Introduction
Ras mutation has been found in many types of human
malignancies including hematologic malignancies with 15% point
mutation in acute myeloid leukemia (AML). Ras-associated protein 1
(Rap1), a close member of Ras-family, has attracted increased
attention in tumorigenesis, progression and metastasis (1,2). Rap1
regulates two important cellular processes including B-RAF/MEK/ERK
activation and integrin-mediated cell adhesion (3,4). Like
other Ras family proteins, Rap1 functions as a molecular switch
between the active form binding GTP and the inactive form binding
GDP. The GDP-GTP cycle is regulated by guanine nucleotide exchange
factors (GEFs) and GTPase-activating proteins (GAPs) (5). GEFs promote the release of GDP,
rendering Rap1 to rebind a new GTP to stimulate the downstream
signal cascade. On the contrary, GAPs stimulate the low intrinsic
GTPase activity of Rap1 to hydrolyze GTP resulting in the
inactivation of Rap1.
Rap1GAP gene locating at chromosome 1p36.1-p35 is a
member of GAPs family which also including SPA-1, E6TP1
(E6-targeted protein 1) and SPAR (spine-associated Rap1GAP)
(6). Little is known about the
function of Rap1GAP in tumorigenesis although the gene is receiving
increased attention. Loss of heterozygosity (LOH) of Rap1GAP was
found in pancreatic cancer and promoter hypermethylation was
recently reported in melanoma cells (7,8).
Decreased expression of Rap1GAP was also reported in papillary
thyroid cancer (9,10). Also upregulated expression of
Rap1GAP exhibited proliferation suppression in vitro as well
as in vivo models. These data led to the proposal that
Rap1GAP may act as a putative tumor suppressor gene in these
tumors.
Previous data of our laboratory showed that Rap1GAP
may play important functions in myelodysplastic syndromes (MDS)
(11,12). Here, we investigated whether Rap1GAP
plays a role, and how, in the pathogenesis and progression of
hematologic malignancies.
Materials and methods
Cell culture
Human 293T cells as the packaging cell for
lentivirus was kindly provided by Dr Yun Zhao (Cyrus Tang
Hematology Research Center, Soochow University). HL-60, NB-4, U937
and SHI-1 human leukemic cell lines were cultured in Iscove’s
modified Dulbecco’s medium (IMDM) (Gibco) with 10% fetal bovine
serum (FBS) (Gibco) in 5% CO2 humidified atmosphere at
37°C.
Patients and sample preparation
After approved by Hospital Ethics Committee and
signing the informed consent, bone marrow mononuclear cells (BMNCs)
were isolated from patients including 8 cases of M2, 7 cases of M3,
6 cases of M5 (according to FAB classification). Eight cases of
non-malignant blood disease patients (including 5 iron deficiency
anemia, 2 hypercellular anemia and 1 idiopathic thrombocytopenia)
were chosen as controls. Non-parametric tests in SPSS were used for
statistical analyses.
Rap1GAP-expressing lentiviral vector and
virus production
A human full-length of Rap1GAP was amplified by PCR
from pcDNA3.1-Rap1GAP (generously offered by Dr P. Stork, Oregon
Health Sciences University, Portland, OR), using Pfu polymerase
with forward primer 5′-GCGGGATCCATGAT TGAGAAGATGCAG-3′ and reverse
primer 5′-GCGGGATC CCTAACAGCCCAGCTGG-3′. To the subcloned
lentivirus pRRL-Venus vector (gifted from Dr Yun Zhao),
BamHI digestion sites were added. All plasmids were
conformed by DNA sequencing.
After packaged according to the protocol of
literature (13), the virus in the
supernatant of transfected cells was centrifuged and kept in -80°C
for later use.
Transduction of human leukemia cells and
FACS sorting
Human leukemic cells HL-60 and NB4 were transfected
with lentivirus. Briefly, 1×106 cells were seeded into
6-well dishes within 2 ml IMDM medium with 10% FBS, then 100 μl
Rap1GAP-virus or control virus was added to each well. After 8–10 h
of incubation, cells were washed twice with sterile
phosphate-buffered saline (PBS) and transferred into 45
cm2 flask for a 3–4 day expansion. Both the
Rap1GAP-transducted cells and control cells were sorted by YFP
signal using FACS (BD Biosciences) and expanded for one more week.
The stable monoclonal cells transfected with Rap1GAP were then
obtained by limited dilution.
Western blot analysis
For western blot analysis, cell pellets were lysed
in 100 μl lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton
X-100, 1% Nonidet P-40, 1% SDS, 1 mM phenylmethylsulfonyl fluoride,
5 μg/ml aprotinin, 5 μg/ml leupeptin) for 60 min on ice. After
centrifugation for 15 min at 12,000 g (4°C), the supernatants were
collected for western blot analysis. Primary antibody
concentrations were as follows: rabbit anti-Rap1GAP monoclonal
antibody (Santa Cruz Biotechnology), 1:1,000; rabbit anti-Rap1
monoclonal antibody (Pierce), 1:500; mouse
anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal
antibody, 1:1,000; rabbit anti-ERK monoclonal antibody 1:1,000 and
rabbit anti-p-ERK monoclonal antibody 1:500 (Cell Signaling).
Rap1 activity assay
Active Rap1 was detected by glutathione
S-transferase (GST)-tagged RalGDS according to the protocol
(Pierce). Briefly, 1×107 cells were collected and lysed
by lysis/binding/washing buffer in which protease inhibitors was
previously added. GST-RalGDS-RBD (20 μg) was added to a spin cup
containing 100-μl glutathione resin within a collection tube.
Immediately, 700 μl cell lysate containing about 500 μg proteins
was transferred into the spin cup. For the prevention of leakage,
the spin cup was closed and sealed with laboratory film. The
reaction mixture was incubated at 4°C for 1 h with gentle rocking.
After incubation, the spin cup was transferred into a new
collection tube and washed by 400 μl lysis/binding/washing buffer
thrice. Finally, 50 μl 2X reducing sample buffer was added to the
resin and centrifuged at 6,000 g for 2 min following a 2-min
incubation at room temperature. The eluted samples containing
active Rap1 were heated for 5 min at 95–100°C and stored at −20°C
for later immunoblot assay.
Reverse transcriptase PCR and
quantitative real-time PCR
Total RNA was extracted from aliquots of cells with
TRIzol reagent according to the manufacturer’s procedure, cDNA was
synthesized using MMLV reverse transcriptase (Gibco). Expression
level was measured by PCR or qRT-PCR. Primers were as follows:
Human Rap1GAP: Forward 5′-CA GAGGAGGACTACATTCCATACCCG-3′ and
reverse 5′-CCAACTTTGCCATCTGGACAACATT-A-3′; β-actin: forward
5′-AGGCCGGCTTCGCGGGCGAC-3′ and reverse 5′-CTCGGGAGCCACACGCAGCTC-3′;
p21: forward 5′-AT GTCAGAACCGGCTGGGGAT-3′ and reverse 5′-GGGC
TTCCTCTTGGAGAAGAT-3′.
For qRT-PCR, all reactions were performed in 96-well
MJ white optical plates. β-actin was used as an endogenous control.
The primers and Taqman probes were designed by the software Primer
Express 2.0. Human Rap1GAP: forward 5′-CAAGAACAGAGCGGAGACC-3′,
reverse 5′-GCCACGT GCTATAGATGAAG-3′, probe 5′-AGCAGAGGCGCTCAA
GGACTTCTCC-3′; Human β-actin: forward 5′-TCACCC
ACACTGTGCCCATCTACGA-3′, reverse 5′-CAGCGGAAC CGCTCA TTGCCAATGG-3′,
probe 5′-ATGCCCTCCCCCA TGCCATCCTGCGT-3′; Human MMP9: forward
5′-GCTGG GCTTAGATCATTCCTCA-3′, reverse 5′-AGGGCGAGGA CCATAGAGGT-3′,
probe 5′-CCCGGCCTTCTGTTCCTGA TAAACCC-3′.
Each reaction with a final volume of 25 μl was
composed of the sample cDNA, TaqMan Universal PCR Mastermix
(Applied Biosystems), primers and probe. Amplification was
performed by a denaturation step at 95 °C for 10 min; followed by
40 cycles of 95°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec.
The relative levels of Rap1GAP and MMP9 were evaluated by
comparative CT value against β-actin and expressed as
2−ΔCT and 2−ΔΔCT. All samples were measured
in triplicate.
Cell differentiation determination
Cells (5×105/ml) were seeded in 6-well
plates and incubated with ATRA (final concentration 10−6
M) or TPA (final concentration 20 nM) for 24, 48 and 72 h. Cells
were collected then washed with PBS for flow cytometry analysis,
PE-conjugated CD11b was used. Nitroblue tetrazolium test (NBT) was
done at the same time.
Detection of apoptosis by fluorescence
microscope
After induced by arsenic trioxide (final
concentration HL-60 cells 5×10−6 M, NB4 cells
2×10−6 M) for 24 h, cells were collected and washed
twice with binding buffer before the next step. Because the
lentiviral vector contained YFP protein, resuspended cells were
stained with PE-conjugated Annexin V to visualize the apoptosis
cells with red fluorescence.
Cell invasion assay
In vitro cell invasion were measured by the
costar transwell chamber system. The membrane filters were coated
with 60 μl Matrigel (BD Biosciences) which was reconstituted with
IMDM at a dilution of 1:10, then dried at 37°C under sterile
conditions overnight. HL-60 cells were suspended in 200 μl IMDM
with 10% FBS at the final density of 2.0×105 cells/well
and seeded into the inserts. The outside of the insert was filled
with 500 μl IMDM with 10% FBS to form the lower chamber. After
24–48 h of incubation, the cells that had migrated into the lower
chamber were collected and counted. The invasion rate was
calculated by the ratio of the number of cells in the lower chamber
to the total number of leukemic cells. Each invasion experiment was
performed in triplicate.
Gelatin zymography
Cells were suspended in 100 μl serum-free medium at
the final density of 2.0×105 cells/well and seeded into
96-well plates. After 24 h of incubation, the culture supernatants
were harvested and centrifuged to remove cellular debris for
zymography. Briefly, 10–15 μl supernatants mixed with loading
buffer were subjected to SDS-PAGE using 10% acrylamide gels with
0.1% gelatin. The gels were washed three times for 30 min in 2.5%
Triton X-100, then incubated overnight in activation buffer (50 mM
Tris-Cl pH 7.5, 10 mM CaCl2, 150 mM NaCl, 1 mM
ZnCl2, 0.02% NaN3) at 37°C. The gelatinolytic
activity was observed as transparent bands after the gels were
stained with 0.1% Coomassie Blue R-250 and destained in 10% acetic
acid and 30% alcohol.
Results
Rap1GAP1 is downregulated in acute
myeloid leukemia
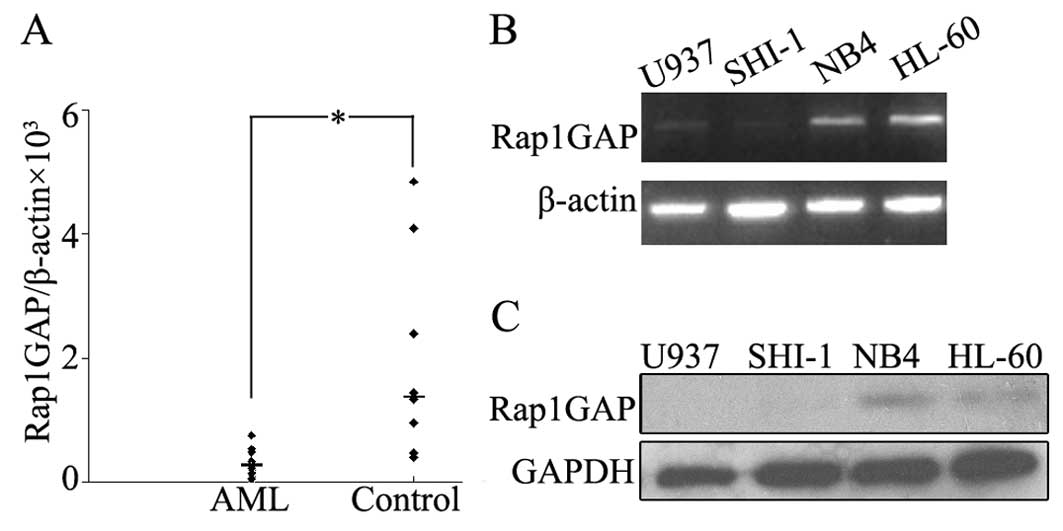
We detected the expression level of Rap1GAP in BMNCs
from patients with acute myeloid leukemia and patients with
non-malignant blood diseases by using qRT-PCR. The median levels of
Rap1GAP transcripts in patients with AML and non-malignant blood
diseases were 0.0002 (ranging from 0 to 0.0025) and 0.0014 (ranging
from 0.0004 to 0.005), respectively. The Rap1GAP expression was
significantly lower in AML than in control groups (P<0.05)
(Fig. 1A). In leukemic cell lines
(HL-60, NB4, U937 and SHI-1) Rap1GAP expression was very low, both
in RNA and protein level (Fig. 1B and
C). Collectively, decreased Rap1GAP expression was observed in
BMNCs of AML patients as well as in leukemic cell lines (i.e.
HL-60, NB4, U937 and SHI-1).
Upregulated Rap1GAP blocks Rap1
activation in leukemic cells
To explore the function of Rap1GAP in hematologic
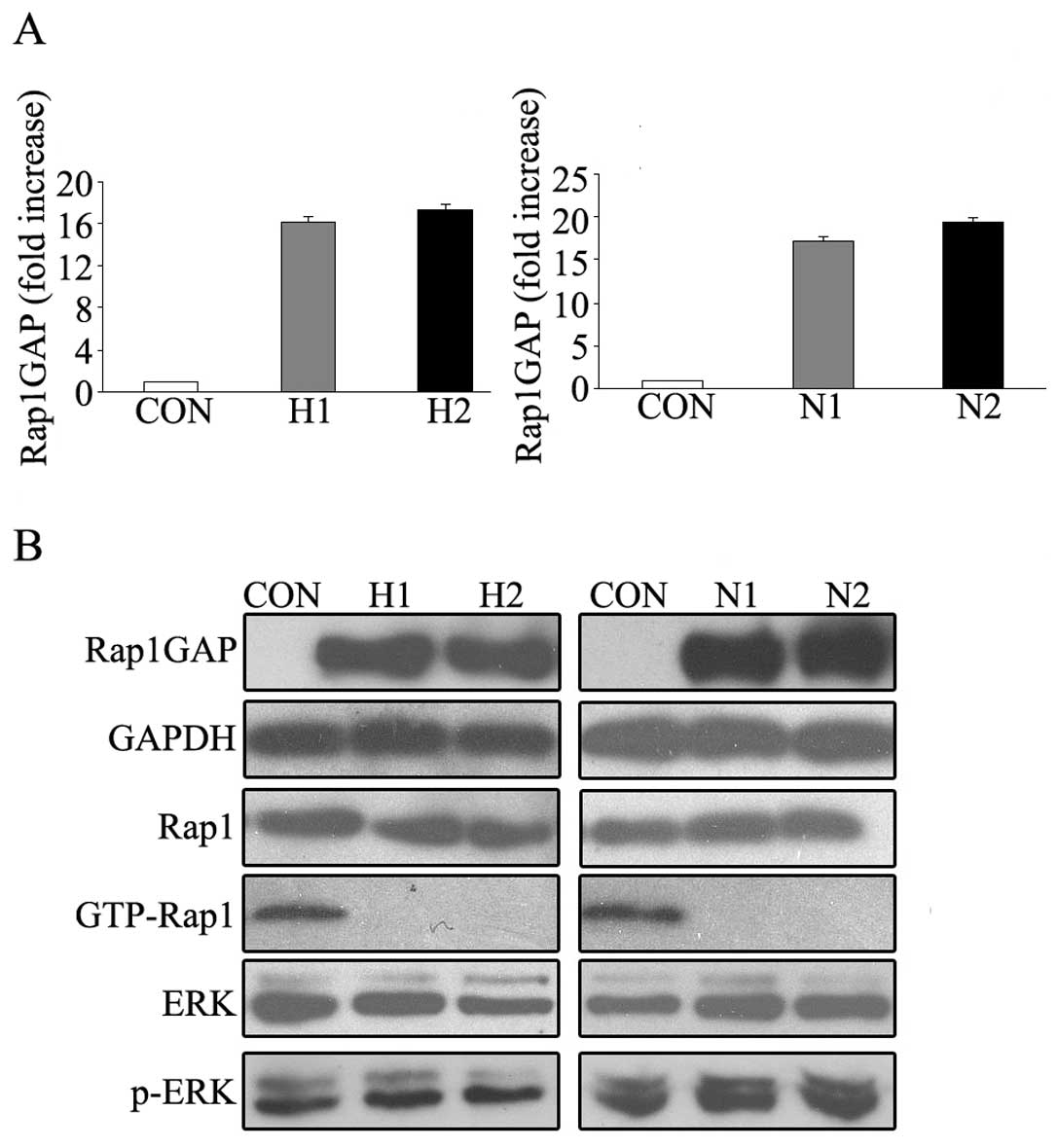
malignancies, two stable Rap1GAP-transfected subclones of both
HL-60 and NB4 cells were identified by qRT-PCR and western blot
analysis. In the results of qRT-PCR, the mRNA level of Rap1GAP
increased about 16.2, 17.3, 17.5 and 19.3 fold in HL-60 (H1 and
H2), NB4 (N1 and N2) stable cell lines, respectively (Fig. 2A). Western blot analysis showed
similar results. As expected, Rap1 activity was almost undetectable
in these Rap1GAP-overexpressed cells. In contrast, high basal Rap1
activity was observed in control cells (Fig. 2B). These results suggested that
increased expression of Rap1GAP in leukemic cells lead to the
abolishment of Rap1 activity.
Upregulated Rap1GAP do not affect myeloid
leukemic cell growth
A large number of studies have reported that the
expression of Rap1GAP was downregulated in a variety of human
malignancies and upregulated Rap1GAP inhibited the proliferation of
malignancies in vivo and in vitro models. But in
hematologic malignancies, increased expression of Rap1GAP in AML
cell lines including HL-60 and NB4 does not influence the
proliferation capability compared to empty control or wild-type
cells (data not shown). Neither was the difference of p-ERK or
total ERK levels between Rap1GAP-transfected cells and their
counterparts observed (Fig.
2B).
Upregulated Rap1GAP promotes the
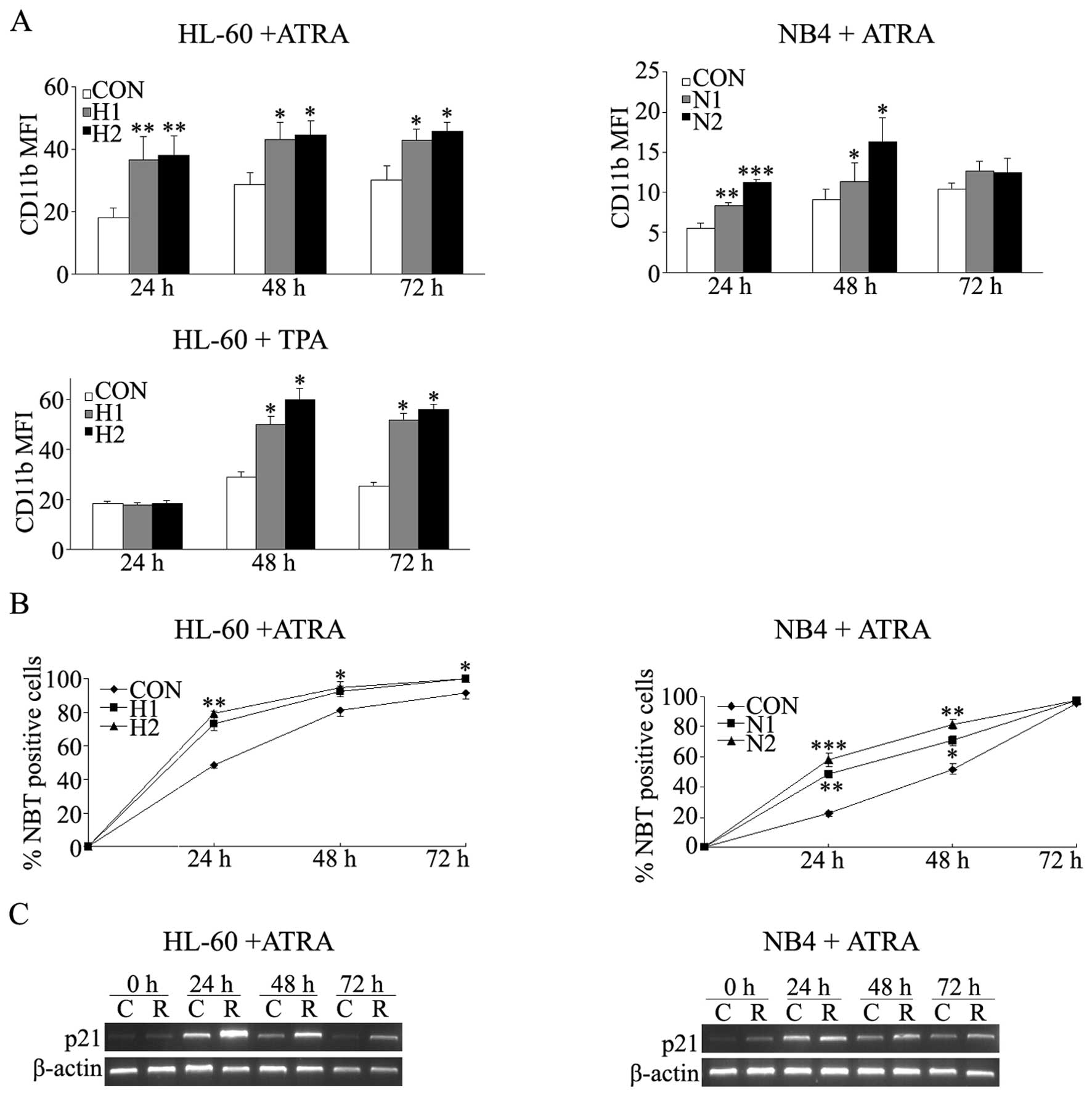
differentiation of NB4 and HL-60 cells induced by ATRA or TPA
HL-60 cells, previously considered as a
promyelocytic leukemia cells, can be induced to differentiate
towards more mature stage by some inducers. All-trans-retinoic acid
(ATRA) induces HL-60 differentiate into granulocyte and phorbol
12-myristate 13-acetate (TPA) stimulates cells to differentiate
into a macrophage-like phenotype. Here, two stable
Rap1GAP-transfected subclones were examined. During ATRA induction,
the CD11b mean fluorescence index (MFI) of HL-60 cells increased
for 1.4- to 2.1-fold in two transfected monoclones than empty
control each day (P<0.05) (Fig.
3A). NBT assay also demonstrated a similar result (Fig. 3B). As shown in Fig. 3A, TPA-induction also resulted in
much higher CD11b MFI of HL-60 cells in Rap1GAP-transfected
monoclones than the counterpart cells at 48 and 72 h (P<0.05),
but the difference was not observed at 24 h. Moreover, TPA-induced
morphological changes of HL-60 cells were observed more obviously
in Rap1GAP-transfected cells than in the empty control cells (data
not shown).
ATRA can also induce NB4 cells to differentiate into
mature granulocyte. The CD11b MFI increased for 1.2- to 2.0-fold in
two transfected monoclones at 24 and 48 h, but the difference was
not observed at 72 h (Fig. 3A). NBT
assay was consistent with the CD11b results (Fig. 3B). During induction, the mRNA level
of p21 of both HL-60 and NB4 cells were measured by RT-PCR (30
cycles) at each time point. The expression of p21 showed higher
level in the Rap1GAP-transfected cells compared to the control
cells (Fig. 3C).
Upregulated Rap1GAP promotes HL-60 and
NB4 cells apoptosis
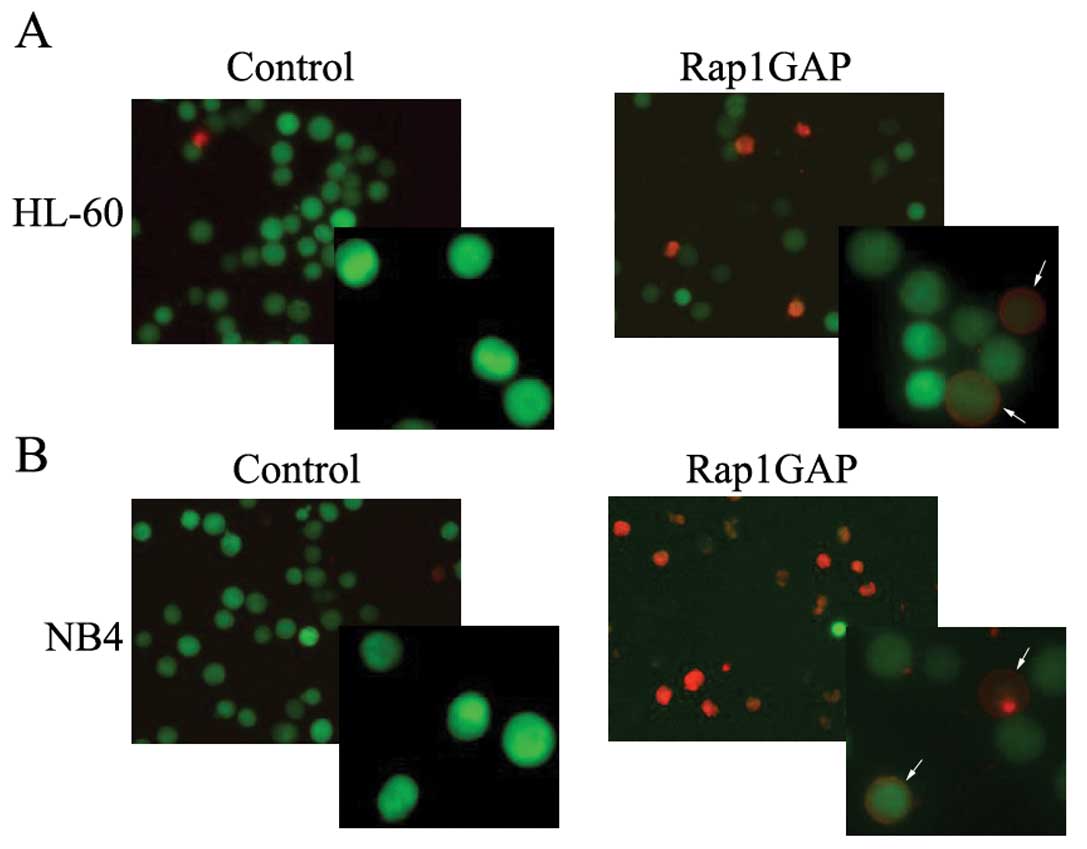
To determine the effect of Rap1GAP on leukemic cell
apoptosis, we use arsenic trioxide to induce HL-60 and NB4 cells to
undergo apoptosis. HL-60 and NB4 cells were transfected with
lentivirus vector carrying YFP protein whose emitting wave
interfere with other fluorescence and can not be analyzed by flow
cytometry without light filter, so the apoptosis cells were
observed directly through fluorescence microscope. For both HL-60
cells in Fig. 4A and NB4 cells
shown in Fig. 4B, we have observed
that the number of cells stained with Annexin V-PE were much
greater in Rap1GAP-transfected cells than that in the counterparts.
These results may indicate that increased expression of Rap1GAP
render leukemic cells to undergo apoptosis.
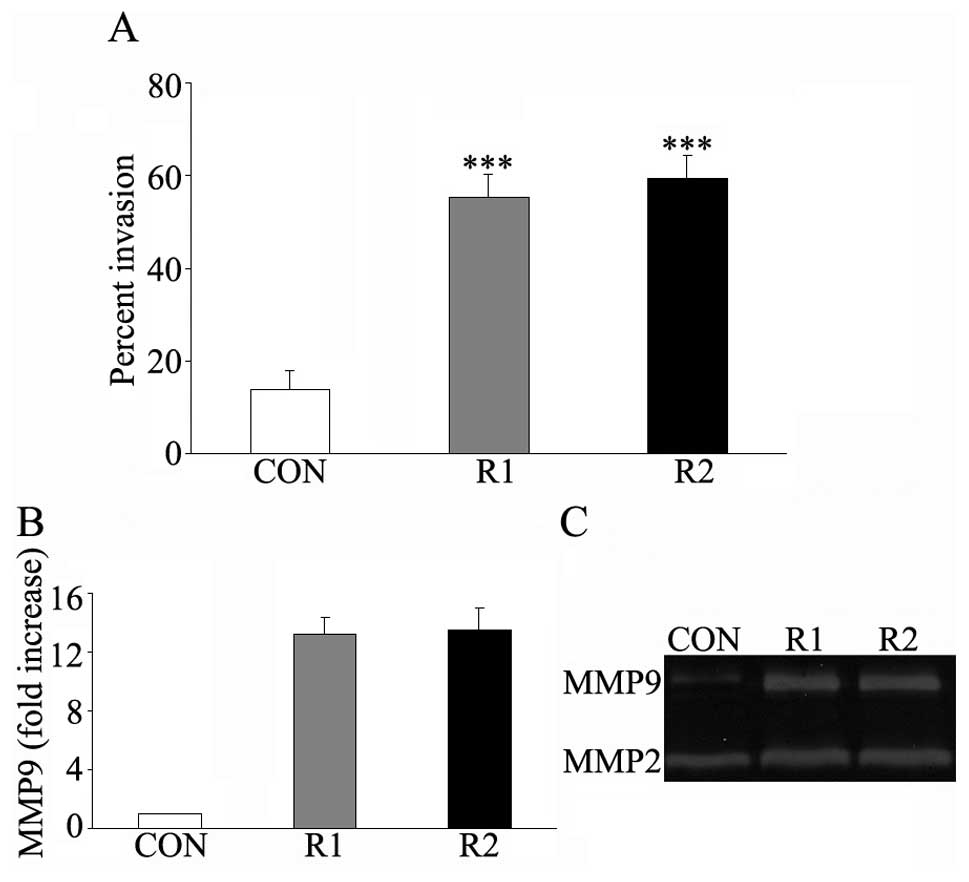
Increased expression of Rap1GAP promotes
leukemic cell invasion in vitro
Cell invasion ability was measured by the transwell
system. After 24–48 h of incubation, the cells migrated into the
lower chamber were collected and counted. The invasion rate of
Rap1GAP-transfected HL-60 cells was about 4-fold that of the empty
control cells (Fig. 5A). The filter
of the upper chamber was coated with Matrigel which was composed of
laminin, collagen type IV, and proteoglycans to imitate human
basement membranes. To invade into the lower chamber, leukemic
cells secreted some matrix metalloproteinases (MMPs) to resolve the
barrier. We therefore investigated the MMP secretion, mainly MMP9
and MMP2 in control- and Rap1GAP-transfected cells. As shown in the
gelatin zymogram, Rap1GAP-transfected cells showed higher MMP9
secretion than the control cells, but changes in MMP2 secretion was
not obvious (Fig. 5C). qRT-PCR was
also performed to determine the mRNA level of MMP9 in
Rap1GAP-transfected cells and the control cells. The mRNA level of
MMP9 was upregulated in Rap1GAP-transfected HL-60 cells (about
12-fold increase) in comparison with the corresponding control
cells (Fig. 5B). These results
suggested that upregulated expression of Rap1GAP enhance the
invasion ability of the leukemic cells probably due to the
expression and secretion of MMP9.
Discussion
In the present study, we demonstrate that Rap1GAP
expression is downregulated in acute myeloid leukemia compared with
non-malignant blood diseases. Upregulated Rap1GAP is associated
with increased differentiation and apoptosis of the leukemic cells.
We also show that increased expression of Rap1GAP promotes leukemic
cell invasion in vitro and increased secretion of MMP9.
The earliest report of Rap1 by Kitayama et al
identified Rap1 as a protein which could counteract the activation
of Ras (14). However, later
studies found that in different cell lines Rap1 activation may
exert contradictory functions not just a counteracting molecule of
Ras (2,3,15).
Although it was unknown whether Rap1 played a ‘good role’ or was a
‘bad actor’ in cancer development, Rap1 was reported to regulate a
variety of cellular processes such as the maturation of
megakaryocytes (16), neuronal
differentiation (17) and T-cell
anergy (18). Some other molecules
which regulate the Rap1 activation were also reported. Tuberin
which shares structural similarity with Rap1GAP was subject to loss
in tuberous sclerosis (19) and
E6TP1 was believed to involved in the development of cervical
cancer (20,21). More recently, Rap1GAP was reported
as a tumor suppressor gene in pancreatic cancer (7), thyroid cancer (9,10),
melanoma (1,8) and oropharyngeal squamous cell
carcinoma (22). According to the
SPA-1 knock-out B6 murine model, which presented a highly activated
Rap1 and ultimately developed myeloid leukemia, SPA-1 was thought
of as an important GAP in hematologic malignancies (23). However, little was known about the
function and effect of Rap1GAP in hematologic malignancies. To
verify if Rap1GAP also acts as a tumor suppressor gene in
hematologic malignancies, we detected the expression of Rap1GAP in
AML and leukemic cell lines. Based on the results that the
expression of Rap1GAP was downregulated in AML as well as in
leukemic cell lines, we thus upregulated the expression of Rap1GAP
in HL-60 and NB4 cells to study the function of Rap1GAP in
vitro.
Rap1GAP regulates GTP-GDP cycles to influence the
B-RAF/MEK/ERK signaling pathway which is involved in cell
proliferation, differentiation and apoptosis (24). Some studies reported that the
upregulated expression of Rap1GAP reduced the level of both
Rap1-GTP and p-ERK resulting in the suppression of cell
proliferation (8–10), while others reported that
upregulated Rap1GAP suppressed the cell proliferation with lower
Rap1-GTP but without change of p-ERK (7). In our study, increased Rap1GAP can
abolish Rap1-GTP, but we did not find suppressed growth or the
change of p-ERK. We do not exactly know why Rap1GAP has no effect
on leukemic cell proliferation and we postulate that it may due to
different cell types. Rap1GAP is mainly expressed in linage
marker-positive BMCs according to a study on B6 mice (23). We believe that Rap1GAP may play an
important role in hematologic cell differentiation and apoptosis.
Interestingly, our results proved this consideration. We found that
increased expression of Rap1GAP promoted the differentiation of
HL-60 and NB4 cells induced by TPA or ATRA and render the cells to
apoptosis in response to arsenic trioxide.
During ATRA treatment, we found the induced
expression of p21 in both NB4 and HL-60 cells by semi-quantitative
RT-PCR. p21 is a cdk (cyclin dependent kinase) inhibitor which
suppresses Rb protein phosphorylation to inhibit cell division.
Enhanced p21 expression may play a critical role in cell cycle
arrest and differentiation (25,26).
In fact, we have observed a much higher expression of p21 in the
Rap1GAP-transfected cells than that in the control cells especially
at 24 h. Upregulated Rap1GAP may shut off the GTP-GDP energy cycles
of Rap1 via p21 to influence the cell differentiation and the exact
process must be more complicated. In addition to the traditional
pathway, Rap1GAP also interacted with the Gz member of trimeric G
and may exert different cell activities (27,28).
Tumor invasion and metastasis involved several
events including tumor cell adhesion, extracellular matrix (ECM)
proteolysis and cell migration (29). In some reports, increased expression
of Rap1GAP reduced tumor invasion by altering cell adhesion or
migration (7,22). Zheng et al demonstrated that
Rap1GAP suppressed two integrin-dependent events, focal adhesion
formation and filamentous actin (F-actin) assembly, which resulting
in the inhibition of melanoma cell migration (8). However, Mitra et al reported
that Rap1GAP promoted invasion of squamous cell carcinomas via
induction of MMP9 secretion and was associated with poor survival,
which was the first report regarding the link between Rap1GAP and
MMP9 (30). MMPs, particularly MMP9
and MMP2, play an important part in the degradation of ECM to
render the spreading of tumor cells. Here, we also observed the
increased invasion rate of Rap1GAP-transfected HL-60 cells as well
as the higher expression and secretion of MMP9 compared to the
control cells. The secretion and activation of MMPs were a
complicated process which involved several genes including tissue
inhibitor of metalloproteinase 2 (TIMP-2), membrane type 1 MMP
(MT1-MMP) and other genes. Presently, we do not know why an
increased expression of Rap1GAP promotes invasion of leukemic cells
and whether it affects the prognosis of AML. This was the first
report on the relationship between Rap1GAP and MMPs in
hematopoietic cells.
In conclusion, our results demonstrated for the
first time that the expression of Rap1GAP was downregulated in AML
compared with the control group. Upregulated expression of Rap1GAP
promoted leukemic cells (i.e. HL-60 or NB4 cells) differentiation
and apoptosis while also promoted cell invasion. This study
provides initial results on the function of Rap1GAP in leukemia
cell lines in vitro. However, the exact mechanism of the
effect of Rap1GAP in leukemogenesis and the relationship between
Rap1GAP and MMP9 needs further exploration.
Acknowledgements
This study has been supported by the National Key
projects of China (2011CB933501), the National Natural Scientific
Foundation of China (#81070402, #81170468) and the project was
funded by the Priority Academic Program for Development of Jiangsu
Higher Education Institutions.
References
|
1
|
Gao L, Feng Y, Bowers R, et al:
Ras-associated protein-1 regulates extracellular signal-regulated
kinase activation and migration in melanoma cells: two processes
important to melanoma tumorigenesis and metastasis. Cancer Res.
66:7880–7888. 2006. View Article : Google Scholar
|
|
2
|
York RD, Yao H, Dillon T, et al: Rap1
mediates sustained MAP kinase activation induced by nerve growth
factor. Nature. 392:622–626. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hattori M and Minato N: Rap1 GTPase:
functions, regulation, and malignancy. J Biochem. 134:479–484.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bos JL, de Bruyn K, Enserink J, et al: The
role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans.
31:83–86. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bos JL, Rehmann H and Wittinghofer A: GEFs
and GAPs: critical elements in the control of small G proteins.
Cell. 129:865–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Daumke O, Wittinghofer A and Weyand M:
Purification, crystallization and preliminary structural
characterization of human Rap1GAP. Acta Crystallogr D Biol
Crystallogr. 60:752–754. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Chenwei L, Mahmood R, et al:
Identification of a putative tumor suppressor gene Rap1GAP in
pancreatic cancer. Cancer Res. 66:898–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng H, Gao L, Feng Y, Yuan L, Zhao H and
Cornelius LA: Down-regulation of Rap1GAP via promoter
hypermethylation promotes melanoma cell proliferation, survival,
and migration. Cancer Res. 69:449–457. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nellore A, Paziana K, Ma C, et al: Loss of
Rap1GAP in papillary thyroid cancer. J Clin Endocrinol Metab.
94:1026–1032. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong X, Korch C and Meinkoth JL: Histone
deacetylase inhibitors upregulate Rap1GAP and inhibit Rap activity
in thyroid tumor cells. Endocr Relat Cancer. 18:301–310. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi X, Chen Z, Qian J, Cen J and Gu M:
Expression of Rap1GAP in human myeloid disease following microarray
selection. Genet Mol Res. 7:379–387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ika SA, Qi XF and Chen ZX: Protein RAP1GAP
in human myelodysplastic syndrome detected by flow cytometry and
its clinical relevance. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
17:612–617. 2009.PubMed/NCBI
|
|
13
|
Dull T, Zufferey R, Kelly M, et al: A
third-generation lentivirus vector with a conditional packaging
system. J Virol. 72:8463–8471. 1998.PubMed/NCBI
|
|
14
|
Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa
Y and Noda M: A Ras-related gene with transformation suppressor
activity. Cell. 56:77–84. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stork PJ: Does Rap1 deserve a bad Rap?
Trends Biochem Sci. 28:267–275. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Delehanty LL, Mogass M, Gonias SL, Racke
FK, Johnstone B and Goldfarb AN: Stromal inhibition of
megakaryocytic differentiation is associated with blockade of
sustained Rap1 activation. Blood. 101:1744–1751. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Wang PY and Ghosh A: Regulation of
cortical dendrite development by Rap1 signaling. Mol Cell Neurosci.
28:215–228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishida D, Yang H, Masuda K, et al:
Antigen-driven T cell anergy and defective memory T cell response
via deregulated Rap1 activation in SPA-1-deficient mice. Proc Natl
Acad Sci USA. 100:10919–10924. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wienecke R, König A and DeClue JE:
Identification of tuberin, the tuberous sclerosis-2 product.
Tuberin possesses specific Rap1GAP activity. J Biol Chem.
270:16409–16414. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh L, Gao Q, Kumar A, et al: The
high-risk human papillomavirus type 16 E6 counters the GAP function
of E6TP1 toward small Rap G proteins. J Virol. 77:1614–1620. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao Q, Kumar A, Singh L, et al: Human
papillomavirus E6-induced degradation of E6TP1 is mediated by E6AP
ubiquitin ligase. Cancer Res. 62:3315–3321. 2002.PubMed/NCBI
|
|
22
|
Zhang Z, Mitra RS, Henson BS, et al:
Rap1GAP inhibits tumor growth in oropharyngeal squamous cell
carcinoma. Am J Pathol. 168:585–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishida D, Kometani K, Yang H, et al:
Myeloproliferative stem cell disorders by deregulated Rap1
activation in SPA-1 deficient mice. Cancer Cell. 4:55–65. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsygankova OM, Ma C, Tang W, et al:
Downregulation of Rap1GAP in human tumor cells alters cell/matrix
and cell/cell adhesion. Mol Cell Biol. 30:3262–3274. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hass R, Gunji H, Datta R, et al:
Differentiation and retrodifferentiation of human myeloid leukemia
cells is associated with reversible induction of cell
cycle-regulatory genes. Cancer Res. 52:1445–1450. 1992.PubMed/NCBI
|
|
26
|
Bürger C, Wick M and Müller R:
Lineage-specific regulation of cell cycle gene expression in
differentiating myeloid cells. J Cell Sci. 107:2047–2054.
1994.PubMed/NCBI
|
|
27
|
Mochizuki N, Ohba Y, Kiyokawa E, et al:
Activation of the ERK/MAPK pathway by an isoform of rap1GAP
associated with G alpha(i). Nature. 400:891–894. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng J, Glick JL, Polakis P and Casey PJ:
Functional interaction between Galpha(z) and Rap1GAP suggests a
novel form of cellular cross-talk. J Biol Chem. 274:36663–36669.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Foda HD and Zucker S: Matrix
metalloproteinases in cancer invasion, metastasis and angiogenesis.
Drug Discov Today. 6:478–482. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mitra RS, Goto M, Lee JS, et al: Rap1GAP
promotes invasion via induction of matrix metalloproteinase 9
secretion, which is associated with poor survival in low N-stage
squamous cell carcinoma. Cancer Res. 68:3959–3969. 2008. View Article : Google Scholar
|