on mitochondrial abnormalities in
HPV-related cervical pathology (19,20).
Statistical analysis
Statistical analyses were performed using R (version
4.1.3; R Foundation for Statistical Computing; http://www.r-project.org/) via the RStudio interface
(Posit Software, PBC; http://posit.co/). Descriptive statistics are
presented as the mean ± standard deviation or median and
interquartile range, as appropriate. One-way analysis of variance
was applied to normally distributed numerical values, Associations
between categorical variables were evaluated using Fisher's exact
test.
In the model-building process, the normal and LSIL
categories were merged to form a low-risk group, while the HSIL and
invasive cancer categories were combined to form a high-risk group,
yielding a binary outcome variable. Logistic regression with a
logit function was used to predict the binomial target variable. As
only one numerical predictor was available, univariate methods were
used to identify potential outliers and extreme values prior to
model fitting. Overall, seven models were constructed and used for
prediction. The performance of all models was assessed using
sensitivity, specificity, accuracy and receiver operating curve
(ROC) analysis, and the area under the curve was determined.
Optimal thresholds were determined using the Youden index.
Statistical significance was defined as P<0.05, with an a set at
0.05 and β at 0.80 (24).
Results
Descriptive statistics
Descriptive characteristics of the full cohort of
100 patients according to disease status are presented in Table I, including age, HPV and smoking
status. The mean age of all patients was 53.45±5.74 (range, 45–65)
years. Patients positive for HPV16 or HPV18 were classified as the
high-risk oncogenic HPV group, in accordance with
established cervical cancer risk stratification. Patients positive
for other HPV genotypes detected by the assay (including non-16/18
high-risk types) as well as HPV-negative individuals were analyzed
together as a non-HPV16/18 group for comparative purposes.
No statistically significant differences were observed between the
groups with respect to age, HPV category or smoking status.
 | Table I.Comparison of demographic and
clinical variables of the full cohort according to cervical disease
group. |
Table I.
Comparison of demographic and
clinical variables of the full cohort according to cervical disease
group.
| Variables | Total (n=100) | Normal (n=38) | LSIL (n=25) | HSIL (n=26) | Cancer (n=11) | P-value |
|---|
| Age, mean ± SD | 53.45±5.74 | 53.9±5.84 | 54.5±5.30 | 51.7±5.58 | 53.5±6.55 | 0.316a |
| (min-max),
years | (45–65) | (45–65) | (45–63) | (45–62) | (45–64) |
|
| HPV, n (%) |
|
|
|
|
| 0.057b |
|
High-risk | 55 (55.00) | 15 (39.47) | 15 (60.00) | 16 (61.54) | 9 (81.82) |
|
|
Low-risk | 45 (45.00) | 23 (60.53) | 10 (40.00) | 10 (38.46) | 2 (18.18) |
|
| Smoking status, n
(%) |
|
|
|
|
| 0.51b |
|
Smoker | 18 (18.00) | 5 (13.16) | 7 (28.00) | 4 (15.38) | 2 (18.18) |
|
|
Non-smoker | 82 (82.00) | 33 (86.84) | 18 (72.00) | 22 (84.62) | 9 (81.82) |
|
Comparison of mtCN results according
to cervical disease category, HPV status and smoking status
Subsequent mtCN analyses were performed only in
samples with adequate DNA yield and quality (n=84), after exclusion
of 16 samples; Table I summarizes
the baseline characteristics of the full enrolled cohort (n=100).
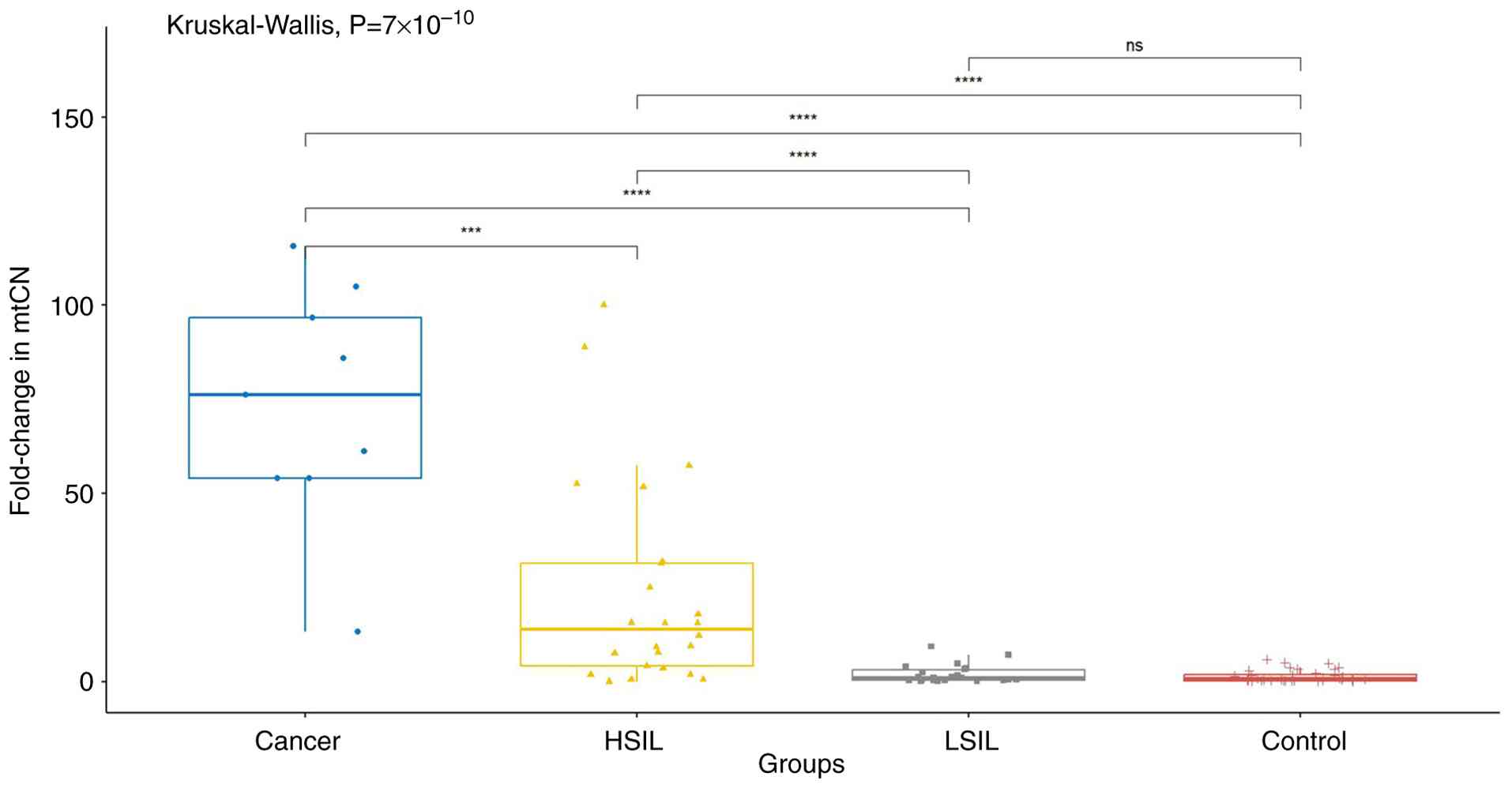
In the first analysis, the groups were compared according to
disease category. A statistically significant difference was
identified among the groups in terms of mtCN (P<0.001). Post hoc
analysis revealed significant differences between the cancer and
control groups, the cancer and HSIL groups, the control and HSIL
groups, the cancer and LSIL groups, and the HSIL and LSIL groups
(P<0.001). However, no significant difference was detected
between the control and LSIL groups. In addition, when HSIL and
cancer cases were combined into a high-risk group and LSIL and
control group patients were combined into a low-risk group, a
statistically significant difference in mtCN was observed between
these groups (P<0.001; Fig.
1).
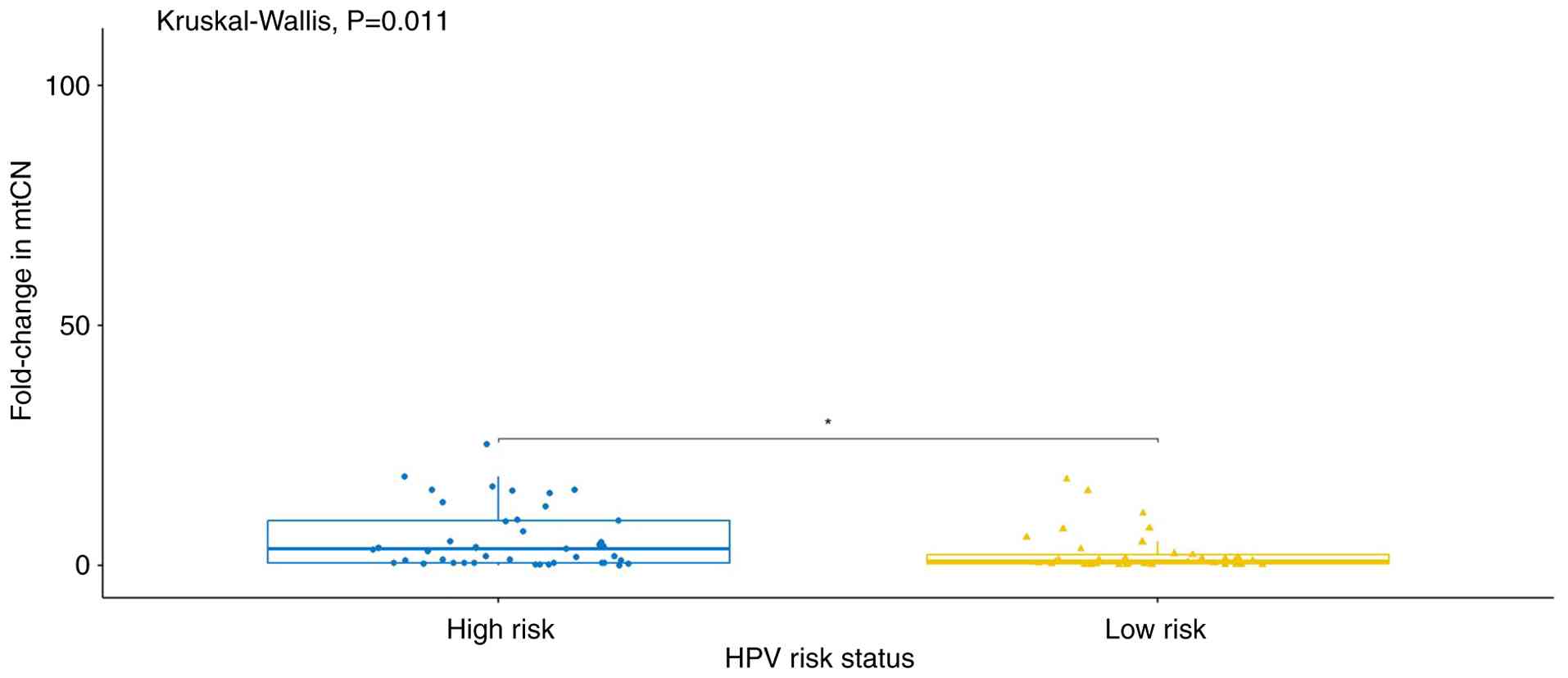
In the second analysis, the participants were
categorized according to their HPV status. Patients positive for
HPV16 and/or HPV18 were classified as the high-risk HPV group,
while patients positive for other HPV types or with no detectable
HPV infection were classified as the low-risk HPV group. A
statistically significant difference in mtDNA copy number was
observed between these groups (P<0.05; Fig. 2).
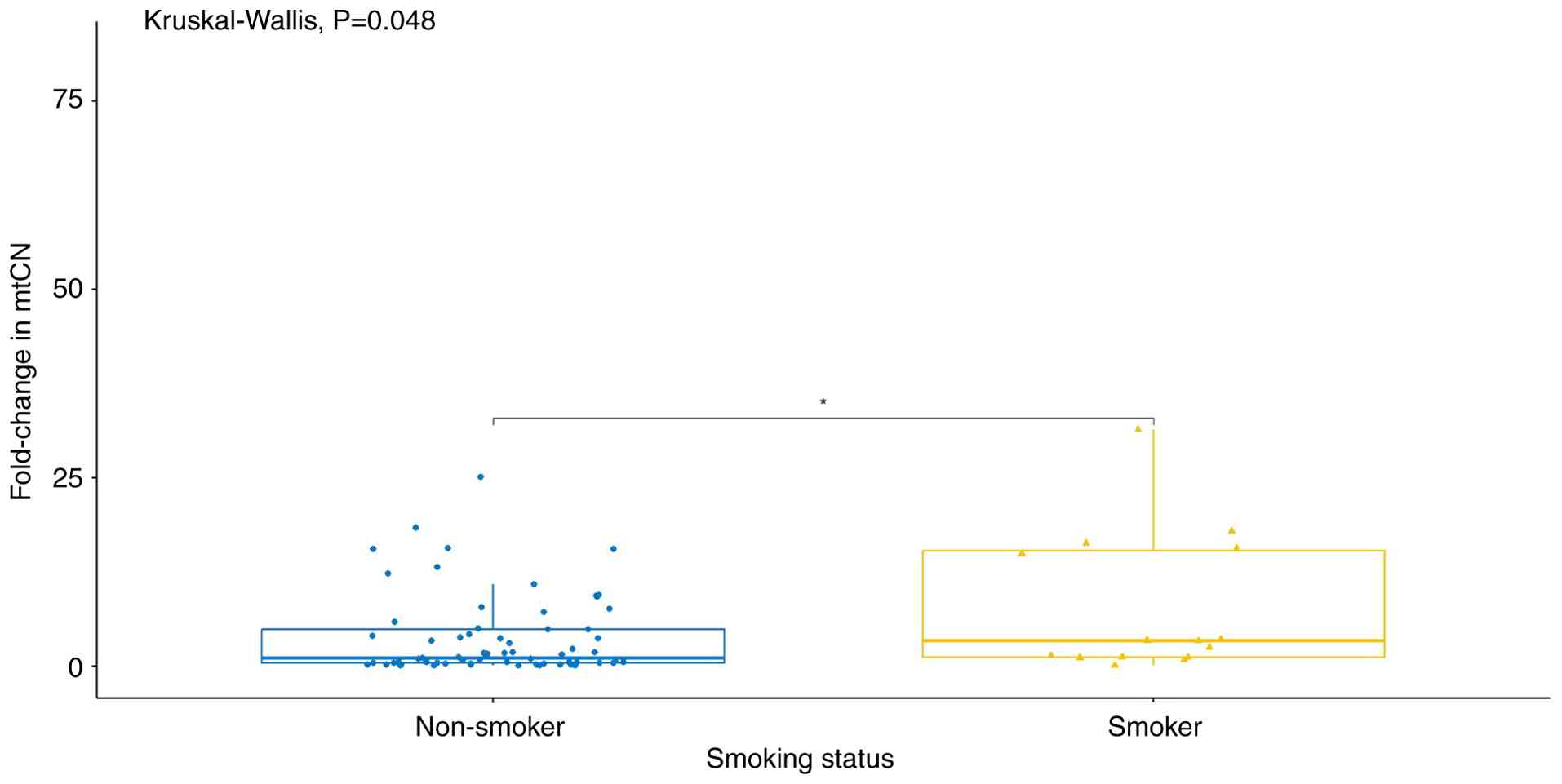
In the third analysis, the mtCN was compared
according to smoking status. A statistically significant difference
was detected between non-smokers and smokers (P<0.05; Fig. 3).
Development of a model for prediction
of high-risk and low-risk cervical lesions
A prediction model was developed to distinguish
between high-risk and low-risk cervical disease groups. Prior to
model development, outlier detection was performed using the local
outlier factor method, and extreme values were excluded from the
analysis. Logistic regression analysis was implemented for model
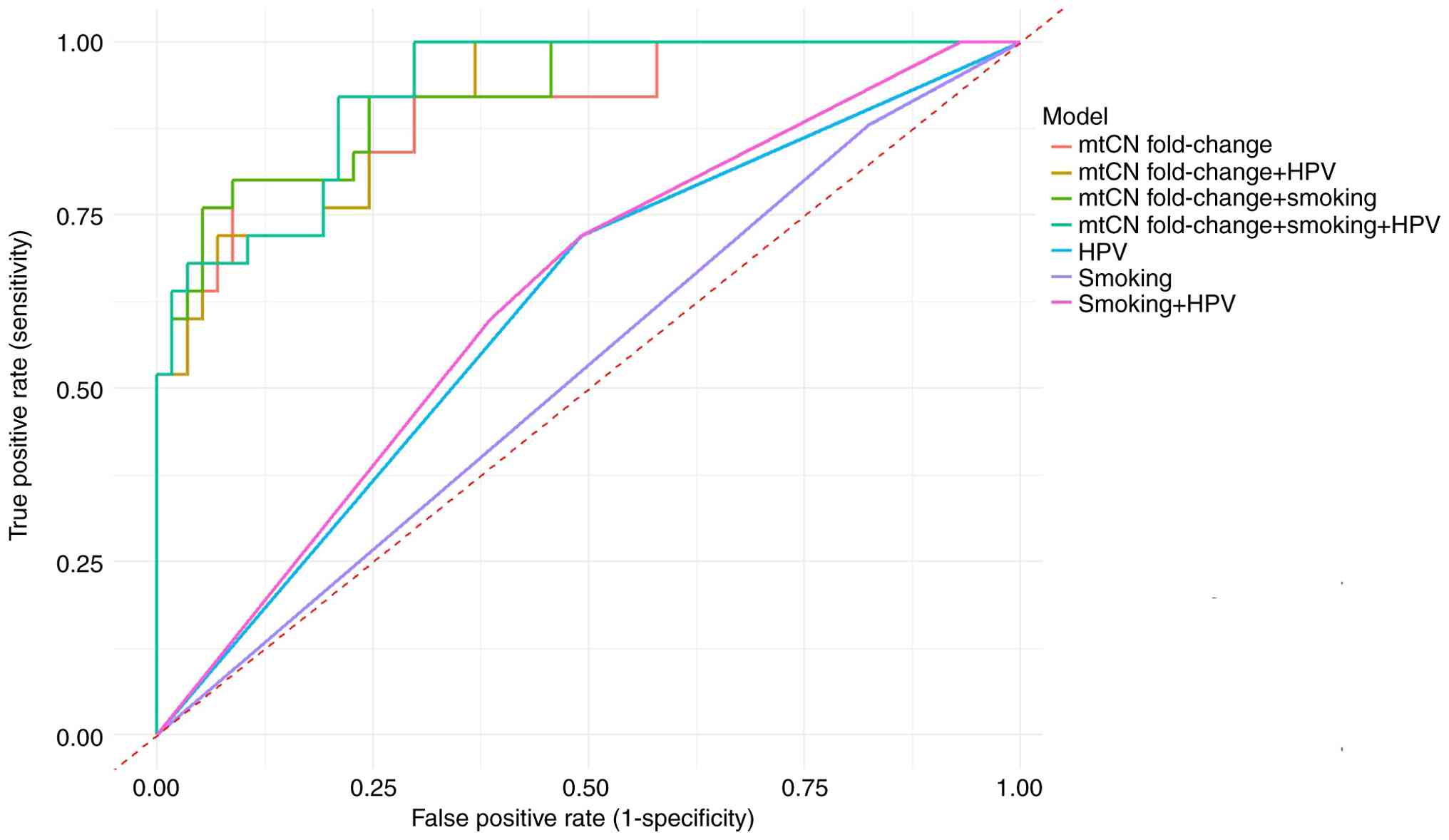
development. Among the seven tested models, the triple model
incorporating mtCN fold change, smoking status and HPV status
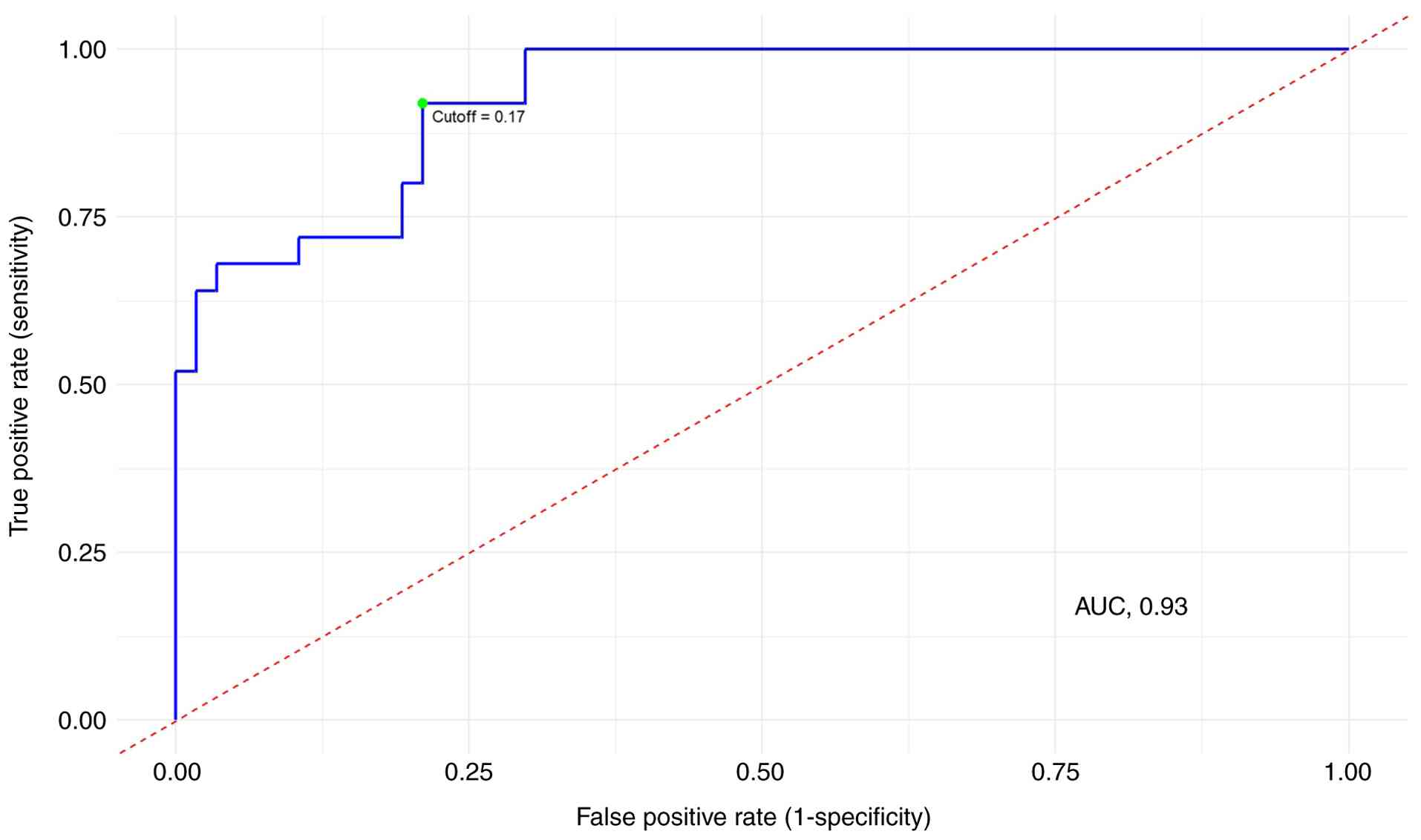
showed the highest discriminative performance (AUC=0.93; Figs. 4 and 5). As the study restricted the ages of the
participants to a narrow range of between 45 and 65 years, age was
excluded from the modeling process.
Among the predictors included in the model, mtCN
changes were identified as the strongest predictor of risk status.
Smoking status and HPV status were evaluated as independent
variables, demonstrated weaker associations and did not reach
statistical significance (Table
II). The triple model, including all three variables,
demonstrated a superior overall performance. Consequently, this
specific model was selected as the final predictive model. The
optimal threshold value for successful differentiation using the
triple model was determined, and a cutoff value of 0.17 was found
to be optimum (Fig. 5). At this
cutoff, the triple model exhibited a sensitivity of 79% and
specificity of 92% (Table
III).
 | Table II.Triple logistic regression model
results. |
Table II.
Triple logistic regression model
results.
|
|
|
| 95 CI% |
|---|
|
|
|
|
|
|---|
| Variables | P-values | Odds ratio | Lower | Upper |
|---|
| Intercept | <0.001 | 1.146 | 1.038 | 1.471 |
| mtCN fold
change | 0.003 | 3.551 | 3.013 | 4.548 |
| HPV | 0.083 | 1.426 | 1.062 | 5.108 |
| Smoking | 0.203 | 1.081 | 1.002 | 2.522 |
 | Table III.Performance metrics outcomes of all
models. |
Table III.
Performance metrics outcomes of all
models.
| Model
variables | Accuracy | Sensitivity | Specificity | Positive predictive
value |
|---|
| mtCN fold
changea | 88 | 91 | 80 | 91 |
| HPVb | 58 | 51 | 72 | 81 |
| mtCN fold change +
smokinga | 88 | 92 | 80 | 91 |
| mtCN fold change +
HPVa | 81 | 76 | 92 | 96 |
| mtCN fold change +
HPV + smokinga | 83 | 79 | 92 | 95 |
Discussion
In the present study, the relationship between mtCN
variations and the progression of CIN to cervical cancer was
investigated. The findings provide important insights and are
consistent with previous studies on mtDNA alterations in various
cancer types. They also expand upon previous studies by providing a
comprehensive understanding of the role of mtCN changes across the
CIN spectrum. Notably, statistically significant increases in mtCN
were observed with increasing disease severity, culminating in
cervical cancer.
Previous studies have reported increased mtCN in
cervical disease. In a cohort consisting exclusively of high-risk
HPV-positive women, Sun et al (18) observed that mtCN levels in
exfoliated cervical cells were higher in women with cervical cancer
compared with cancer-free controls, supporting a potential role for
mtCN alterations in cervical carcinogenesis. Similarly, based on
tissue-based analyses, Warowicka et al (19) reported that the mtCN in HSIL and
carcinoma is elevated compared with that in LSIL (19). In the cohort of the present study,
mtCN increased in a stepwise manner with increasing histological
severity, which was consistent with these previous
observations.
Al-Awadhi et al (20) reported elevated mtCN in cervical
abnormalities irrespective of HPV status, suggesting an adaptive
response to mitochondrial stress. The present study extends the
results provided in the literature in the following three aspects:
i) mtCN was evaluated across the full CIN spectrum in tissue, that
is, in normal, CIN1, CIN2/3 and cancerous tissues, rather than by
the comparison of only extreme disease categories; ii) HPV status
was explicitly incorporated, demonstrating that HPV positivity
amplifies the stepwise increase in mtCN; and iii) these findings
were integrated into a joint predictive framework combining mtCN
with HPV status and smoking, with defined operating characteristics
(threshold, 0.17; sensitivity, 79%; specificity, 92%), thereby
positioning mtCN within a clinically oriented risk-stratification
context (Fig. 5).
High-risk HPV oncoproteins establish a persistent
pro-oxidant state that perturbs mitochondrial homeostasis.
E6/E7-driven oxidative stress increases ROS production and DNA
damage in infected epithelia; since mtDNA is in close proximity to
the respiratory chain, lacks protective histones and has limited
repair capacity, it is particularly susceptible to damage. Such
mitochondrial damage can activate mitochondrial biogenesis
pathways, including the peroxisome proliferator-activated receptor
g coactivator-1a (PGC-1α)/nuclear respiratory factor 1
(NRF-1)/mitochondrial transcription factor A (TFAM) axis, leading
to an increase in mtCN as a compensatory response to maintain
bioenergetic function. These mechanisms, described in previous
reviews of HPV-related oxidative-stress and mitochondrial biology,
provide a biologic rationale for the stepwise increase in mtCN
observed with increasing cervical disease severity and for the
higher mtCN detected in HPV-positive samples in the present cohort
(18,25,26).
Previous studies have reported both increased and
decreased mtCN across different tumor types (27). Elevated mtCN has been observed in
breast, bladder, esophageal, head and neck squamous, kidney and
liver cancers, with lung carcinoma as a notable exception, when
compared with non-tumorous tissues (28). By contrast, reduced mtCN has been
reported in kidney clear cell carcinoma, hepatocellular carcinoma
and myeloproliferative tumors (29). The association between mtCN
variations and the risk of cancer progression has also been
documented. A meta-analysis by Hu et al (30) demonstrated a significant association
between increased mtCN and the risk of lymphoma, melanoma and
breast cancer, whereas an inverse relationship was observed for
hepatic carcinoma. These divergent results suggest that mtCN may
increase as a compensatory response to mitochondrial damage via
increased mitochondrial biogenesis, whereas decreased mtCN may
occur when mitochondrial damage becomes excessive and damaged cells
are eliminated by mitophagy and increased cellular turnover. Which
of these occurs depends on tissue context, tumor metabolic demands
and disease stage. In cervical pathology, syntheses of recent
evidence indicate a tendency toward mtCN elevation in high-grade
lesions and cervical cancer, particularly in HPV-positive settings,
consistent with compensatory mitochondrial biogenesis under chronic
ROS-induced stress. The findings of the present study align with
this context-dependence and position cervical disease on the
‘increased mtCN’ side of this bidirectional framework (20,26).
Given the high prevalence of smoking in Türkiye, a
comparison between cervical tissue samples from smokers and
non-smokers was performed in the current study. Consistent with
findings from large epidemiologic cohorts, smoking has been
associated with a reduction in leukocyte mtCN (31,32),
generally interpreted as systemic depletion associated with
oxidative stress. However, in the present tissue-based analysis,
upregulated mtCN was observed in the cervical samples of smokers.
This apparent discrepancy supports a tissue-blood dissociation,
whereby localized oxidative stress and increased bioenergetic
demands in pre-neoplastic or tumor tissue may promote compensatory
mitochondrial biogenesis via the PGC-1α/NRF-1/TFAM axis (33). This interpretation is consistent
with previous literature on cervical cancer, which has reported the
elevation of mtCN in high-grade cervical lesions or cervical cancer
(20,26). It is important to note that this
stratified, within-tissue observation, when combined with the
CIN-graded analysis and the integrated predictive model combining
mtCN, HPV status and smoking, constitutes a novel contribution of
the present study. Together, these findings refine the
interpretation of mtCN as a biomarker for risk stratification in
HPV-related cervical disease.
The potential of mtCN as a biomarker for cancer
prognosis is well-documented. Distinct mtCN levels have been shown
to differentiate control populations from patients with solid
tumors and other diseases (9,23).
Despite this, the routine clinical assessment of mtCN is not
possible due to the lack of a standardized method (34). Although the qPCR technique remains
the gold standard for mtCN quantification, variations in its
application among different laboratories contribute to inconsistent
results. In particular, differences in reference genes, which often
vary based on the sample type and disease context, complicate
standardization (28,34). An additional technical limitation is
that the nuclear genome can replicate a significant portion of the
mtDNA genome, creating nuclear mtDNA segments that may interfere
with mtCN measurement (35,36). This genetic overlap necessitates the
careful selection of mtDNA target regions to avoid the
co-amplification of these pseudogenes and ensure accurate mtCN
quantification (36,37). In the present study, mtCN was
demonstrated to be a significant biomarker for the progression of
CIN to cervical cancer. The observed mtCN alterations in cervical
tissues, notably the increased mtCN in HPV-positive cases and among
smokers, further highlight its potential utility in clinical
practice. However, the lack of standardized methods for mtCN
assessment underscores the requirement for the development of
consistent and reproducible techniques for clinical implementation.
The present study contributes to the growing body of evidence
supporting the importance of mtCN as a biomarker and emphasizes
that standardized protocols are necessary to enhance its utility in
cancer prognosis and screening.
The interaction between mtCN and HPV positivity was
evaluated using the Wald test within logistic regression models,
which is similar to the analytical approach previously described by
Sun et al (18). Logistic
regression models have also been employed in other studies
investigating mtCN in various cancer types, for example, in a study
on head and neck squamous cell carcinoma in which mtCN modeling in
leukocytes was performed (38). In
the present study, ROC curve analysis was used to compare all
possible model combinations. Among these, the triple model
incorporating mtCN fold-change, smoking status and HPV status
demonstrated the strongest classification performance. Notably,
mtCN fold-change alone was the strongest contributor to the model.
The ROC curve analysis for the triple model indicates a
discrimination threshold of 0.17, corresponding to a sensitivity
and specificity of 79 and 92%, respectively.
Several limitations of the present study must be
acknowledged. Firstly, the sample size was relatively small, which
may limit the generalizability of the findings. Therefore, larger
studies are required to validate these results and provide more
robust evidence. Second, the cross-sectional design of the study
with single-time-point sampling precludes the determination of
whether mtCN alterations are a cause or a consequence of CIN
progression. Prospective longitudinal cohort studies with serial
cervical tissue sampling are required to clarify the temporal and
causal relationship between mtCN dynamics and cervical disease
development. Furthermore, although qPCR is considered the gold
standard for mtCN quantification, the lack of standardized
protocols across laboratories may introduce variability and limit
comparability. The development of standardized methods for mtCN
assessment will be essential to ensure consistency and
reproducibility in clinical settings. In addition, the cancer group
included only 8 patients; consequently, the power analysis was
conducted by combining the HSIL and cancer groups into a single
high-risk category and comparing this with a low-risk category
combining the LSIL and normal groups. While this approach ensured
sufficient statistical power to detect overall differences in mtCN,
it may obscure variations specific to invasive cancer. Therefore,
the results should be interpreted with caution, as this grouping
limits the ability to draw distinct conclusions with regard to the
cancer subgroup alone. Finally, the study did not account for all
potential confounding factors, such as environmental exposures and
genetic susceptibility, that may influence mtCN levels. Future
studies should consider these variables to provide a more
comprehensive understanding of the factors affecting mtCN in
cervical cancer.
In conclusion, the present study provides compelling
evidence that alterations in mtCN are significantly associated with
the progression of CIN to cervical cancer. The findings highlight
the potential of mtCN as a biomarker for cervical cancer risk
assessment, particularly in HPV-positive cases. However, larger
longitudinal studies with standardized protocols are essential to
validate these results and facilitate their clinical translation.
Overall, the insights gained from the study contribute to the
growing body of evidence supporting the importance of mtCN in
cancer biology and underscore that it is necessary to continue to
investigate its role in tumorigenesis and its potential use as a
diagnostic tool.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Miss Merve Ulak
(Genetic Diseases Evaluation Center, Ankara Etlik City Hospital)
for their assistance with sample processing and laboratory
workflows; Miss Esra Cesur (Genetic Diseases Evaluation Center,
Ankara Etlik City Hospital), Miss Seyda Gurevin (Genetic Diseases
Evaluation Center, Ankara Etlik City Hospital) and Miss Merve
Karademir (Genetic Diseases Evaluation Center, Ankara Etlik City
Hospital) for their support in DNA extraction and qPCR
procedures.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
IO and HBE contributed to conceptualization, formal
analysis, investigation and drafting the original manuscript. MTA
performed statistical analysis, data curation (systematic data
retrieval, clinical record verification, database management),
designed scientific figures and assisted with graphical data
presentation. OO contributed to clinical methodology, patient
recruitment, clinical evaluation and tissue acquisition. FSD
contributed to formal analysis and methodology. ECD contributed to
analysis and interpretation of data and manuscript review and
editing. AYT contributed to patient recruitment, clinical
evaluation and tissue acquisition. HLK contributed to
conceptualization, and manuscript review and editing. IO and HBE
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Ankara Etlik City Hospital (approval No: AEŞH-EK1-2023-015; date,
22.03.2023). All participants provided written informed consent
prior to inclusion in the study. The study was conducted in
accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
İzzet Özgürlük, ORCID: 0000-0002-9553-9265; Haktan
Bağış Erdem, ORCID: 0000-0002-4391-1387; Mustafa Tarik Alay, ORCID:
0000-0002-1563-2292; Okan Oktar, ORCID: 0000-0002-9696-7886;
Firdevs Şahin-Duran, ORCID: 0000-0001-9244-9590; Ezgi Çevik-Demir,
ORCID: 0000-0003-2612-3861; Aysu Yeşim Tezcan, ORCID:
0000-0002-4763-6546; Hüseyin Levent Keskin, ORCID:
0000-0002-2268-3821.
Glossary
Abbreviations
Abbreviations:
|
CIN
|
cervical intraepithelial neoplasia
|
|
LSIL
|
low-grade squamous intraepithelial
lesion
|
|
HSIL
|
high-grade squamous intraepithelial
lesion
|
|
mtDNA
|
mitochondrial DNA
|
|
mtCN
|
mitochondrial DNA copy number
|
|
nDNA
|
nuclear DNA
|
|
HPV
|
human papillomavirus
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
ΔCq
|
difference in quantification cycle
|
|
ND1
|
NADH dehydrogenase subunit 1
|
|
HBB
|
hemoglobin subunit β
|
|
ROS
|
reactive oxygen species
|
|
ROC
|
receiver operating characteristic
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Lei J, Ploner A, Elfström KM, Wang J, Roth
A, Fang F, Sundström K, Dillner J and Sparén P: HPV vaccination and
the risk of invasive cervical cancer. N Engl J Med. 383:1340–1348.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coquillard G, Palao B and Patterson BK:
Quantification of intracellular HPV E6/E7 mRNA expression increases
the specificity and positive predictive value of cervical cancer
screening compared to HPV DNA. Gynecol Oncol. 120:89–93. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: unresolved issues. Nat
Rev Cancer. 7:11–22. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maver P and Poljak M: Primary HPV-based
cervical cancer screening in Europe: implementation status,
challenges, and future plans. Clin Microbiol Infect. 26:579–583.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papa S and Skulachev V: Reactive oxygen
species, mitochondria, apoptosis and aging. Mol Cell Biochem.
197:305–319. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gustafsson CM, Falkenberg M and Larsson
NG: Maintenance and expression of mammalian mitochondrial DNA. Annu
Rev Biochem. 85:133–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johns DR: Seminars in medicine of the Beth
Israel Hospital, Boston. Mitochondrial DNA and disease.
Mitochondrial DNA and disease. N Engl J Med. 333:638–644. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wallace DC: Mitochondria and cancer. Nat
Rev Cancer. 12:685–698. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yakes FM and Van Houten B: Mitochondrial
DNA damage is more extensive and persists longer than nuclear DNA
damage in human cells following oxidative stress. Proc Natl Acad
Sci USA. 94:514–519. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Erdem HB, Ceylan AC, Sahin I, Sever-Erdem
Z, Citli S and Tatar A: Mitochondrial DNA copy number alterations
in familial mediterranean fever patients. Bratisl Lek Listy.
119:425–428. 2018.PubMed/NCBI
|
|
12
|
Öğütlü H, Esin İS, Erdem HB, Tatar A and
Dursun OB: Mitochondrial DNA copy number is associated with
attention deficit hyperactivity disorder. Psychiatr Danub.
32:168–175. 2020. View Article : Google Scholar
|
|
13
|
Öğütlü H, Esin IS, Erdem HB, Tatar A and
Dursun OB: Mitochondrial DNA copy number may be associated with
attention deficit/hyperactivity disorder severity in treatment: A
one-year follow-up study. Int J Psychiatry Clin Pract. 25:37–42.
2021. View Article : Google Scholar
|
|
14
|
Lee HC and Wei YH: Mitochondrial
biogenesis and mitochondrial DNA maintenance of mammalian cells
under oxidative stress. Int J Biochem Cell Biol. 37:822–834. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shokolenko I, Venediktova N, Bochkareva A,
Wilson GL and Alexeyev MF: Oxidative stress induces degradation of
mitochondrial DNA. Nucleic Acids Res. 37:2539–2548. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abd Radzak SM, Mohd Khair SZN, Ahmad F,
Patar A, Idris Z and Yusoff AA: Insights regarding mitochondrial
DNA copy number alterations in human cancer. Int J Mol Med.
50:1042022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cruz-Gregorio A, Manzo-Merino J,
Gonzaléz-García MC, Pedraza-Chaverri J, Medina-Campos ON, Valverde
M, Rojas E, Rodríguez-Sastre MA, García-Cuellar CM and Lizano M:
Human papillomavirus types 16 and 18 early-expressed proteins
differentially modulate the cellular redox state and DNA damage.
Int J Biol Sci. 14:21–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun W, Qin X, Zhou J, Xu M, Lyu Z, Li X,
Zhang K, Dai M, Li N and Hang D: Mitochondrial DNA copy number in
cervical exfoliated cells and risk of cervical cancer among
HPV-positive women. BMC Women's Health. 20:1392020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Warowicka A, Kwasniewska A and
Gozdzicka-Jozefiak A: Alterations in mtDNA: A qualitative and
quantitative study associated with cervical cancer development.
Gynecol Oncol. 129:193–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Al-Awadhi R, Alroomy M, Al-Waheeb S and
Alwehaidah MS: Altered mitochondrial DNA copy number in cervical
exfoliated cells among high-risk HPV-positive and HPV-negative
women. Exp Ther Med. 26:5212023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fleider LA, de Los Ángeles Tinnirello M,
Gómez Cherey F, García MG, Cardinal LH, García Kamermann F and
Tatti SA: High sensitivity and specificity rates of
cobas® HPV test as a primary screening test for cervical
intraepithelial lesions in a real-world setting. PLoS One.
18:e02797282023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mambo E, Chatterjee A, Xing M, Tallini G,
Haugen BR, Yeung SCJ, Sukumar S and Sidransky D: Tumor-specific
changes in mtDNA content in human cancer. Int J Cancer.
116:920–924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwertman NC, Owens MA and Adnan R: A
simple more general boxplot method for identifying outliers.
Computational statistics & data analysis. 47:165–174. 2004.
View Article : Google Scholar
|
|
25
|
Letafati A, Taghiabadi Z, Zafarian N,
Tajdini R, Mondeali M, Aboofazeli A, Chichiarelli S, Saso L and
Jazayeri SM: Emerging paradigms: unmasking the role of oxidative
stress in HPV-induced carcinogenesis. Infect Agent Cancer.
19:302024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pereira IOA, Silva NNT, Lima AA and da
Silva GN: Qualitative and quantitative changes in mitochondrial DNA
associated with cervical cancer: A comprehensive review. Environ
Mol Mutagen. 65:143–152. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin CS, Wang LS, Tsai CM and Wei YH: Low
copy number and low oxidative damage of mitochondrial DNA are
associated with tumor progression in lung cancer tissues after
neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg.
7:954–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moraes CT: What regulates mitochondrial
DNA copy number in animal cells? Trends Genet. 17:199–205. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Y, Schon EA, Wilichowski E,
Vazquez-Memije ME, Davidson E and King MP: Rearrangements of human
mitochondrial DNA (mtDNA): New insights into the regulation of
mtDNA copy number and gene expression. Mol Biol Cell. 11:1471–1485.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu L, Yao X and Shen Y: Altered
mitochondrial DNA copy number contributes to human cancer risk:
Evidence from an updated meta-analysis. Sci Rep. 6:358592016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Çakir M: Evaluation of smoking and
associated factors in Turkey. Iran J Public Health. 52:766–772.
2023.PubMed/NCBI
|
|
32
|
Wu S, Li X, Meng S, Fung T, Chan AT, Liang
G, Giovannucci E, De Vivo I, Lee JH and Nan H: Fruit and vegetable
consumption, cigarette smoke, and leukocyte mitochondrial DNA copy
number. Am J Clin Nutr. 109:424–432. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abu Shelbayeh O, Arroum T, Morris S and
Busch KB: PGC-1α is a master regulator of mitochondrial lifecycle
and ROS stress response. Antioxidants (Basel). 12:10752023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Longchamps RJ, Castellani CA, Yang SY,
Newcomb CE, Sumpter JA, Lane J, Grove ML, Guallar E, Pankratz N,
Taylor KD, et al: Evaluation of mitochondrial DNA copy number
estimation techniques. PLoS One. 15:e02281662020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hazkani-Covo E, Zeller RM and Martin W:
Molecular poltergeists: Mitochondrial DNA Copies (numts) in
sequenced nuclear genomes. PLoS Genet. 6:e10008342010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li B, Kalinowski P, Kim B, Pauls AD and
Poburko D: Emerging methods for and novel insights gained by
absolute quantification of mitochondrial DNA copy number and its
clinical applications. Pharmacol Ther. 232:1079952022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Filograna R, Mennuni M, Alsina D and
Larsson NG: Mitochondrial DNA copy number in human disease: The
more the better? FEBS Lett. 595:976–1002. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Lv H, Ji P, Zhu X, Yuan H, Jin G,
Dai J, Hu Z, Su Y and Ma H: Mitochondrial DNA copy number is
associated with risk of head and neck squamous cell carcinoma in
Chinese population. Cancer Med. 7:2776–2782. 2018. View Article : Google Scholar : PubMed/NCBI
|