Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common malignancy and the third most common cause of cancer-related
deaths worldwide, claiming over one million lives annually
(1). The highest incidence rates
are reported in East Asia (2). The
prognosis of patients with HCC is poor, with a 5-year survival rate
after diagnosis of ~10% (3).
Ets1 is the founding member of the Ets family of
transcription factors and has been shown to promote invasive
behavior in multiple cell types (4–10). The
regulation of matrix metalloproteases MMP-1, MMP-3, MMP-9 and
urokinase type plasminogen activator (uPA) expression have been
ascribed to Ets1 (4–10). Expression of Ets1 is also associated
with poor prognosis in patients with tumors including breast
cancer, ovarian tumor, and hepatocellular carcinoma.
Various molecular alterations occur in
pre-neoplastic nodules and escalate in HCC, including dysregulation
of well-known molecular pathways in carcinogenesis (11–14).
The important role of microRNAs (miRNAs) in regulating these
pathways has been emphasized. MiRNAs are small (18–24 nucleotides),
evolutionarily conserved, endogenous, single-stranded, non-coding
RNA molecules, that negatively modulate gene expression in animals
and plants. Mature miRNAs operate via sequence-specific
interactions with the 3′ untranslated region (UTR) of cognate mRNA
targets, causing suppression of translation and mRNA decay
(15,16). A large body of evidence suggests
that the multigene regulatory capacity of miRNAs is dysregulated
and exploited in cancer. Indeed, miRNA loci are often targeted by
genetic and epigenetic defects, and miRNA signatures facilitating
tumor classification and the prediction of clinical outcome have
been reported (17,18). A global reduction of miRNA abundance
appears to be a general trait of human cancers, playing a causal
role in the transformed phenotype (19–21).
Aberrant expression of miRNA has also been linked to a variety of
cancers, including HCC (22–25).
In the current study, we show that the ets1
proto-oncogene, which is highly expressed in HCC (26), is targeted by miR-1 and
miR-499. MiR-1 and miR-499 specifically inhibit the
expression of Ets1. Overexpression of miR-1 and
miR-499 in the HepG2 HCC cell line inhibited cellular
invasion and migration. Taken together, these results suggest that
Est1 is negatively regulated by miR-1 and miR-499 in
HepG2 cells, which may contribute to the invasive and migratory
potential of hepatocellular carcinoma.
Materials and methods
Cell Culture
HepG2 and HEK 293 cell lines (American Type Culture
Culture Collection, Manassas, VA, USA) were maintained in
Dulbecco’s modified Eagle medium (DMEM) (Gibco-BRL, Grand Island,
NY, USA) containing 10% (v/v) fetal bovine serum (FBS) supplemented
with 100 U/ml penicillin and 100 μg/ml streptomycin, at 37°C with
5% CO2.
Vector construction
For construction of miRNA expression plasmids,
miR-1 and miR-499 precursors were amplified from
human genomic DNA by PCR using the primer pairs: miR-1, F,
5′-TAGAAGCTTGCCTCTGAGCTGCCTTCTCTA-3′ and R,
5′-TATCTCGAGCACCACAGCCGCCTGGCTGGC-3′; miR-499, F,
5′-TAGAAGCTTGTGTCCCAGCTGCACA AGGTA-3′ and R,
5′-TATCTCGAGTGTCTCCCATCACCA CCACCA-3′. PCR products were cloned
into pcDNA3.0 (Invitrogen, Carlsbad, CA, USA). In the same way, the
miRNA expression plasmids of miR-139, miR-181a,
miR-200b, miR-221, miR-365 and miR-429
had been constructed in our laboratory. For the construction of
luciferase reporter vectors, 3′UTR segments of ets1
(Ets1-3′UTR-1 and Ets1-3′UTR-2) were amplified from human genomic
DNA using the primer pairs: Ets1-3′UTR-1, F,
5′-ACGTCTAGACTGTGAGTATA ACTCCTGCAG-3′ and R, 5′-GATCATATGATATGAAA
TCAGGCTACAGTA-3′; Ets1-3′UTR-2, F, 5′-ACGTCTAG
AGCAAGTGACATTGTCACATCA-3′ and R, 5′-GATCATA
TGCACCAATCAGAAAGCCGTACA-3. Mutant inserts containing substitutions
in the miRNA complementary sites were generated by PCR using the
primers: Ets1-3′UTR-1mut, F, 5′-TTGTTGAACTCTTACCTCGCCGGGCAAGAATTT
CAAGGAACC-3′ and R, 5′-GGTTCCTTGAAACTTCTT
GCCCGGCGAGGTAAGAGTTCAACAA-3′; Ets1-3′UTR-2mut, F,
5′-TTTTTTTCTTAAAAATCCGGCCGGGCTCTA AGGTGGTCTCAG-3′ and R,
5′-CTGAGACCACCTTAGAG CCCGGCCGGATTTTTAAGAAAAAAA-3′. PCR products
were cloned into the modified pGL3 control vector (Promega,
Madison, WI, USA) immediately downstream of the stop codon of the
luciferase gene. Wild-type and mutant inserts were confirmed by
sequencing.
miRNAs, small interfering RNA (siRNA) and
transfection
The miR-1 and miR-499 duplexes,
ets1 and negative control siRNAs were designed and
synthesized by GenePharma (Shanghai, China). The sequences are as
follows (sense/antisense): miR-1,
5′-UGGAAUGUAAAGAAGUAUGUAU-3′/5′-AUACAUACUUCUUUACAUUCCA-3′;
miR-499, 5′-UUA AGACUUGCAGUGAUGUUU-3′/5′-AAACAUCACUGCAA
GUCUUAA-3′; ets1 siRNA, 5′-ACUUGCUACCAUCCCGU
ACTT-3′/5′-GUACGGGAUGGUAGCAAGUTT-3′; negative control siRNA,
5′-UUCUCCGAACGUGUCACGUTT -3′/5′-ACGUGACACGUUCGGAGAATT-3′.
Transfection was performed using Lipofectamine 2000 (Invitrogen).
In brief, cells were seeded in six-well plates to reach an optimum
density of 50% confluency after 24 h. For transfections, siRNA (20
μM) or miRNA (20 μM) was combined with 5 μl of Lipofectamine 2000
and 250 μl of Opti-MEM medium (Gibco-BRL). This mixture was added
to cells and incubated for 6 h before replacing with fresh medium.
Total-RNA and protein were extracted 48 h after transfection for
use in qRT-PCR and western blot analysis.
Luciferase reporter assays
HEK 293 cells were plated in 24-well plates to reach
80–90% confluency. Cells were co-transfected with luciferase
reporter vectors (100 ng) containing the ets1 3′UTR
(pGL3m-Ets1-3′UTR-1 and pGL3m-Ets1-3′UTR-2) or
ets1 3′UTR mutant (pGL3-Ets1-3′UTR-1mut and
pGL3-Ets1-3′UTRmut-2mut) and pRL-TK control Renilla luciferase
vector (Promega) (8 ng) using Lipofectamine 2000 (Invitrogen).
Luciferase activity was measured by Dual luciferase assays
(Promega) 48 h after transfection.
RNA extraction and qRT-PCR
Total-RNA was extracted with TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions. cDNA was
synthesized using oligo(dt) primers and Impro-II reverse
transcriptase (Promega) according to the manufacturer’s
instructions. qRT-PCR reactions were prepared using SYBR Premix Ex
Taq (Takara, Kyoto, Japan). Reactions were performed in triplicate
using an Mx3000P real-time PCR instrument (Agilent Technologies,
Santa Clara, CA, USA). The PCR primers were: ets1, F,
5′-TGGAGTC AACCCAGCCTATC-3′ and R, 5′-TCTGCAAGGTGTCTGTC TGG-3′;
GAPDH, F, 5′-TCAGTGGTGGACCTGACCTG-3′ and R,
5′-TGCTGTAGCCAAATTCGTTG-3′. Expression of ets1 was
calculated according to the delta-delta Ct method, normalizing to
GAPDH.
Western blot analysis
Total cell lysates were extracted using sodium
dodecyl sulfate (SDS) buffer. Proteins were resolved by 12%
SDS-PAGE and transferred onto polyvinylidene fluoride membranes.
Membranes were probed with monoclonal antibodies to Ets1 (sc-55581;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and β-actin
(sc-47778; Santa Cruz Biotechnology). Detection was performed with
Supersignal (Pierce, Rockford, IL, USA) chemiluminescence reagent.
Quantitative analysis was performed using Quantity One software
(Bio-Rad, Hercules, CA, USA).
Transwell cell invasion and migration
assays
For invasion assays, transfected HepG2 cells were
serum-starved for 18 h in DMEM containing 0.1% (v/v) FBS. Cells
were trypsinized and resuspended in the same medium and
2×105 cells were added to the upper chamber of each well
(6.5 mm in diameter, 8 μm pore size; Corning, Inc., Corning, NY,
USA) coated with 30 mg/cm2 matrigel extracellular matrix
(ECM) gel (Sigma-Aldrich, St. Louis, MO, USA). Medium containing
0.1% (v/v) FBS, supplemented with hepatocyte growth factor (HGF)
(20 ng/ml) (ProSpec-Tany TechnoGene, Ltd., East Brunswick, NJ, USA)
was placed in the lower compartment of the chamber. After
incubation for 24 h at 37°C, cells on the upper membrane surface
were removed by carefully wiping with a cotton swab, and the
filters were fixed by treatment with 95% (v/v) ethanol for 30 min.
Cells were stained with 0.2% (w/v) crystal violet solution for 30
min. Cells adhering to the undersurface of the filter were counted
(five high-power fields/chamber) using an inverted microscope.
Migration assays were performed as described above, excluding the
use of matrigel and with an incubation time of 12 h.
Statistical analysis
All values are reported as the means ± standard
deviation. Differences were assessed by two-tailed Student’s t-test
of Excel software. P<0.05 was considered statistically
significant.
Results
Interaction of miR-1 or miR-499 with the
3′UTR of Ets1 mRNA
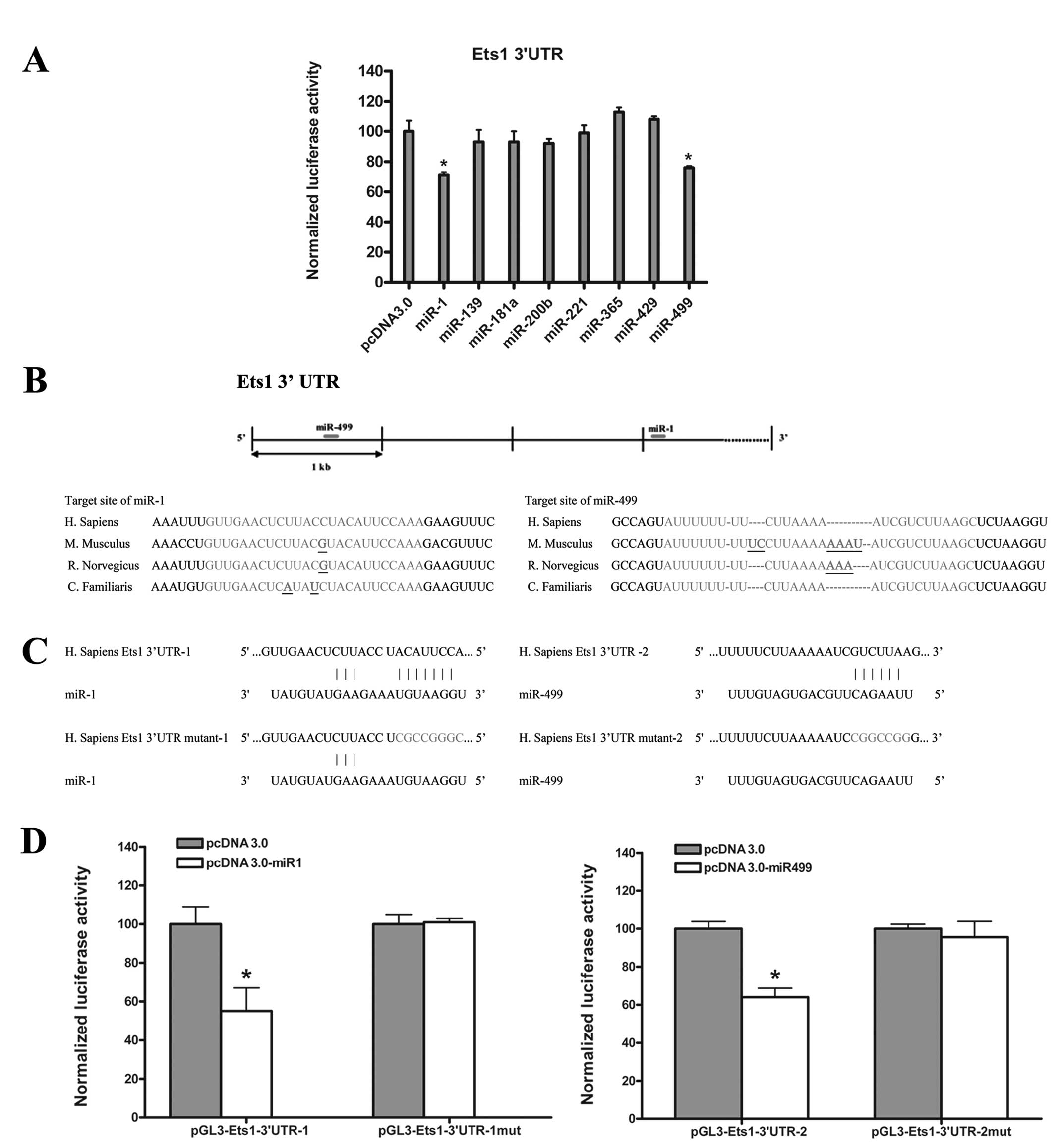
In order to identify miRNAs regulating Ets1, we used
TargetScan (www.targetscan.org), an online software
program, to predict miRNAs targeting 3′UTR of the ets1 mRNA.
This analysis revealed that the 3′UTR of ets1 contains
putative sites for >20 miRNAs. We tested the ability of a subset
of these miRNAs (miR-1, miR-139, miR-181a,
miR-200b, miR-221, miR-365, miR-429 and
miR-499) to target the ets1 3′UTR using luciferase
reporter assays. Our analysis showed that only miR-1 and
miR-499 induced an obvious decrease in relative luciferase
activity (Fig. 1A). Further
investigation showed that the putative target sites for
miR-1 and miR-499 are conserved in mammalian species
(Fig. 1B). The target sites for
miR-499 and miR-1 locate in the front and rear
fragment of 3′UTR of ets1 mRNA, respectively. To investigate
this potential interaction experimentally, the 3′UTR of ets1
mRNA was divided into two fragments, Ets1-3′UTR-1 and Ets1-3′UTR-2,
and sub-cloned into a modified pGL3 control plasmid (pGL3m), as
previously described (27). We then
tested the ability of miR-1 or miR-499 to inhibit
luciferase activity of pGL3m-Ets1-3′UTR-1 or pGL3m-Ets1-3′UTR-2
following co-transfection into HEK 293 cells. Our analysis
demonstrated that both miR-1 and miR-499 induced a
significant decrease in relative luciferase activity (~40%)
compared with vector transfected cells (Fig. 1D). To test the specificity of this
interaction, miR-1 and miR-499 overexpression
constructs were co-transfected with ets1 luciferase reporter
constructs containing substitutions disrupting the miRNA target
sites (Fig. 1C). We did not observe
any decrease in relative luciferase activity in miRNA transfected
cells compared with vector control (Fig. 1D).
miR-1 and miR-499 downregulate Ets1
expression
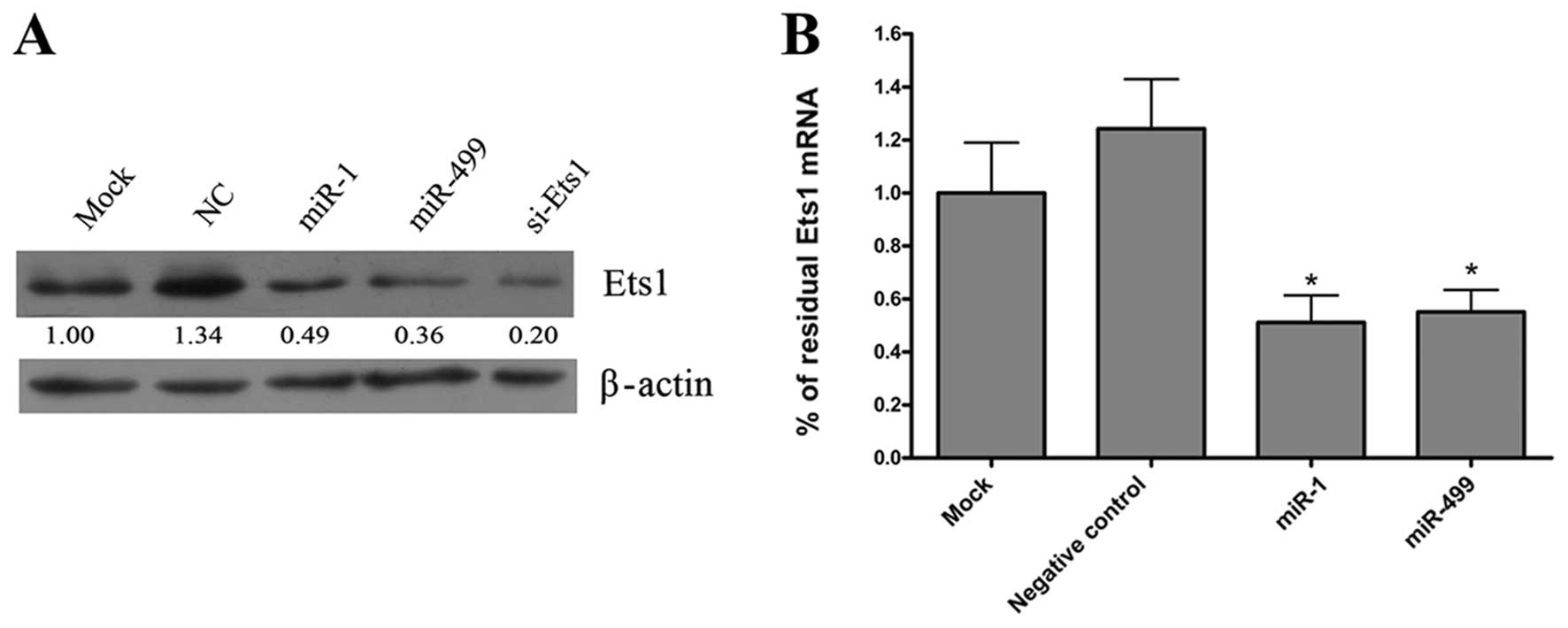
To investigate whether miR-1 or
miR-499 affect endogenous Ets1 protein expression, we
transfected miR-1 and miR-499 duplexes into HepG2
cells and analyzed Ets1 expression after 48 h by western blot
analysis. As a positive control, cells were also transfected with
Ets1 siRNA. We found that miR-1 and miR-499
dramatically reduced the expression of Ets1 protein compared
to negative control (Fig. 2A).
qRT-PCR analysis of ets1 mRNA expression 48 h after
transfection with miR-1 and miR-499 duplexes,
revealed that the level of Ets1 mRNA was also significantly
reduced compared to negative control (Fig. 2B). There were no significant
differences on Ets1 protein and mRNA levels between overexpression
of individual miRNAs or co-transfection of both miR-1 and
miR-499.
miR-1 and miR-499 negatively regulate
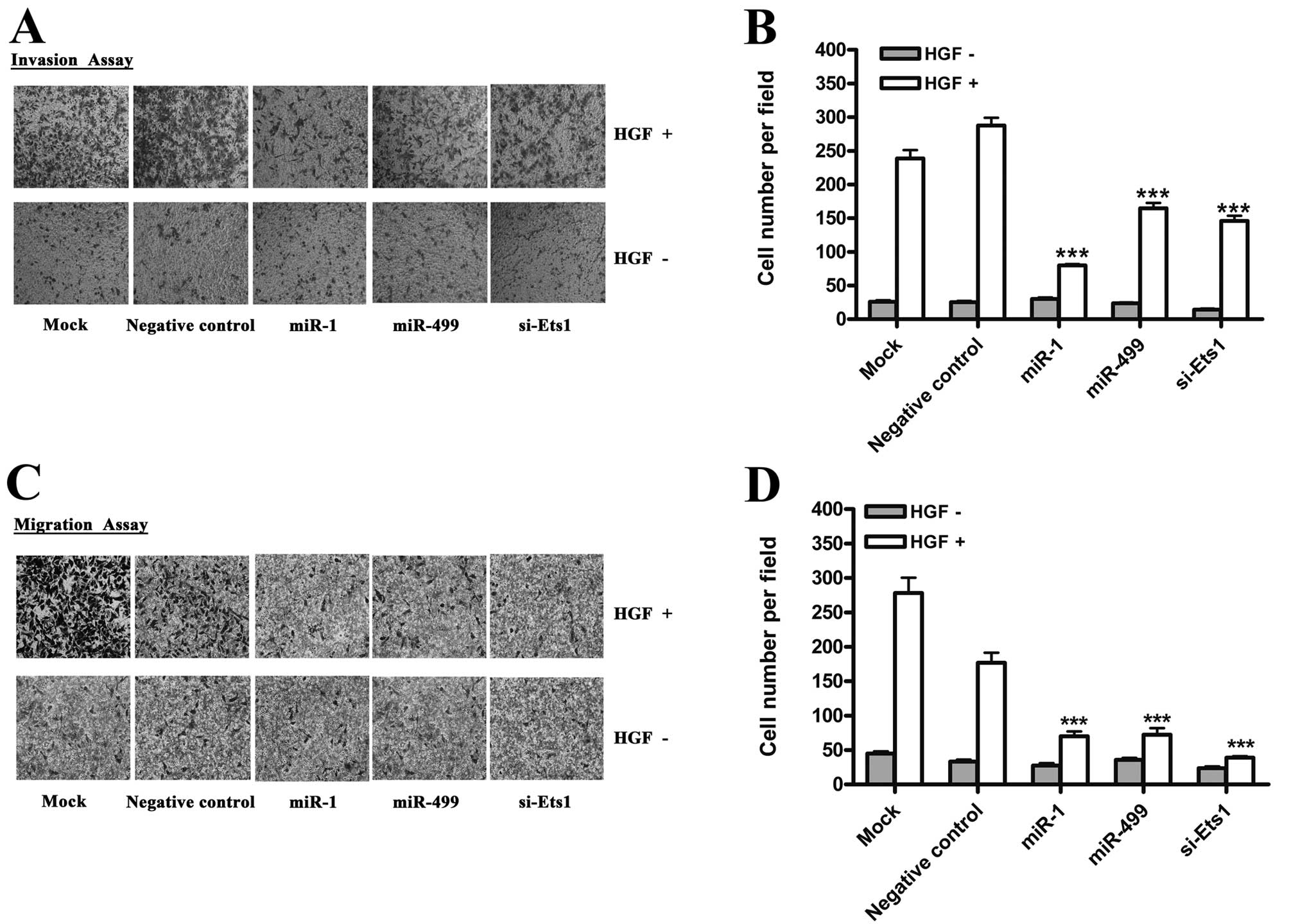
cell invasion and migration in vitro
HGF (hepatocyte growth factor), a cytokine also
known as scatter factor (SF), can significantly promote the
invasion of hepatoma carcinoma cells (28). Moreover, Ets1 has been shown to play
a key role in the acquisition of invasive behavior by inducing the
expression of MMP-1, MMP-3, MMP-9 and
uPA(4). Given that
miR-1 and miR-499 are capable of affecting Ets1
expression, we examined the effect of overexpressing these miRNAs
on HGF-induced invasiveness of HepG2 cells using the matrigel
invasion assay system. As shown in Fig.
3A, miR-1 and miR-499 significantly reduced
HGF-induced invasion of HepG2 cells. Next, we examined the effect
of miR-1 and miR-499 on HGF-induced migration of
HepG2 cells using transwell migration assays. Similar to the
invasion assays, miR-1 and miR-499 also inhibited the
migration behavior of HepG2 cells (Fig.
3C). To eliminate the possibility of off-target effects, we
transfected cells with Ets1 siRNA. In a similar manner to miRNA
overexpression, we found that knockdown of Ets1 inhibited the
invasion and migration induced by HGF (Fig. 3A and C).
Discussion
Growing evidence indicates that Ets1 plays a key
role in the invasive behavior of many mammalian tumors. In this
study, we demonstrate that miR-1 and miR-499
negatively regulate the ets1 proto-oncogene at the
post-transcriptional level, via conserved sites within the 3′UTR.
Furthermore, overexpression of miR-1 or miR-499
inhibited the invasion and migration of HepG2 cells in
vitro, emphasizing the essential role of these two miRNAs in
hepatic oncogenesis and tumor behavior.
In general, miRNAs may function as both tumor
suppressors and oncogenes in tumors. The correlation between the
expression of specific miRNAs and cancer has been widely observed.
It has also been shown that a global reduction of miRNA abundance
may be a general trait of human cancers (19–21).
This suggests that miRNAs may have a crucial function in cancer
progression (29).
Overexpression of Ets1 is highly associated with
many types of cancer. Ets1 expression is generally higher in
invasive tumors than in benign tumors (6,9), and
is indicative of poor prognosis (7,9,26,30).
The expression of Ets1 is also correlated with histological
differentiation of HCC (26). This
suggests that Ets1 is higher in poorly differentiated HCC and may
yield relative biological information to HCC.
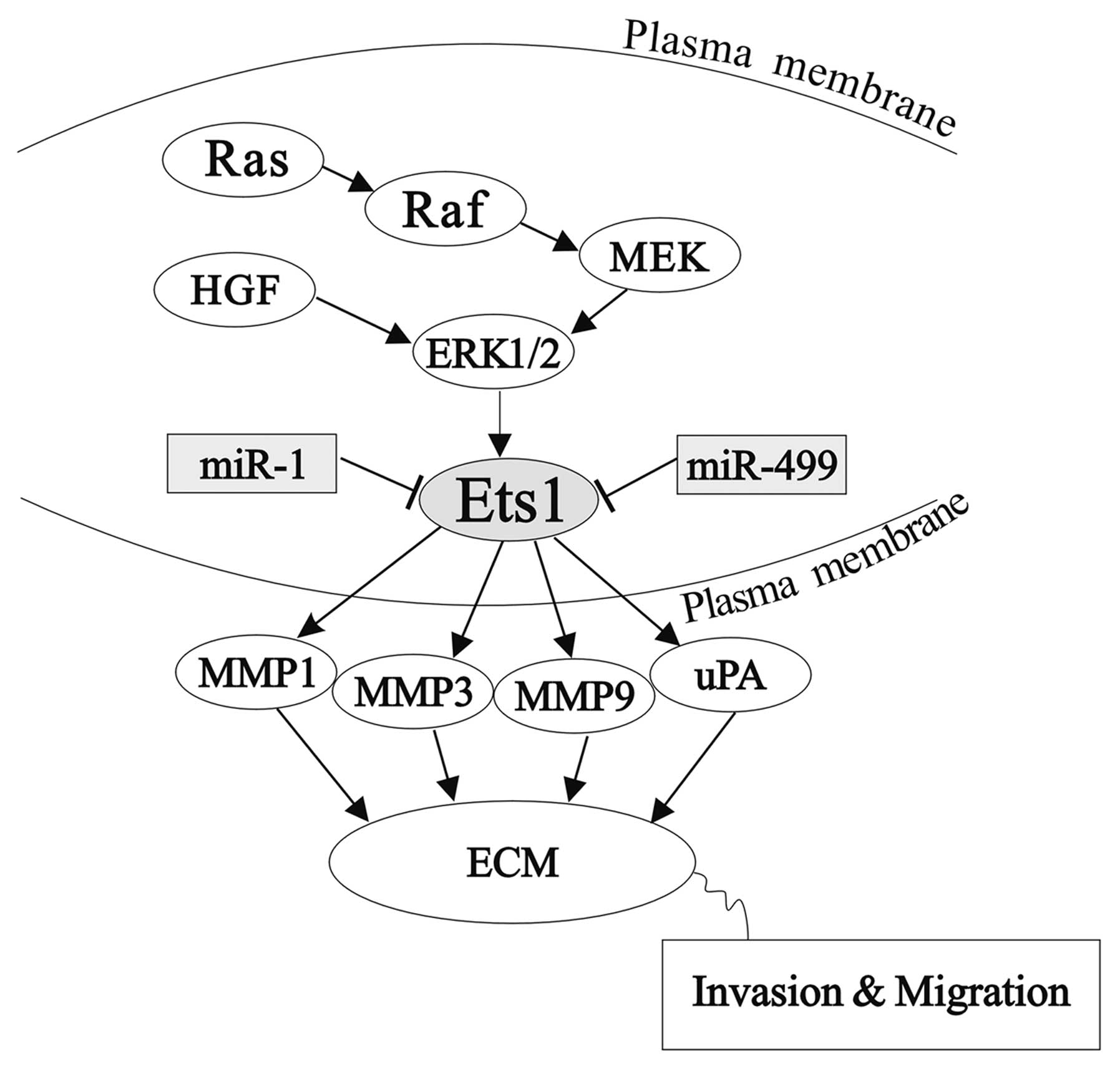
Ets1 responsive genes include those encoding certain
proteases, including the matrix metalloproteases, MMP-1,
MMP-3, MMP-9 and uPA(4). A schematic representation is shown in
Fig. 4. These proteases are
involved in ECM-degradation, a key event in invasion. Ets1
expression positively correlates with MMP-1 in angiosarcoma of the
skin (31), and with MMP-1 and
MMP-9 in ovarian carcinoma cells and stromal fibroblasts in breast
and ovarian cancer, respectively (7,9). Ets1
expression also correlates with expression of uPA in lung and brain
tumors (6,8,10).
Targeted knockdown of Ets1 leads to a decrease in the expression of
MMP-1 and MMP-9. Correspondingly, overexpression of Ets1 induced
the production of MMP-1, MMP-3 and MMP-9 or MMP-1, MMP-9 and uPA,
in hepatoma cells and endothelial cells respectively (32–34).
There is growing evidence to show that Ets1 may also
be involved in the regulation of c-Met, the receptor for HGF/SF,
which induces migration (35).
Furthermore, c-Met may also activate Ets1, as HGF/SF has
been shown to stimulate Ets1 activity through the
Ras/Raf/MEK1/ERK1/2 pathway in MDCK cells (36).
In our study, we found that miR-1 and
miR-499 inhibit HGF-induced invasion and migration in HCC
HepG2 cells, by repressing the expression of the ets1
proto-oncogene. These results indicate that miR-1 and
miR-499 may represent candidates for anticancer therapy. In
conclusion, miR-1 and miR-499 inhibit Ets1 expression
by binding to the 3′UTR of the ets1 mRNA, thereby reducing
HGF-induced cell invasion and migration.
Acknowledgements
This study was partially supported by the Chinese
State Key Projects for Basic Research (2010CB912801, 2009CB521804),
the Chinese National Natural Science Foundation Projects (81072021)
and the Beijing Natural Science Foundation Project (7101007).
References
|
1
|
WHO. The global burden of disease 2004
update. World Health Organization; Geneva: (part 2): pp. 12–13.
2004
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
London WT and McGlynn KA: Liver cancer.
Cancer Epidemiology and Prevention. Schottenfeld D and Fraumeni JF
Jr: 3rd edition. Oxford University Press; New York: pp. 763–786.
2006, View Article : Google Scholar
|
|
4
|
Sementchenko VI and Watson DK: Ets target
genes: past, present and future. Oncogene. 19:6533–6548. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naito S, Shimizu S, Matsuu M, et al: Ets-1
upregulates matrix metalloproteinase-1 expression through
extracellular matrix adhesion in vascular endothelial cells.
Biochem Biophys Res Commun. 291:130–138. 2002. View Article : Google Scholar
|
|
6
|
Kitange G, Tsunoda K, Anda T, et al:
Immunohistochemical expression of Ets-1 transcription factor and
the urokinase-type plasminogen activator is correlated with the
malignant and invasive potential in meningiomas. Cancer.
89:2292–2300. 2000. View Article : Google Scholar
|
|
7
|
Behrens P, Rothe M, Wellmann A, Krischler
J and Wernert N: The Ets-1 transcription factor is up-regulated
together with MMP 1 and MMP 9 in the stroma of pre-invasive breast
cancer. J Pathol. 194:43–50. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takanami I, Takeuchi K and Karuke M:
Expression of ETS-1 is correlated with urokinase-type plasminogen
activator and poor prognosis in pulmonary adenocarcinoma. Tumour
Biol. 22:205–210. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Behrens P, Rothe M, Florin A, Wellmann A
and Wernert N: Invasive properties of serous human epithelial
ovarian tumors are related to Ets-1, MMP-1 and MMP-9 expression.
Int J Mol Med. 8:149–154. 2001.PubMed/NCBI
|
|
10
|
Nakada M, Yamashita J, Okada Y and Sato H:
Ets-1 positively regulates expression of urokinase-type plasminogen
activator (uPA) and invasiveness of astrocytic tumors. J
Neuropathol Exp Neurol. 58:329–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wurmbach E, Chen YB, Khitrov G, et al:
Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee JS and Thorgeirsson SS: Comparative
and integrative functional genomics of HCC. Oncogene. 25:3801–3809.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lemmer ER, Friedman SL and Llovet JM:
Molecular diagnosis of chronic liver disease and hepatocellular
carcinoma: the potential of gene expression profiling. Semin Liver
Dis. 26:373–384. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Budhu A, Jia HL, Forgues M, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li LM, Hu ZB, Zhou ZX, et al: Serum
microRNA profiles serve as novel biomarkers for HBV infection and
diagnosis of HBV-positive hepatocarcinoma. Cancer Res.
70:9798–9807. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou J, Yu L, Gao X, et al: Plasma
microRNA panel to diagnose hepatitis B virus-related hepatocellular
carcinoma. J Clin Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ladeiro Y, Couchy G, Balabaud C, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanda K, Nakayama T, Onizuka S, Tomioka T
and Kanematsu T: Expression of the Ets-1 proto-oncogene is linked
to cell differentiation of human hepatocellular carcinoma.
Hepatogastroenterology. 49:747–751. 2002.PubMed/NCBI
|
|
27
|
Cui J, Fu H, Feng J, Zhu J, Tie Y, Xing R,
Wang C and Zheng X: The construction of miRNA expression library
for human. Progr Biochem Biophys. 34:389–394. 2007.
|
|
28
|
Nakamura T, Nishizawa T, Hagiya M, et al:
Molecular cloning and expression of human hepatocyte growth factor.
Nature. 342:440–443. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Span PN, Manders P, Heuvel JJ, et al:
Expression of the transcription factor Ets-1 is an independent
prognostic marker for relapse-free survival in breast cancer.
Oncogene. 21:8506–8509. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Naito S, Shimizu K, Nakashima M, et al:
Overexpression of Ets-1 transcription factor in angiosarcoma of the
skin. Pathol Res Pract. 196:103–109. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oda N, Abe M and Sato Y: ETS-1 converts
endothelial cells to the angiogenic phenotype by inducing the
expression of matrix metalloproteinases and integrin beta3. J Cell
Physiol. 178:121–132. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sato Y, Abe M, Tanaka K, et al: Signal
transduction and transcriptional regulation of angiogenesis. Adv
Exp Med Biol. 476:109–115. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang Y, Xu W, Lu J, He F and Yang X:
Invasiveness of hepatocellular carcinoma cell lines: contribution
of hepatocyte growth factor, c-met, and transcription factor Ets-1.
Biochem Biophys Res Commun. 286:1123–1130. 2001. View Article : Google Scholar
|
|
35
|
Tamagnone L and Comoglio PM: Control of
invasive growth by hepatocyte growth factor (HGF) and related
scatter factors. Cytokine Growth Factor Rev. 8:129–142. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Paumelle R, Tulasne D, Kherrouche Z, et
al: Hepatocyte growth factor/scatter factor activates the
ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling
pathway. Oncogene. 21:2309–2319. 2002.PubMed/NCBI
|