Introduction
Gliomagenesis, like development of other
malignancies, involves the accumulation of a series of genetic
alterations (1). Many of the genes
altered during glioma development were identified using standard
molecular approaches, and these genes normally participate in a
range of cellular functions (e.g., governing cellular
proliferation, cell infiltration, angiogenesis, and cell death).
Genetic aberrations are frequently found in human glioma: gene
amplification of epidermal growth factor receptor (EGFR) (2) and murine double minute 2 (MDM2)
(3,4); overexpression of platelet-derived
growth factor receptor (PDGFR) (5);
gene mutation of retinoblastoma (Rb), p53 (6) and phosphatase and tensin homolog
deleted on chromosome ten (PTEN) (7); deletion of cyclin-dependent kinase
inhibitor 2A (CDKN2A/p16INK4A) (3,4).
Despite this information, mechanism of tumorigenesis and
progression in glioblastoma (GB) have not been understood in detail
because malignant gliomas, including GB, have significant
morphological heterogeneity in each tumor; individual tumors are
genetically and histopathologically very heterogeneous. In order to
overcome this complexity in glioma phenotypes and identify putative
therapeutic targets, more global and systematic approaches,
including proteomic (8),
transcriptomic (5,6), and comparative genomic hybridization
analyses, have been performed. Under these circumstances, we
performed proteomic analysis to compare protein expression profiles
in diffuse astrocytoma (DA) and GB, and we found that total
expression of SIRT2 was lower in GB than in DA (8).
Sirtuin 2 (SIRT2) is a NAD-dependent deacetylase,
and is a member of the human sirtuin family that was initially
identified based on structural homology to the Saccharomyces
cerevisiae Sir2 protein (silent information regulator)
(9). In human, there are seven
proteins of the sirtuin family (SIRT1–7) (10,11).
Among all sirtuins, SIRT2 was the most highly express in brain
tissue, and SIRT2 expression was particularly prominent in the
postnatal hippocampus (12), and it
has been suggested that SIRT2 has neuronal functions, including
cytoskeletal growth cone dynamics (11), neurite outgrowth, and
oligodendrocyte arborization in vitro(13). It has been suggested that SIRT2 may
have tumor-suppressor activity because SIRT2 suppressed colony
formation in glioma cell lines and controlled cell cycle
progression by acting as a regulator of mitotic exit (14–16).
Additionally, we reported that subcellular localization of SIRT2
was translocated from cytoplasm to nucleus when cells were exposed
to ionizing radiation in the human fibroblast cell line TIG-1
(16). In the present study, we
evaluated of the expression and subcellular localization of SIRT2
in samples from patients with GB and/or DA using
immunohistochemistry, and we assessed the prognostic significance
of SIRT2 expression pattern in GB patients. We demonstrated that
although nuclear SIRT2 expression was seen in all gliomas examined,
SIRT2 localization was predominantly nuclear in GB samples but
predominantly cytoplasmic in control samples; moreover, the
percentage of GB cells with SIRT2-positive nuclei was negatively
correlated with survival time of patients with GB.
Materials and methods
Cell culture
As a control, we analyzed primary glial cells
isolated from brain tissue of C57BL/6 mice. Cells were grown in
Dulbecco's modified Eagle's medium (DMEM, Gibco Invitrogen Corp.,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, at
37°C, under 5% CO2, and in 12-well chamber slides
(17).
Tissue collections
This study used surgically resected samples from 16
patients with glioma being treated at Tottori University and from
15 patients with glioma whose samples were stored at the tissue
archive of Toyama University Hospital (Table I). The samples were fixed with 10%
formalin and embedded in paraffin. The glioma specimens were
classified according to the World Health Organization (WHO)
International Histological Classification of Tumors (18). We also examined autopsy specimens of
brain tissue from 5 neurologically and neuropathologically normal
individuals (causes of death: acute heart failure, squamous cell
carcinoma, acute myocardial infarction, disseminated intravascular
coagulation, or pneumonia). Among 31 brain tumor samples, 15
samples of GB (patient nos. 10–24 in Table I) were subjected to the tissue
microarray (TMA) method, in which tissue cylinders with a diameter
of 0.6 mm were punched from GB areas of each tissue blocks
(19) (Table I). Clinical data, including age,
gender, and survival time from the initial operation, were obtained
from the hospital records. Multiple 5-μm sections were prepared
from each specimen. One section was stained with hematoxylin and
eosin (H&E), and the others were used for the
immunohistochemical tests. This study was approved by the Ethics
Committer of Tottori University (Permission: no. 1434) and Toyama
University Hospital (Permission: no. 19–12).
 | Table ICharacteristics of 23 patients with
glioblastoma, eight patients with astrocytoma, and five normal
individuals. |
Table I
Characteristics of 23 patients with
glioblastoma, eight patients with astrocytoma, and five normal
individuals.
| Patient no. | Age | Gender | Diagnosis (WHO
grade) | Tissue sample | SIRT2 labeling index
(%) | SIRT2 cytoplasm | Survival
(months) |
|---|
| Brain tumor |
| 1 | 70 | Female | GB (lV) | Biopsy | 72.3 | − | 9 |
| 2 | 26 | Female | GB (lV) | Biopsy | 75.3 | − | 30 |
| 3 | 56 | Male | GB (lV) | Biopsy | 69.4 | + | 17 |
| 4 | 57 | Male | GB (lV) | Biopsy | 60.6 | + | 17 |
| 5 | 58 | Female | GB (lV) | Biopsy | 68.8 | + | 34 |
| 6 | 55 | Male | GB (lV) | Biopsy | 75.2 | + | 4 |
| 7 | 65 | Male | GB (lV) | Biopsy | 91.8 | + | 16 |
| 8 | 76 | Female | GB (lV) | Biopsy | 74.9 | − | 5 |
| 9 | 71 | Male | GB (lV) | TMA | 39.9 | − | 26 |
| 10 | 50 | Male | GB (lV) | TMA | 63.1 | + | 4 |
| 11 | 72 | Female | GB (lV) | TMA | 89.1 | − | 5 |
| 12 | 62 | Male | GB (lV) | TMA | 66.3 | + | 14 |
| 13 | 78 | Female | GB (lV) | TMA | 69.5 | + | 15 |
| 14 | 32 | Female | GB (lV) | TMA | 74.4 | − | 12 |
| 15 | 58 | Female | GB (lV) | TMA | 22.8 | + | 8 |
| 16 | 58 | Female | GB (lV) | TMA | 90.4 | − | 6 |
| 17 | 58 | Male | GB (lV) | TMA | 96.6 | − | 24 |
| 18 | 67 | Male | GB (lV) | TMA | 51.7 | − | 46 |
| 19 | 49 | Male | GB (lV) | TMA | 56.1 | − | 17 |
| 20 | 69 | Male | GB (lV) | TMA | 49.7 | + | 8 |
| 21 | 69 | Male | GB (lV) | TMA | 28.8 | − | 27 |
| 22 | 63 | Male | GB (lV) | TMA | 52.0 | + | 12 |
| 23 | 71 | Male | GB (lV) | TMA | 75.1 | − | 6 |
| 24 | 51 | Female | DA (ll) | Biopsy | 40.5 | − | 103 |
| 25 | 68 | Male | DA (ll) | Biopsy | 34.8 | + | 7 |
| 26 | 25 | Male | DA (ll) | Biopsy | 26.7 | + | 56 |
| 27 | 20 | Male | DA (ll) | Biopsy | 19.4 | + | 36 |
| 28 | 25 | Male | DA (ll) | Biopsy | 34.2 | − | 58 |
| 29 | 18 | Female | DA (ll) | Biopsy | 17.4 | + | 41 |
| 30 | 30 | Female | DA (ll) | Biopsy | 73.0 | + | 25 |
| 31 | 74 | Male | DA (ll) | Biopsy | 83.8 | + | 9 |
|
| Normal | | | Cause of death | | | | |
|
| 32 | 70 | Female | AHF | Autopsy | 25.5 | + | − |
| 33 | 76 | Male | SCC | Autopsy | 12.8 | + | − |
| 34 | 77 | Male | AMI | Autopsy | 25.2 | + | − |
| 35 | 78 | Male | DIC | Autopsy | 29.0 | + | − |
| 36 | 76 | Male | Pn | Autopsy | 50.6 | + | − |
Immunofluorescence
Primary glial cells grown in 12-well chamber slides
were washed twice in phosphate-buffered saline, pH 7.4 (PBS); fixed
in 4% paraformaldehyde for 15 min; and permeabilized in 0.2%
Nonidet P-40 (Nacalai Tesque, Kyoto, Japan) in PBS for 2 min. After
two sequential 5-min washes in PBS, cells were incubated in PBS
with 5% skim milk (Difco, Detroit, MI, USA) for 30 min. Normal
serum served as blocking reagent. A rabbit polyclonal antibody
raised against purified, recombinant human SIRT2 protein was used
at a 1:100 dilution; the antibody was diluted in PBS containing 1%
bovine serum albumin. The specificity and affinity of the
polyclonal anti-human-SIRT2 antibody (anti-SIRT2) (Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA) for use in primary cell
cultures was established previously by using the anti-SIRT2
antibody to detect SIRT2 in mouse cells (20). Herein, cells were incubated with
anti-SIRT2 antibody for 1 h at room temperature. Thereafter, they
were washed three times for 5 min each in PBS with 0.2% Nonidet
P-40, then incubated in PBS with 5% skim milk for 15 min, and
finally incubated with Alexa Flour 488 goat anti-rabbit IgG
(Invitrogen Corp., Carlsbad, CA, USA) diluted 1:1,000 for 30 min.
Stained cells were washed three times for 5 min each in PBS with
0.2% Nonidet P-40 and then counterstained with 1 μg/ml Hoechst
33258 (Sigma-Aldrich Inc., St. Louis, MO, USA). PBS replaced the
anti-SIRT2 antibody in parallel negative-control experiments.
Immunohistochemistry
Sections were deparaffinized, and endogenous
peroxidase activity was quenched by incubation for 30 min with 0.3%
hydrogen peroxide, and samples were washed with PBS. Normal serum
served as blocking reagent. The anti-SIRT2 antibody was diluted in
PBS with 1% bovine serum and used at a dilution of 1:250. The
specificity and affinity of anti-SIRT2 for use in sectioned tissue
samples was established previously by using the anti-SIRT2 antibody
for immunohistochemical detection of SIRT2 in paraffin sections
(20). Sections were incubated with
the anti-SIRT2 antibody for 18 h at 4°C. PBS replaced the antibody
in parallel negative-control samples. The EnVision kit (Dako,
Glostrup, Denmark) was used according to the manufacturer's
protocol to detect the bound antibody. 3,3′-Diaminobenzidene
tetrahydrochloride (DAB) was the final chromogen. Sections were
counterstained with hematoxylin. More than 200 tumor cells in the
tumor area or astrocytes in normal brain were scored, and the
percentage of cells showing positive staining in nuclei was
designated as the SIRT2 labeling index (SIRT2-LI), as a percentage
(%). SIRT2 expression in cytoplasm was also evaluated and
classified into two groups; negative (−), when no immunoreactivity
was observed in cytoplasm in tumor cells in glioma specimens or
astrocytes in normal brain specimens, positive (+), when
immunoreactivity was observed in the cytoplasm without regard to
percentage of positive cells. SIRT2 cytoplasm positive rate (%) was
calculated as follows: positive case number/total case number in
GB, DA and normal control, respectively.
Statistical analysis
Mann-Whitney's U-test was used to compare nuclear
SIRT2-LI in GB, DA, and normal control samples. The survival curve
was estimated by the Kaplan-Meier method and log-rank test.
P<0.05 was considered significant.
Results
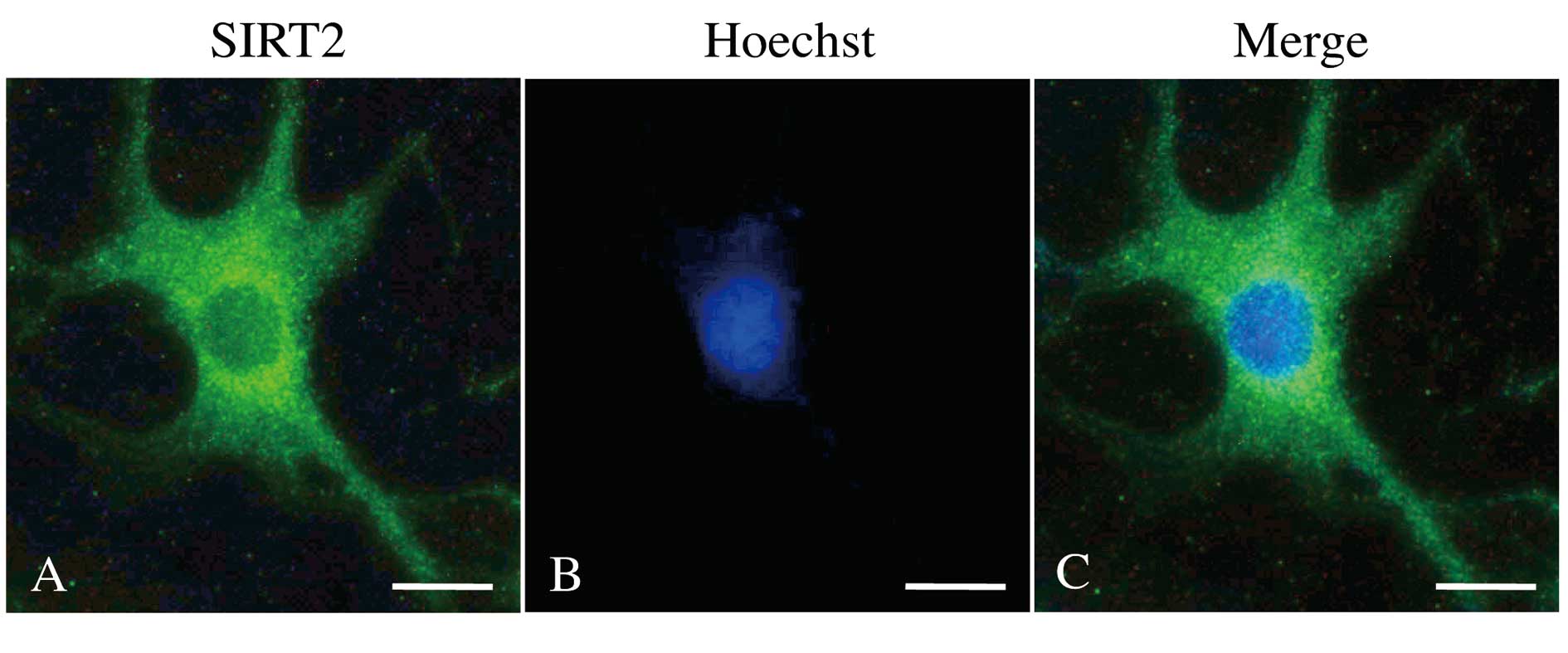
Immunofluorescence and
immunohistochemistry of SIRT2
No antibody staining was seen in cells treated with
PBS rather than anti-SIRT2 antibody (negative controls) in the
immunofluorescent or immunohistochemical studies. As expected
(10), anti-SIRT2 antibody staining
localized to the cytoplasm in astrocytic cells from primary
cultures of normal mouse brain, but no significant reaction was
seen in the nucleus of these cells (Fig. 1). Similarly, anti-SIRT2 antibody
staining was observed in the cytoplasm of some astrocytes in
autopsy samples from the normal individuals, although the
cytoplasmic staining varied from cell to cell (Fig. 2A-C). Nuclear staining was also seen
in a small percentage of astrocytes in autopsy samples (Fig. 2C). The signal intensity and
proportion of positively stained glioma cells varied with
histological grade. A representative stained section from a DA
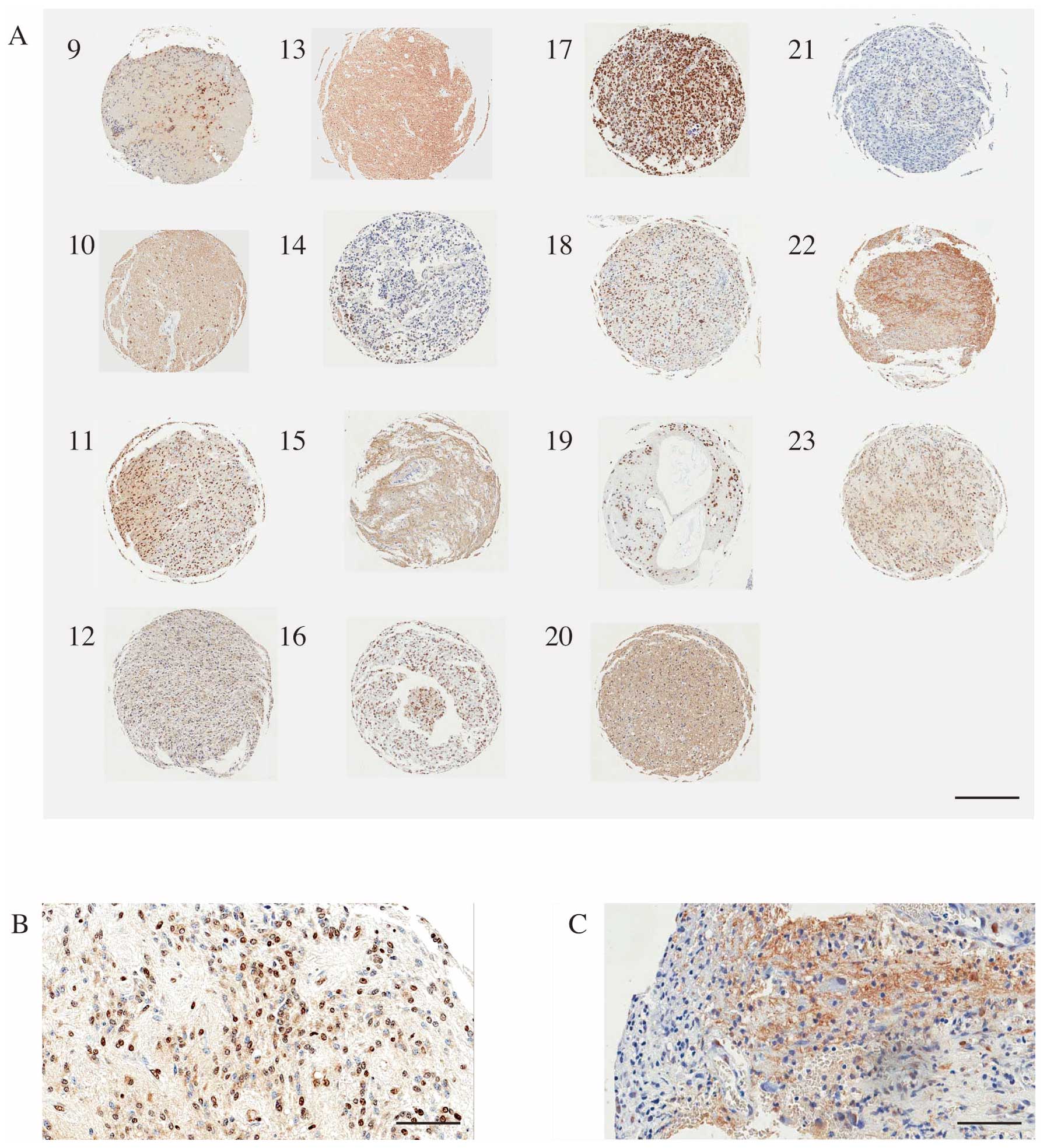
(grade II) is shown in Fig. 2D-F,
and a specimen from a GB (grade IV) is shown in Fig. 2G-I and Fig. 3, respectively. Clear
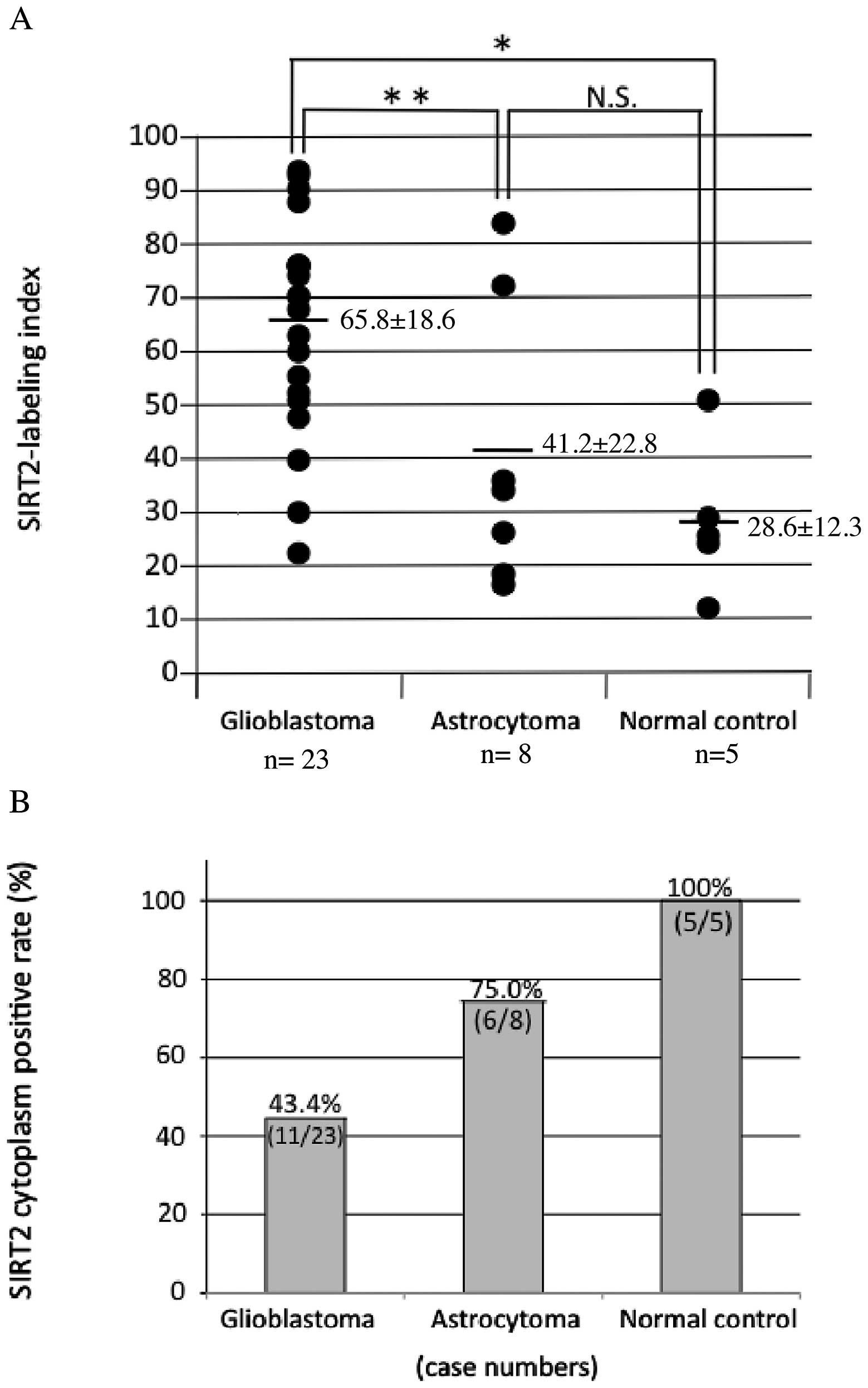
immunoreactivity was also observed in TMA specimens (Fig. 3). The mean SIRT2-LI for all
specimens within each group was 65.8±18.6 for GB (grade IV)
specimens, 41.2±22.8 for DA (grade II) specimens, and 28.6±12.3 for
normal control specimens (mean ± SD, Fig. 4A and Table I). The mean SIRT2-LI of the GB
specimens was significantly higher than that of normal control
specimens (P=0.003, Mann-Whitney's U-test) and significantly higher
than that of DA specimens (P=0.021) (Fig. 4A and Table I). However, there was no significant
difference in mean SIRT2-LI between DA and normal control specimens
(P=0.31). Conversely, SIRT2 cytoplasm positive rate was 43.4% for
GB specimens (11/23), 75.0% DA specimens (6/8), 100% normal control
specimens (5/5) (Fig. 4B and
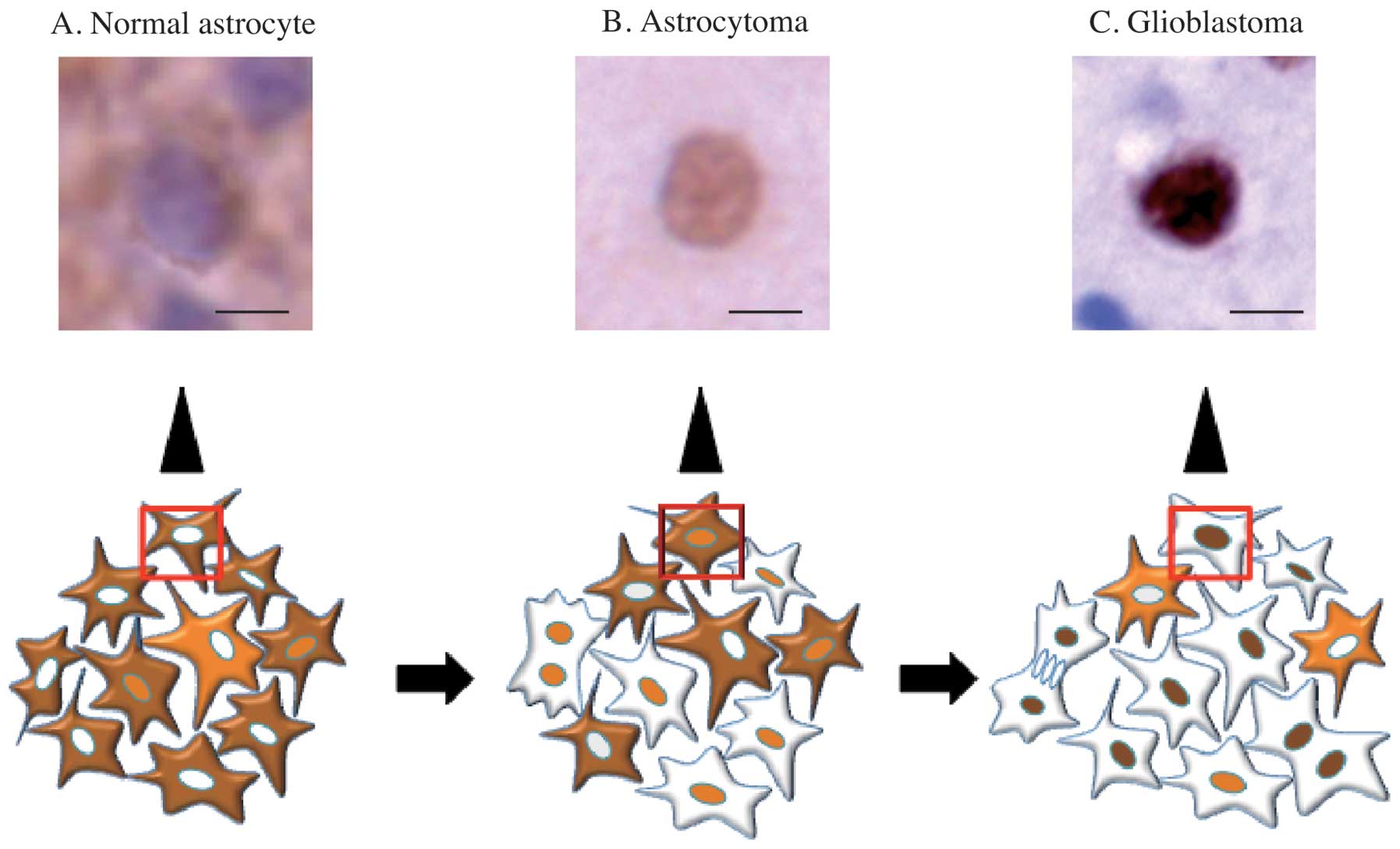
Table I). In this analysis, SIRT2
nuclear localization was observed more frequent in the more
malignant specimens, and, conversely, cytoplasmic localization was
less frequent in the more malignant samples (Fig. 5).
 | Figure 2Immunohistochemistry of normal brain
tissue and glioma using the anti-SIRT2 antibody. (A-C) Normal brain
tissue. Serial sections of normal frontal white matter stained with
hematoxylin and eosin (H&E, A) and anti-SIRT2 antibody (B). At
high-power magnification B, ~28.6% of the astrocytic cells have
positively-stained cytoplasm (C). (D-F) Diffuse astrocytoma (grade
II). Contiguous sections of a diffuse astrocytoma stained with
H&E (D) and anti-SIRT2 antibody (E). At high-power
magnification of panel E, ~37% of the astrocytic tumor cells have
nuclear staining (F). (G-I) Glioblastoma (GB, grade IV). Contiguous
sections of GB stained with H&E (G) and anti-SIRT2 antibody
(H). At high-power magnification H, ~74% glioblastoma cells have
positively-stained nuclei (I). Bars in panel A, B, D, E, G and H =
160 μm; Bar in panel C, F and I = 40 μm. |
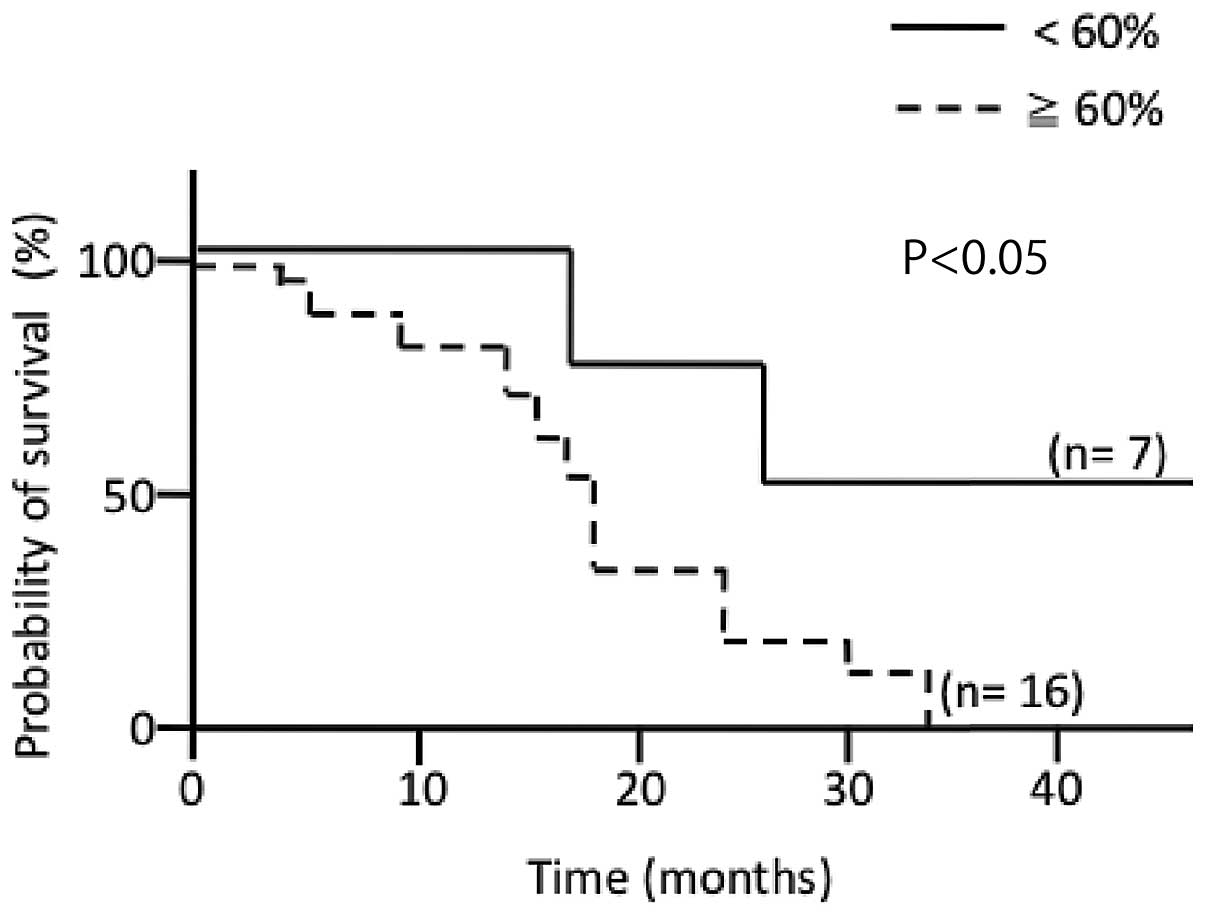
Prognostic significance of SIRT2-LI for
glioblastoma
In general, it seemed that SIRT2-LI value was
negatively related to survival time in patients with glioma
(Table I). To evaluate the
prognostic significance of the SIRT2-LI and this apparent
relationship, the samples from patients with GB were divided in two
groups, low SIRT-LI (<60%, n=7) and high SIRT2-LI (≥60%, n=16),
and survival curve of the patients represented in each group was
calculated using the Kaplan-Meier method and log-rank test. The
patients represented in the low SIRT2-LI group had a significantly
longer survival time than the patients represented in the high
SIRT2-LI group (Fig. 6, P<0.05,
Kaplan-Meier method and log-rank test). These findings indicated
that SIRT2-LI might be a useful marker for the prognosis of GB
patients.
Discussion
Glioma is the most common brain tumor in humans, and
it represents ~25% of primary brain tumors. According to WHO
International Histological Classification of Tumors, glioma is
divided into four grades based on histology (19). High grade glioma, glioblastoma (GB,
grade IV), is the most malignant and has a median survival time of
~1 year, even after surgical resection, radiation therapy, and
chemotherapy. By contrast, patients with low-grade DA (grade II)
have a better prognosis and a median survival time of 10–15 years
(21). To develop new and useful
prognostic markers for GB, it is necessary to understand more
precisely the process of gliomagenesis. The proportion of tumor
cells with abnormal p53 protein expression increases in gliomas as
they undergo malignant progression (7,22). As
in the case of p53, aberrant SIRT2 protein expression may
contribute to malignant progression in glioma.
Reportedly, SIRT2 protein mainly localizes to the
nucleus during the mitotic phase of the cell cycle in normal cells,
and the protein mainly localized to the cytoplasm during all other
phases of the cell cycle (10). In
neoplastic tissues, the percentage of cells showing mitotic phase
increases according to malignancy progresses. Thus, the high
SIRT2-LI and the low SIRT2 cytoplasm positive rate in GB samples
might have reflected a larger percentage of cells in mitosis in
gliomas.
SIRT2 protein mainly localizes to the centrosome in
nucleus of the HeLa cells (10).
Moreover, overexpression of SIRT2 in the nucleus of HeLa cells
causes multinucleation (10). Based
on these observations, we suggest that nuclear accumulation of
SIRT2 in glioma might cause multinucleation, a morphological marker
of malignancy in gliomas. In cytoplasm of SAOS2 cells, SIRT2
protein binds to histone deacetylase 6 (HDAC6) (23), and activation of cytoplasmic HDAC6
is reportedly related to oncogenic tumorigenesis (24). Moreover, a SIRT2 and HDAC6
(SIRT2-HDAC6) complex binds to the spindle apparatus at mitosis in
SAOS2 cells (23). SIRT2, together
with HDAC6, plays a role in regulating microtubule dynamic
instability and the deacetylation of tubulin to control progression
of mitotic phase (23). Therefore,
the high SIRT2-LI and low SIRT2 cytoplasm positive rate that we
observed in some glioma samples might have reflected the phenomena
of tumorigenesis itself, and SIRT2 protein might translocate from
cytoplasm to the nucleus as gliomas become more malignant.
The immunohistochemical determination of
proliferative activity using the monoclonal antibody MIB-1, which
recognizes Ki-67 a nuclear antigen, has been widely demonstrated to
be clinically useful in distinguishing malignancies from benign
tumor cells (25,26). However, MIB-1-LI did not reliability
correlate with patient survival in cases of GB (27). To date, there are few established
diagnostic markers for GB or useful prognostic markers for patients
with GB (28,29), although expression of WT1 (Wilms'
tumor gene) and nestin, and IDH-1/2 gene mutation are reported as
diagnostic or prognostic markers (30,31).
Our study demonstrated that SIRT2-LI was a marker of malignancy for
GB and that SIRT2-LI was significantly correlated with the survival
time of patients with GB, indicating that SIRT2-LI could predict
the prognosis of GB patients.
Acknowledgements
We thank Ms. Atsuko Iwata and Ms. Tomomi Araoka
(Divisions of Neuropathology, Department of Brain and
Neurosciences, Tottori University Faculty of Medicine) for their
excellent technical assistance.
References
|
1
|
Huse JT and Holland EC: Targeting brain
cancer: advances in the molecular pathology of malignant glioma and
medulloblastoma. Nat Rev Cancer. 10:319–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frederick L, Wang XY, Eley G and James CD:
Diversity and frequency of epidermal growth factor receptor
mutations in human glioblastomas. Cancer Res. 60:1383–1387.
2000.PubMed/NCBI
|
|
3
|
Shete S, Hosking FJ, Robertson LB, et al:
Genome-wide association study identifies five susceptibility loci
for glioma. Nat Genet. 41:899–904. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wrensch M, Jenkins RB, Chang JS, et al:
Variants in the CDKN2B and RTEL1 regions are associated with
high-grade glioma susceptibility. Nat Genet. 41:905–908. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Rocco F, Carroll RS, Zhang J and Black
PM: Platelet-derived growth factor and its receptor expression in
human oligodendrogliomas. Neurosurgery. 42:341–346. 1998.PubMed/NCBI
|
|
6
|
Xiao A, Wu H, Pandolfi PP, Louis DN and
Van Dyke T: Astrocyte inactivation of the pRb pathway predisposes
mice to malignant astrocytoma development that is accelerated by
PTEN mutation. Cancer Cell. 1:157–168. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hiratsuka M, Inoue T, Toda T, et al:
Proteomics-based identification of differentially expressed genes
in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys
Res Commun. 309:558–566. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Michan S and Sinclair D: Sirtuins in
mammals: insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
North BJ and Verdin E: Interphase
nucleo-cytoplasmic shuttling and localization of SIRT2 during
mitosis. PLoS One. 2:e7842007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Blander G and Guarente L: The Sir2 family
of protein deacetylases. Annu Rev Biochem. 73:417–435. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pandithage R, Lilischkis R, Harting K, et
al: The regulation of SIRT2 function by cyclin-dependent kinases
affects cell motility. J Cell Biol. 180:915–929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harting K and Knoll B: SIRT2-mediated
protein deacetylation: An emerging key regulator in brain
physiology and pathology. Eur J Cell Biol. 89:262–269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Inoue T, Nakayama Y, Yamada H, et al:
SIRT2 downregulation confers resistance to microtubule inhibitors
by prolonging chronic mitotic arrest. Cell Cycle. 8:1279–1291.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inoue T, Hiratsuka M, Osaki M and Oshimura
M: The molecular biology of mammalian SIRT proteins: SIRT2 in cell
cycle regulation. Cell Cycle. 6:1011–1018. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inoue T, Hiratsuka M, Osaki M, et al:
SIRT2, a tubulin deacetylase, acts to block the entry to chromosome
condensation in response to mitotic stress. Oncogene. 26:945–957.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kato M, Brijlall D, Adler SA, Kato S and
Herz F: Effect of hyperosmolarity on alkaline phosphatase and
stress-response protein 27 of MCF-7 breast cancer cells. Breast
Cancer Res Treat. 23:241–249. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fukuoka J, Fujii T, Shih JH, et al:
Chromatin remodeling factors and BRM/BRG1 expression as prognostic
indicators in non-small cell lung cancer. Clin Cancer Res.
10:4314–4324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Werner HB, Kuhlmann K, Shen S, et al:
Proteolipid protein is required for transport of sirtuin 2 into CNS
myelin. J Neurosci. 27:7717–7730. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Holland EC: Gliomagenesis: genetic
alterations and mouse models. Nat Rev Genet. 2:120–129. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu Y, Guignard F, Zhao D, et al: Early
inactivation of p53 tumor suppressor gene cooperating with NF1 loss
induces malignant astrocytoma. Cancer Cell. 8:119–130. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nahhas F, Dryden SC, Abrams J and Tainsky
MA: Mutations in SIRT2 deacetylase which regulate enzymatic
activity but not its interaction with HDAC6 and tubulin. Mol Cell
Biochem. 303:221–230. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YS, Lim KH, Guo X, et al: The
cytoplasmic deacetylase HDAC6 is required for efficient oncogenic
tumorigenesis. Cancer Res. 68:7561–7569. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schiffer D, Cavalla P, Chio A, Richiardi P
and Giordana MT: Proliferative activity and prognosis of low-grade
astrocytomas. J Neurooncol. 34:31–35. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Di X, Nishizaki T, Harada K, Kajiwara K,
Nakayama H and Ito H: Proliferative potentials of glioma cells and
vascular components determined with monoclonal antibody MIB-1. J
Exp Clin Cancer Res. 16:389–394. 1997.
|
|
27
|
Uematsu M, Ohsawa I, Aokage T, et al:
Prognostic significance of the immunohistochemical index of
survivin in glioma: a comparative study with the MIB-1 index. J
Neurooncol. 72:231–238. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johannessen AL and Torp SH: The clinical
value of Ki-67/MIB-1 labeling index in human astrocytomas. Pathol
Oncol Res. 12:143–147. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moskowitz SI, Jin T and Prayson RA: Role
of MIB1 in predicting survival in patients with glioblastomas. J
Neurooncol. 76:193–200. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rushing EJ, Sandberg GD and
Horkayne-Szakaly I: High-grade astrocytomas show increased Nestin
and Wilms' tumor gene (WT1) protein expression. Int J Surg Pathol.
18:255–259. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ducray F, Idbaih A, Wang XW, Cheneau C,
Labussiere M and Sanson M: Predictive and prognostic factors for
gliomas. Expert Rev Anticancer Ther. 11:781–789. 2011. View Article : Google Scholar : PubMed/NCBI
|