Introduction
The incidence of squamous cell carcinoma of the
esophagus and gastroesophageal junction cancer has markedly
increased over the past 30 years in Kazakh people living in
Xinjiang, west of China (1). These
types of cancer are associated with a poor prognosis unless
detected and treated while in the early stages. Conventional
pathologic methods usually play an important role in the diagnosis
of cancer by providing important differential information between
benign and malignant diseases (2,3).
However, pathological evaluation is occasionally only suggestive
and not conclusive. This may lead to a conflict between pathology
and clinical diagnoses. These conflicting findings have stimulated
the search for more objective and quantitative methods for the
diagnoses of the related cancer.
Aneuploidy is the most common feature of many solid
tumors (4), including esophageal
cancer (EC). Solid tumors are characterized by complicated
karyotypes using classical genetic parameters (5,6). Thus,
malignant cells can be diagnosed by detecting aneuploidy usually
found in aneusomic nuclei. A rapid and sensitive method for
detecting the aneusomy of a specific chromosome in an individual
cell is fluorescence in situ hybridization (FISH) (7–9). For
this purpose, specific centromeric DNA probes have enumerated the
degrees of aneuploidy.
FISH was originally developed as a method to detect
chromosome aberrations and is now widely used for gene mapping for
the diagnosis of congenital diseases and for detecting specific
gene copy numbers found in malignant cells (4,9–11).
One advantage of the FISH method is that it is an
objective and quantitative evaluation procedure for the detection
of malignant cells. However, the sensitivity and specificity in the
diagnosis of EC in Kazakh people have yet to be determined. We
report the results of an original study in which we compare the
FISH procedure with clinical findings and conventional pathological
methods to detect esophageal cancer cells.
Patients and methods
From January 2009 to January 2010, 20 patients with
suspected esophageal cancer were tested by barium radiological
digital imaging. Biopsy material was further examined
pathologically at the First Affiliated Hospital of Xinjiang Medical
University. The results confirmed that all of the patients had
esophageal cancer and thus were enrolled in this study. The
patients included 15 males and 5 females, with an average age of
58.3 years (range 45–75).
Tissues appearing to be suspicious of neoplasms were
biopsied by fiberoptic esophageal-gastroscopy. These samples were
tested and analyzed by pathology and FISH. Informed consent was
provided by all of the patients.
Each biopsied lesion was treated in duplicate and
subjected to five touch impressions on sterile slides for FISH
analysis and then preserved in formalin for later pathological
examination.
For pathology, cells were stained with hematoxylin
and eosin. Pathologic evaluations were carried out by pathologists
in the Department of Pathology at the First Affiliated Hospital of
Xinjiang Medical University.
The histopathology of the biopsies was classified as
follows: class I indicated the absence of atypical or abnormal
cells; class II indicated atypical pathology with no evidence of
malignancy; class III was suggestive of, but not conclusive for,
malignancy; IIIa indicated mild dysplasia albeit not cancerous;
IIIb indicated advanced dysplasia; class IV was strongly suggestive
of malignancy; and class V was indicative of malignancy.
For FISH, dual color probes were prepared. Briefly,
touch preparations of cells were made on glass slides, air-dried
overnight at 25°C and then stored at −80°C. Centromeric probes
labeled with fluorochrome were used for the visualization and
enumeration of copy numbers. Spectrum orange- and green-labeled
probes were used to visualize centromeric regions of chromosomes 3
and 17. Reagents were purchased from Abbott Molecular, Inc., Des
Plaines, IL, USA.
Frozen touch preparations of cells were denatured
with 70% formamide and then washed twice in standard saline citrate
(SSC) at 74°C and at 25°C each for 2 min in a water bath. Slides
were then dehydrated through a series of graded ethanol dilutions
(70, 85 and 100%, each for 2 min). A hybridization solution (10 μl)
was applied to each microscope slide. The hybridization solution
contained 1 μl of each of the DNA probes, 7 μl of a hybridization
buffer and 1 μl of double distilled water. The solution was covered
with a coverslip and sealed with rubber cement. After incubation
for 16 h at 42°C in a humidity chamber, slides were washed with an
SSC solution for 5 min at 74°C and 25°C for 2 min.
Diamidinophenylindole II (an anti-face solution; 5 μl), was applied
to each spot and covered with a coverslip. The slides were observed
under a fluorescence microscope which was connected to a cooled
charge-coupled device camera and an image analyzer system (Leica
Microsystems, Ltd., Germany).
FISH signal analysis was then carried out. All
cells, apart from damaged cells or those with overlapping nuclei,
were evaluated. A total of 100 nuclei were counted, and the total
numbers of centromeric signals were recorded. If there were <100
nuclei on a slide, as many nuclei as possible were counted and
enumerated for centromeric signals. When the percentage of
hyperdisomic nuclei (i.e., more than three copies for at least one
nucleus) was >10%, we judged the tissue to be malignant.
FISH diagnosis was made without clinical information
or the result of conventional histopathology, nor were the results
of FISH analysis made known to the pathologists. Thus, the two
diagnoses were independently performed in a blinded manner.
The t-test was used to determine the significance
between the number of countable copy number centromeres in
chromosomes 3 and 17. P<0.05 was considered to indicate a
statistically significant difference.
Results
Specific diagnoses made based on histopathology
determined that 15 patients had primary esophageal cancer, 10 cases
had squamous cell carcinoma, 5 cases had adenocarcinoma and 5 cases
had benign lesions (Table I).
 | Table IClinical features of the patients. |
Table I
Clinical features of the patients.
| Case | Age | Gender | Histology |
|---|
| 1 | 71 | M | Sq |
| 2 | 52 | F | Ad |
| 3 | 45 | M | Ad |
| 4 | 70 | M | Ad |
| 5 | 53 | M | Neg |
| 6 | 75 | M | Neg |
| 7 | 45 | F | Sq |
| 8 | 71 | M | Sq |
| 9 | 55 | M | Sq |
| 10 | 58 | M | Sq |
| 11 | 54 | F | Sq |
| 12 | 69 | M | Sq |
| 13 | 55 | M | Neg |
| 14 | 54 | M | Ad |
| 15 | 62 | M | Sq |
| 16 | 56 | F | Neg |
| 17 | 45 | F | Sq |
| 18 | 62 | M | Neg |
| 19 | 54 | M | Ad |
| 20 | 60 | M | Sq |
Classification was as follows: 2 cases were class
II, 3 cases were IIIa; 1 case was IIIb; 2 cases were class IV; 12
cases were class V; and no cases were class I. Classes higher than
IIIb were positive for esophageal cancer. One case was
false-negative, but no cases were false-positive. Thus, using
histopathology as a parameter, sensitivity was 93% and specificity
was 100%.
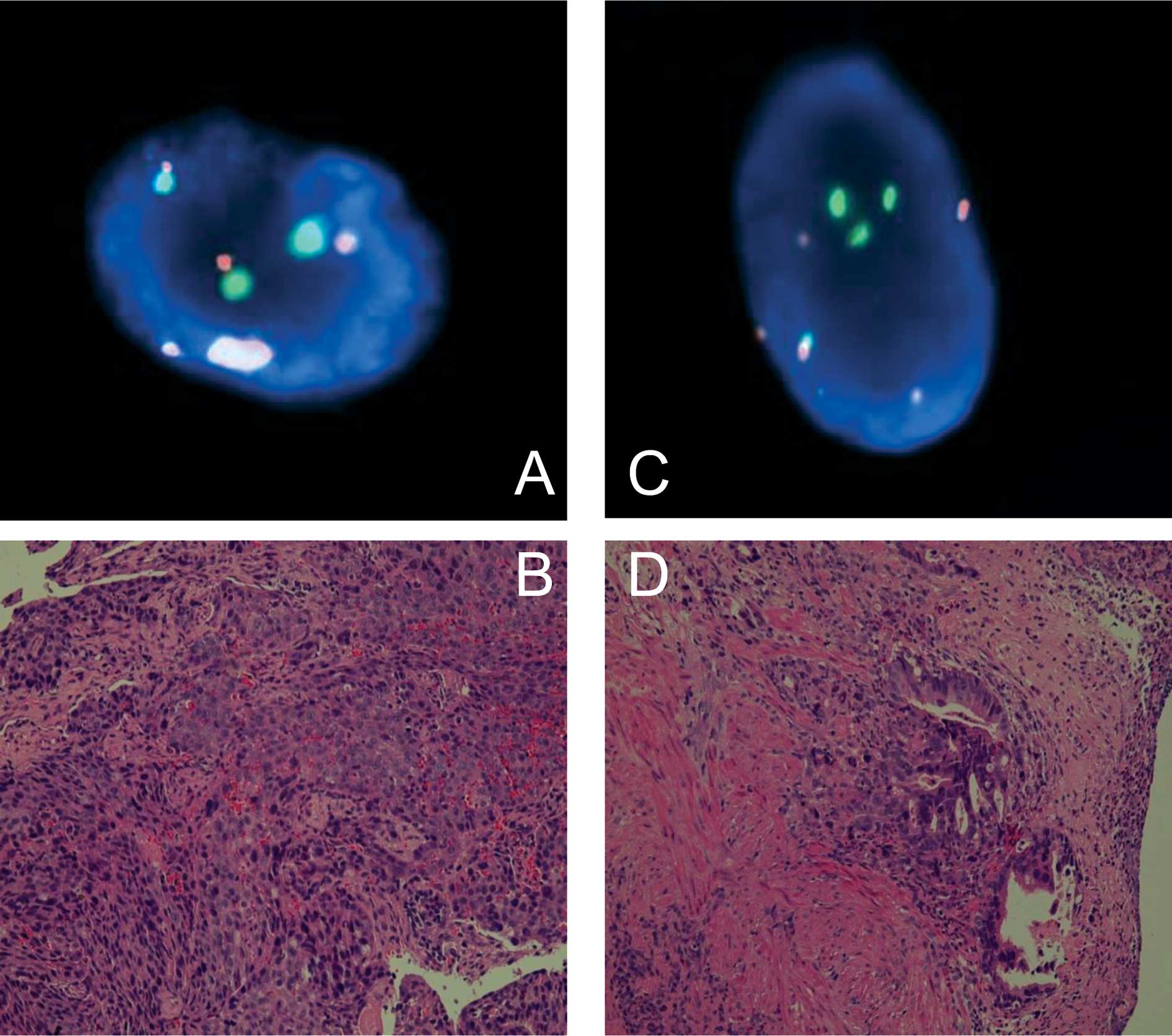
FISH analyses (30–100 countable nuclei) showed that
16 of the 20 cases had abnormal copy numbers in either chromosome 3
or 17. Representative findings comparing biopsy pathology and FISH
are shown in Fig. 1.
FISH did not reveal any false-positive or
false-negative cases with a sensitivity and specificity of 100%
(Table II). The centromeres of
chromosome 3 copy numbers were significantly higher (p=0.035) than
those of chromosome 17.
 | Table IIResults of FISH and conventional
biopsy pathology. |
Table II
Results of FISH and conventional
biopsy pathology.
| Case | Countable cells | 3 copies Chr. 3/Chr.
17 | 4 copies Chr. 3/Chr.
17 | 5 copies Chr. 3/Chr.
17 | ≥6 copies Chr. 3/Chr.
17 | Hyperdisomy (%) | FISH | Pathology |
|---|
| 1 | 33/51 | 8/8 | 3/7 | 8/6 | 0/0 | 57/41 | Positive | Sq, V |
| 2 | 100/100 | 39/51 | 18/12 | 18/7 | 10/7 | 85/77 | Positive | Ad, V |
| 3 | 71/100 | 36/50 | 6/25 | 17/13 | 9/9 | 95/97 | Positive | Ad, V |
| 4 | 90/100 | 14/25 | 18/19 | 35/33 | 11/5 | 86/82 | Positive | Ad, V |
| 5 | 79/82 | 2/1 | 0/1 | 0/0 | 0/0 | 2.5/2.4 | Negative | Negative, II |
| 6 | 30/34 | 1/3 | 1/0 | 0/0 | 0/0 | 6.6/8.8 | Negative | Negative, IIIa |
| 7 | 100/100 | 10/6 | 3/2 | 4/5 | 3/1 | 20/14 | Positive | Sq, IV |
| 8 | 100/100 | 55/36 | 9/13 | 9/26 | 17/8 | 90/83 | Positive | Sq, IV |
| 9 | 100/100 | 41/28 | 7/8 | 12/7 | 9/3 | 69/46 | Positive | Sq, V |
| 10 | 36/46 | 11/12 | 13/21 | 8/1 | 1/1 | 91/76 | Positive | Sq, V |
| 11 | 64/57 | 29/26 | 16/8 | 11/2. | 2/2 | 90/66 | Positive | Sq, V |
| 12 | 100/100 | 40/41 | 20/21 | 24/23 | 7/3 | 91/88 | Positive | Sq, V |
| 13 | 79/98 | 36/20 | 1/12 | 16/33 | 18/8 | 89/74 | Positive | a Negative, IIIa |
| 14 | 100/100 | 30/30 | 25/20 | 20/15 | 2/5 | 77/70 | Positive | Ad, V |
| 15 | 100/100 | 44/35 | 20/20 | 30/10 | | 94/75 | Positive | Sq, V |
| 16 | 98/94 | 2/1 | 1/1 | | | 3/2.1 | Negative | Negative, II |
| 17 | 100/100 | 47/39 | 19/11 | 6/7 | 0/3 | 72/60 | Positive | Sq, V |
| 18 | 100/100 | 3/3 | 1/3 | | | 4/6 | Negative | Negative, IIIa |
| 19 | 100/100 | 32/7 | 13/4 | 3/1 | 1/1 | 38/13 | Positive | Ad, V |
| 20 | 100/100 | 19/8 | 1/4 | 2/3 | 3/1 | 25/16 | Positive | Sq, IIIb |
Discussion
We demonstrated that FISH analysis is more accurate
in diagnosing esophageal cancer than using pathology examinations
of biopsies. Similar results using FISH analyses were reported in a
number of studies (4,8,10,12).
However, our findings contrasted FISH with those of the pathology
of the biopsy specimens. Han et al (13) examined 113 EC patients using
multiplex FISH with chromosome-specific centromere DNA probes 3, 8,
10, 12, 17 and 20. In that study, chromosomal signal numbers and
all of the chromosomes were found to be abnormal copy numbers. The
present study used only two probes which were specific for the
centromeric region of chromosomes 3 and 17.
Fritcher et al (6) investigated esophageal adenocarcinoma
using the FISH method with centromeric region probes C-MYC, P16,
HER2 and 20q13. In that study, the sensitivity of cytology was 45%
for the detection of esophageal adenocarcinoma, but FISH was 100%.
The same study used FISH analysis with centromeric probes 7, 11,
12, 17 and 18. Aneusomy was not found in the normal controls of any
chromosomes. In contrast, chromosomal abnormalities were found in
all carcinoma specimens (14). In
these studies, the FISH method detected esophageal squamous cancer
and adenocarcinoma cells in touch preparations of resected tumors.
Thus, our study compared the pathology of biopsy specimens to the
FISH method.
The cut-off value for the percentage of hyperdisomic
cells was set at 10% (i.e., more than 10% means cancer is present
and less than 10% means absence of cancer), since normal cells are
often less than 6%. This discrepancy probably occurs due to the
counting of sister chromatids as copies. When we set the cut-off
value at 10%, no false-positive or false-negative cases were noted.
Additionally, the sensitivity and specificity was 100%. Centromeres
of chromosome 3 copy numbers were significantly higher than those
of chromosome 17. The reason for this discrepancy is unknown but
Kazakh people have a higher incidence of cancer (1) which may correlate with their higher
copy numbers in chromosome 3.
Using biopsy pathology we found one false-negative
case. In this case, barium radiological digital imaging and
examination by esophageal gastroscopy strongly suggested cancer;
the clinical diagnosis was recurrent EC, but the pathologic
diagnosis was IIIa. These findings suggest that FISH is able to
detect chromosome aberrations that may lead to the commencement of
oncogenesis of EC (i.e., IIIb). This hypothesis has been supported
by other studies (6,13). Additional studies are needed to
further evaluate and support our findings. For example, we propose
using different ethnic groups and increased numbers of patients. We
believe that FISH may provide decisive information for the
detection of malignancies, particularly in cases classified as IIIa
or IIIb. Subsequently, the FISH assay may have a higher sensitivity
of diagnosis than other current methods.
In a previous study, we did not correctly diagnose
malignancy in lung cancer patients using FISH (15,16).
There are two possible reasons for our false-negative FISH results.
One would be the failure to obtain cell material that was absent in
cancer cells. The second reason would be that the cancer cells were
near-diploid. Therefore, we were not able to detect aneusomy in the
target chromosomes.
Previous studies suggest that the sensitivity of
FISH is superior to that of conventional cytology (13,16).
However, there are some disadvantages to the FISH method of
analysis. To begin with, this method does not generate information
about the histopathology or type of esophageal cancer since one
cannot observe morphological features. Furthermore, FISH is
expensive. A further drawback is that FISH signal counting under
fluorescence microscopy is time-consuming and requires trained
personnel.
In the future, we expect to be able to detect more
aneusomic cells using additional probes for other chromosomal
regions as reported (17–19), e.g., EGFR, C-MYC, HER2, P16, P53,
cyclin D1, X and Y, which may be successful.
Taken together, FISH can detect multiple chromosomal
changes and is an ideal prognostic tool, thereby opening the door
to different therapeutic approaches in the future (18).
In conclusion, the FISH method successfully detected
esophageal cancer in Kazakh patients using only two probes. This
method appeared more sensitive than the biopsy pathology diagnostic
method. Centromeres of chromosome 3 showed the most sensitive probe
diagnostically in Kazakh patients with esophageal cancer.
Acknowledgements
This work was supported by a grant from the First
Hospital of Xinjiang Medical University (2008-YFY-06) and the
Chinese Postdoctoral Fund of Xinjiang Medical University
(20080-3014). Thanks are extended to the Department of Hematology,
First Affiliated Hospital of Xinjiang Medical University for the
technical support. We thank Professor Glenn S. Bulmer for reviewing
this manuscript.
Abbreviations:
|
FISH
|
fluorescence in situ
hybridization
|
|
EC
|
esophageal cancer
|
|
SSC
|
standard saline citrate
|
References
|
1
|
Zhang YM: Distribution of esophageal
cancer in the Xinjiang. XMU Med. 11:139–145. 1988.
|
|
2
|
Odze RD: Barrett esophagus: histology and
pathology for the clinician. Nat Rev Gastroenterol Hepatol.
6:478–490. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yerian L: Histology of metaplasia and
dysplasia in Barrett’s esophagus. Surg Oncol Clin N Am. 18:411–422.
2009.
|
|
4
|
Rajagopalan H and Lengauer C: Aneuploidy
and cancer. Nature. 432:338–341. 2004. View Article : Google Scholar
|
|
5
|
Halling KC and Kipp BR: Fluorescence in
situ hybridization in diagnostic cytology. Hum Pathol.
38:1137–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fritcher EG, Brankley SM, Kipp BR, et al:
A comparison of conventional cytology, DNA ploidy analysis, and
fluorescence in situ hybridization for the detection of dysplasia
and adenocarcinoma in patients with Barrett’s esophagus. Hum
Pathol. 39:1128–1135. 2008.PubMed/NCBI
|
|
7
|
Doak SH, Jenkins GJS, Parry EM, Griffiths
AP, Shah V, Baxter JN and Parry JM: Characterisation of p53 status
at the gene, chromosomal and protein levels in oesophageal
adenocarcinoma. Br J Cancer. 89:1729–1735. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakamura H, Saji H, Idiris A, et al:
Chromosomal instability detected by fluorescence in situ
hybridization in surgical specimens of non-small cell lung cancer
is associated with poor survival. Clin Cancer Res. 9:2294–2299.
2003.PubMed/NCBI
|
|
9
|
Dey P: Aneuploidy and malignancy: an
unsolved equation. J Clin Pathol. 57:1245–1249. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Albertson DG, Collins C and McCormick F:
Chromosome aberrations in solid tumors. Nat Genet. 34:369–376.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bahmanyar S and Ye W: Dietary patterns and
risk of squamous-cell carcinoma and adenocarcinoma of the esophagus
and adenocarcinoma of the gastric cardia: a population-based
case-control study in Sweden. Nutr Cancer. 54:171–178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Varshney D, Zhou YY, Geller SA and Alsabeh
R: Determination of HER-2 status and chromosome 17 polysomy in
breast carcinomas comparing Hercep Test and PathVysion FISH assay.
Am J Clin Pathol. 121:70–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han QY, Shun H, Yu PW, et al: [Application
of multicolor fluorescence in situ hybridization to early diagnosis
of esophageal squamous cell carcinoma]. Ai Zheng. 27:396–401.
2008.
|
|
14
|
You XY, Johannes F, Shuang LX and Chiang
JL: Genomic instability in precancerous lesions before inactivation
of tumor suppressors p53 and APC in patients. Cell Cycle.
13:1443–1447. 2006.PubMed/NCBI
|
|
15
|
Idiris A, Nakamura H, Maidiniyeti N, et
al: Detection of lung cancer cells in bronchial washings using
fluorescence in situ hybridization. J Jpn Bronchoesophagol Soc.
56:470–475. 2005. View Article : Google Scholar
|
|
16
|
Nakamura H, Idiris A, Kawasaki N, et al:
Quantitative detection of lung cancer cells by fluorescence in situ
hybridization: comparison with conventional cytology. Chest.
128:906–911. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reichelt U, Duesedau P, Tsourlakis M, et
al: Frequent homogeneous HER-2 amplification in primary and
metastatic adenocarcinoma of the esophagus. Mod Pathol. 20:120–129.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arnold D, Peinert S, Voigt W and Schmoll
HJ: Epidermal growth factor receptor tyrosine kinase inhibitors:
present and future role in gastrointestinal cancer treatment: a
review. Oncologist. 11:602–611. 2006. View Article : Google Scholar
|
|
19
|
Kawaguchi Y, Kono K, Mimura K, Mitsui F,
Sugai H, Akaike H and Fujii H: Targeting EGFR and HER-2 with
cetuximab and trastuzumab-mediated immunotherapy in esophageal
squamous cell carcinoma. Br J Cancer. 97:494–501. 2007. View Article : Google Scholar : PubMed/NCBI
|