Introduction
Lung cancer is the leading cause of cancer mortality
worldwide. Although smoking is known to be a major risk factor of
lung cancer, 25% of lung cancer patients worldwide are never
smokers (1). In Asian countries,
30–40% of non-small cell lung cancer (NSCLC) patients are never
smokers. NSCLC in never smokers tends to be driven by a single
somatic mutation (1). The
identification of activating mutations of the epidermal growth
factor receptor (EGFR) is one of the most important
discoveries in the field of lung cancer. EGFR mutations
which are present primarily in women, in never smokers and in
Asians are sensitive to EGFR-targeted therapy, such as
gefitinib (2). The EML4/ALK
fusion gene, formed by chromosomal rearrangement, has been
identified in NSCLC (3). The
EML4/ALK fusion genes are present primarily in young
patients and in patients with little or no smoking habits (4). Lung cancer identified with the
ALK fusion gene is present in ~5% of NSCLC patients and is
sensitive to the ALK inhibitors (5,6).
A novel fusion gene resulting from a linkage between
the kinesin family member 5B (KIF5B) gene and the rearranged
during transfection (RET) gene was identified in NSCLC
(7–10), including a Japanese group (9,10).
KIF5B and RET are located at 10p11.22 and at
10q11.21, respectively. Since there is 10.6 Mb between KIF5B
and RET, a long inversion event is necessary to form the
fusion gene (7,8). This fusion gene is more frequent in
never smokers and in Asian patients with adenocarcinoma and exists
exclusively with other mutations, such as EGFR, Kras,
Braf, erbB2 or EML4/ALK fusions (8–10).
In the present study, we independently investigated
the KIF5B/RET fusion gene status in surgically-treated
Japanese lung cancer patients, as well as other types of cancers at
a single institution using a RT-PCR based assay. The findings were
analyzed in reference to the clinicopathological features of
NSCLC.
Patients and methods
Patients
The study group included 371 patients with
adenocarcinoma of the lung (270 were diagnosed as adenocarcinoma
and 101 were squamous cell carcinoma) who had undergone surgery at
the Department of Surgery, Nagoya City University Hospital between
1997 and 2011. The lung tumors were classified according to the 7th
edition of the General Rule of Clinical and Pathological Record of
Lung Cancer in Japan. All tumor samples were immediately frozen and
stored at −80°C until assayed. The study was approved by the ethics
committee of the hospital.
The clinical and pathological characteristics of the
270 adenocarcinoma patients were as follows: 172 cases at stage I,
40 at stage II, 51 at stage III and 7 at stage IV. The mean age was
65.8±8.8 years (range, 38–88). Among the 270 adenocarcinoma
patients, 160 were male and 110 were female. One hundred and
eighteen patients were non-smokers. As regards the mutation
statuses of EGFR (2,8,11,12),
Kras (8,13,14),
Braf (8,15), erbB2 (8,12,16)
and EML4/ALK fusion (8), the
samples from these patients were previously analyzed. Sixteen
samples overlapped with a previous study (8). Pathological findings were confirmed by
an independent pathologist (S.S.). The clinical and pathologic
characteristics of the 101 patients with squamous cell carcinoma
were as follows: 57 cases at stage I, 25 at stage II and 19 at
stage III. The mean age was 67.3±8.8 years (range, 29–85). Among
the patients, 88 were male and 13 were female. Four patients were
non-smokers.
Moreover, 60 patients with breast cancer, 1 patient
with papillary adenocarcinoma of the thyroid and 11 patients with
metastatic lung tumors from colorectal adenocarcinoma were also
investigated. All patients had undergone surgery at the Department
of Surgery, Nagoya City University Hospital. Among the 60 patients
with breast cancer, 29 were diagnosed with scirrhous carcinoma, 16
as solid tubular carcinoma, and 15 as papillotubular carcinoma.
PCR assay for KIF5B/RET
Total RNA was extracted from lung cancer tissues
using an Isogen kit (Nippon Gene, Tokyo, Japan) according to the
manufacturer’s instructions. RNA concentration was determined by a
NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc.,
Rockland, DE, USA). RNA (1 μg) was reverse transcribed using the
first strand cDNA synthesis kit with 0.5 μg oligo(dT)16
(Roche Diagnostics GmbH, Mannheim, Germany) according to the
manufacturer’s instructions. The reaction mixture was incubated at
25°C for 15 min, 42°C for 60 min, 99°C for 5 min and then at 4°C
for 5 min. The cDNA concentration was determined by a NanoDrop
ND-1000 Spectrophotometer. Each cDNA (1 μl) was used for the PCR
analysis. The PCR reactions were performed using an Ex Taq kit
(Takara Bio Inc., Shiga, Japan) in a 50-μl reaction volume. The
primer sequences for screening the KIF5B/RET fusion gene
were as follows: forward primer, 5′-AAATGAGCTCAACAGATGGCGTAA-3′ (at
exon 12 of KIF5B gene) and reverse primer,
5′-AGAACCAAGTTCTTCCGAGGAAT-3′ (at exon 12 of RET gene). The
cycling conditions were as follows: initial denaturation at 98°C
for 10 sec, followed by 40 cycles at 98°C for 10 sec and 68°C for 1
min. The products were purified using a Qiagen PCR purification kit
(Qiagen, Valencia, CA). To confirm the variant, further primer sets
for the KIF5B/RET fusion gene were used: forward primers,
5′-TAAGGAAATGACCAACCAACCACCAG-3′ (for variant 1) or
5′-GTGAAACGTTGCAAGCAAGCAGTTAG-3′ (for variant 3), and reverse
primer, 5′-CCTTGACCACTTTTCC AAATTC-3′. The cycling conditions were
as follows: initial denaturation at 94°C for 10 sec, followed by 40
cycles at 94°C for 30 sec, 62°C for 30 sec (variant 1) or 40 sec
(variant 3) and 72°C for 30 sec. Amplified DNAs were separated on
2% agarose gels, and the bands were visualized using ethidium
bromide and an image was captured under ultraviolet
transillumination. Amplified fragments were then subjected to
direct sequence analysis. To confirm the fusion point, a designed
sequencing primer, 5′-TAGTCCAGCTTCGAGCACAA-3′ was also used.
Statistical analysis
The overall survival of patients with lung
adenocarcinoma was examined using the Kaplan-Meier method, and
differences were examined by the log-rank test. The other
clinicopathological characteristics were examined using the
Student’s t-test and χ2 test as appropriate. The
analyses were performed using the Excel software and differences
were considered significant when the P-value was <0.05.
Results
PCR assays
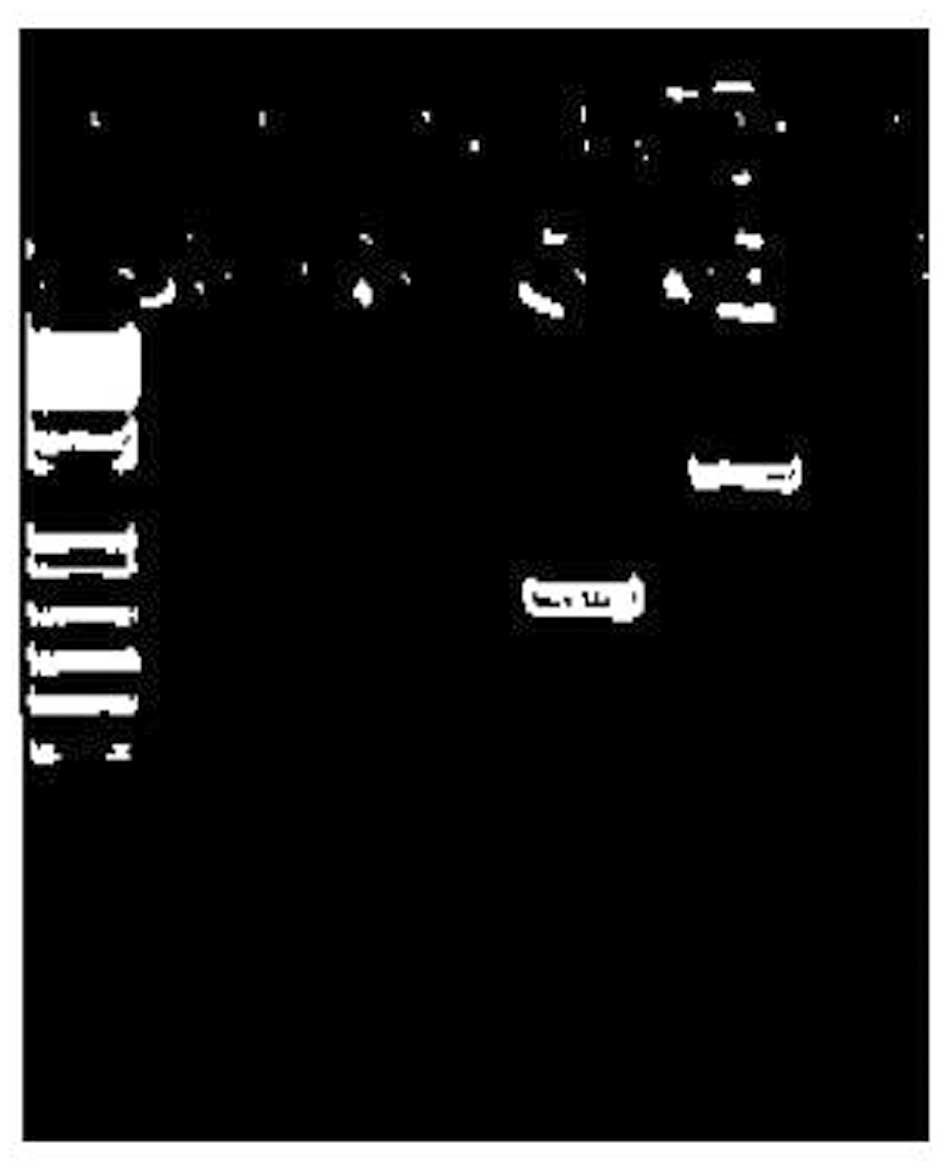
For the screening purpose, we performed a RT-PCR
assay for the KIF5B/RET fusion gene using several primer
sets. Three of 126 adenocarcinoma samples from patients lacking
EGFR, Kras, Braf and erbB2 mutations
had PCR products, suspected as KIF5B/RET fusion genes
(Fig. 1). Matched adjacent normal
lung tissues available in all 3 cases had no bands, suggesting that
the translocations were somatic. As shown in Table I, all 3 patients were female and
non-smokers with mixed subtype adenocarcinomas. Two cases displayed
bands at 681 bp (suggested as variant 1) (8) and 1 case showed a band at 1395 bp
(suggested as variant 3) (8). Lung
adenocarcinomas (n=144) with either EGFR, Kras,
Braf, erbB2 or EML4/ALK gene mutations and 101
patients with squamous cell carcinoma of the lung were also
analyzed but there was no KIF5B/RET fusion. In summary,
3/123 (2.4%) of female and 3/184 (1.6%) of never smoker NSCLC
patients possessed the KIF5B/RET fusion genes.
 | Table IKIF5B-RET fusion found in
patients with wild-type adenocarcinoma. |
Table I
KIF5B-RET fusion found in
patients with wild-type adenocarcinoma.
| Variant |
Differentiation | Gender | Agea | BI | Stage | Acinar | Solid | Pap | Micropap | Lepidic |
|---|
| V1 | Well | Female | 64 | 0 | Ia | - | - | 70 | - | 30 |
| V1 | Moderate | Female | 58 | 0 | IIIa | 40 | - | 50 | 10 | - |
| V3 | Poor | Female | 79 | 0 | Ia | 20 | 40 | 30 | 30 | - |
Of the 270 lung adenocarcinoma patients, 126 lacked
EGFR, Kras, Braf and erbB2 mutations.
To identify the characteristics of KIF5B/RET fusion-positive
patients, these 126 patients were considered as wild-type. Table II summarizes clinicopathological
features of the wild-type adenocarcinoma patients. Wild-type
patients 3/126 (2.4%) had KIF5B/RET fusion genes. The mean
age of the KIF5B/RET fusion-positive patients was 67.0±10.8
years, whereas that of the KIF5B/RET fusion-negative
patients was 65.2±9.3 years. There was a significant association
between KIF5B/RET fusion and gender (P=0.002). There was
also an association between KIF5B/RET fusion and smoking
status (P=0.003). As regards age and stage, there was no
association with KIF5B/RET fusion (Table II). In addition, 60 patients with
breast cancer, 1 patient with papillary adenocarcinoma of the
thyroid, and 11 patients with metastatic lung tumors from
colorectal adenocarcinoma were also analyzed. There was no
KIF5B/RET fusion in all the patients with other types of
cancer.
 | Table IIClinicopathological data of 126
patients with wild-type lung adenocarcinoma. |
Table II
Clinicopathological data of 126
patients with wild-type lung adenocarcinoma.
| | KIF5B/RET
fusion | |
|---|
| |
| |
|---|
| Factors | No. of samples
(%) | (+) n=3 (%) | (−) n=123 (%) | P-value |
|---|
| Mean age
(years) | 126 | 67.0±10.8 | 65.2±9.3 | 0.741a |
| Age |
| ≤65 | 67 (53.2) | 2 (66.7) | 65 (52.8) | 0.911b |
| >65 | 59 (46.8) | 1 (33.3) | 58 (47.2) | |
| Gender |
| Male | 104 (82.5) | 0 (0) | 104 (84.6) | 0.002b |
| Female | 22 (17.5) | 3 (100) | 19 (15.4) | |
| Smoking |
| Never smoker | 23 (18.3) | 3 (100) | 20 (16.3) | 0.003b |
| Smoker | 103 (81.7) | 0 (0) | 103 (83.7) | |
| Stage |
| I | 74 (58.7) | 2 (66.7) | 72 (58.5) | 0.756b |
| II–IV | 52 (41.3) | 1 (33.3) | 51 (41.5) | |
Sequencing analysis for the KIF5B-RET
gene
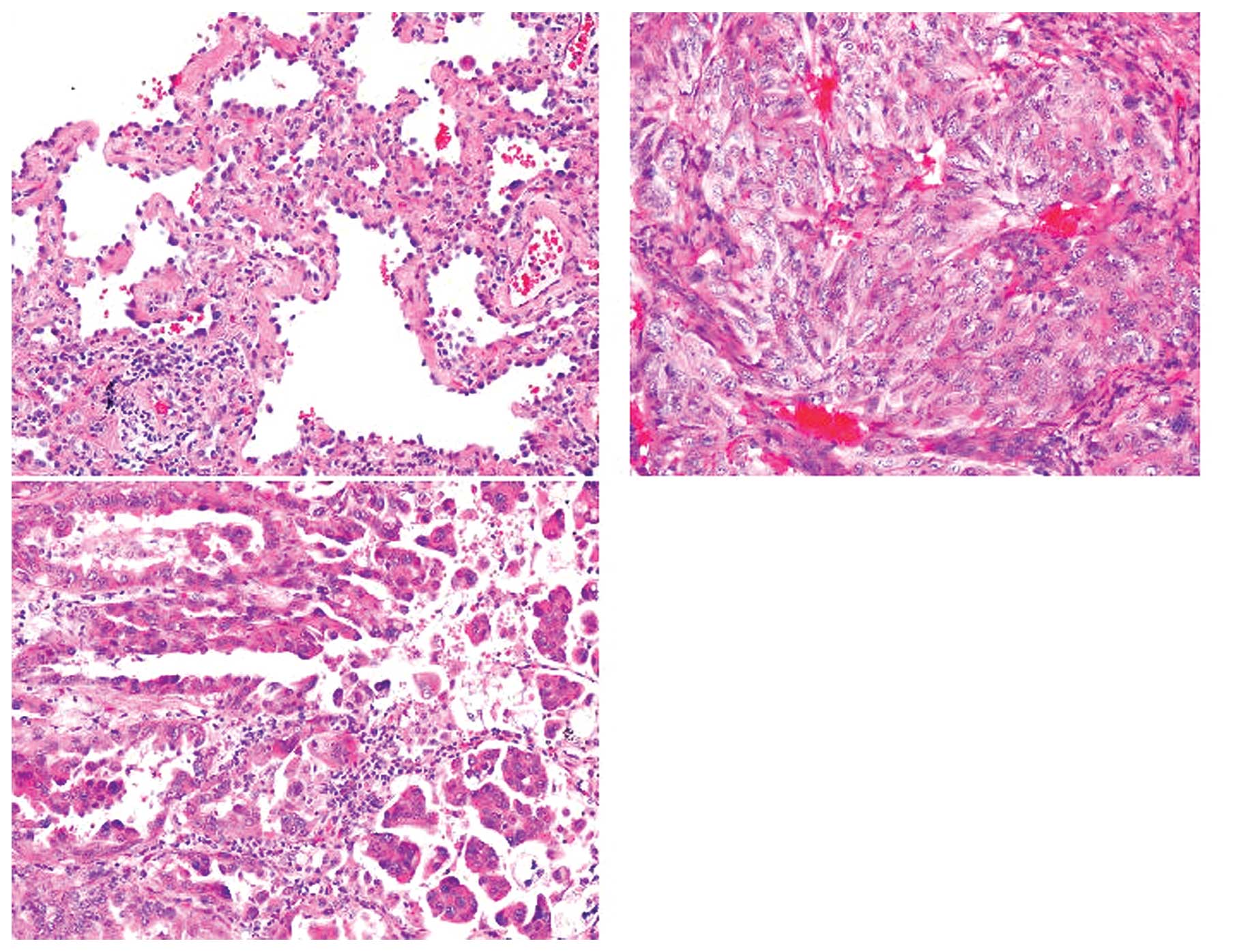
From the direct sequencing of the PCR products (681
bp), 2 cases showed junctions between exon 15 of the KIF5B
gene and exon 12 of the RET gene (Fig. 2), termed variant 1 (V1) (8). These cases were papillary dominant
mixed subtype adenocarcinomas (Fig.
3). However, for the longer product (1395 bp), we were unable
to detect the KIF5B/RET fusion point by direct sequencing.
Thus, we designed a new sequencing primer at exon 17 of the
KIF5B gene. The sequencing results from the PCR product
showed the junction between exon 22 of the KIF5B gene and
exon 12 of the RET gene (Fig.
2), termed variant 3 (V3) (8).
The case was solid dominant mixed subtype adenocarcinoma (Fig. 3). Given the genetic sequence, all
fusions contained both a dimerization unit (coiled-coil domain of
KIF5B) and a tyrosine kinase unit (from RET)
(Fig. 2).
Correlation between KIF5B/RET
translocation and patient outcomes in NSCLC
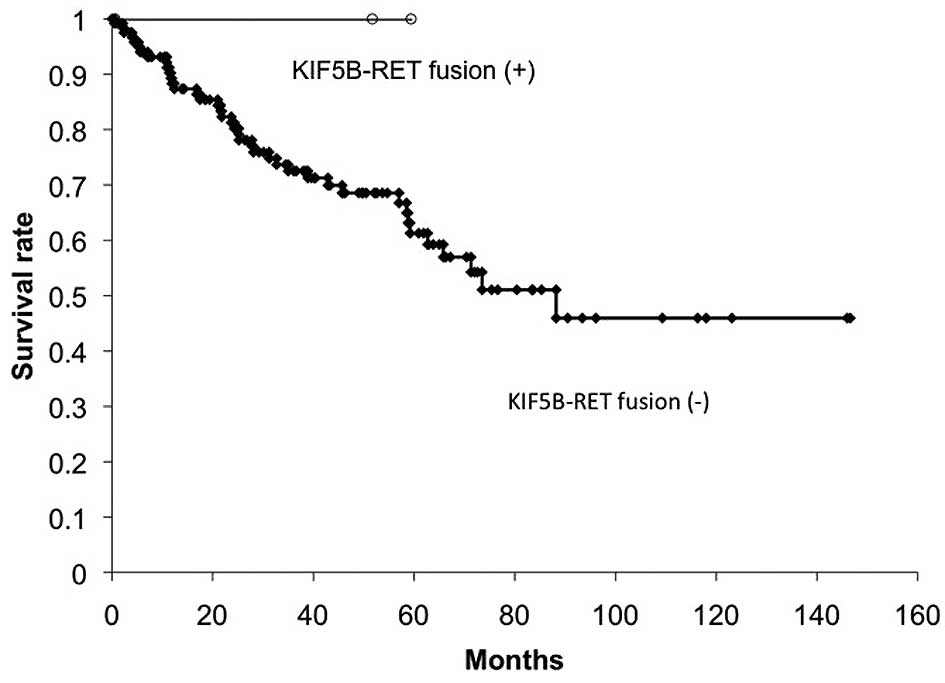
The overall survival of the 126 patients with
wild-type lung adenocarcinoma was analyzed in reference to the
KIF5B/RET fusion gene statuses. The median observation
period of the KIF5B/RET fusion-positive patients was 51.7
months (0.6–60.7 months after the primary operation) and that of
the KIF5B/RET fusion-negative patients was 36.2 months
(0.5–146.6 months after the primary operation). The overall
survival of the KIF5B/RET fusion-positive patients was 100%
at 5 years, whereas that of the KIF5B/RET fusion-negative
patients was 61.3% at 5 years and 46.0% at 10 years (Fig. 4). Although the number of
KIF5B/RET fusion-positive cases was small, there was no
significant difference between the KIF5B/RET fusion statuses
(P=0.352). One KIF5B/RET fusion (variant 1) case showed
recurrence of lung cancer; however, the patient had a good response
to the pemetrexed treatment.
Discussion
In the present study, 371 NSCLC tissues including
270 adenocarcinoma and 101 squamous cell carcinoma were
investigated to identify the clinicopthological characteristics of
KIF5B/RET fusion gene statuses. The KIF5B/RET fusion
genes were detected exclusively with mutations of EGFR,
Kras, Braf, erbB2 and EML4/ALK fusion.
The KIF5B/RET fusion genes were detected in 3 cases of
adenocarcinomas, but not in adjacent normal lung tissues or other
types of cancer, suggesting that the fusions were oncogenic driver
mutations, specific for lung adenocarcinomas.
RET has previously been reported as an
activated oncogene in papillary thyroid carcinoma, where
chromosomal translocations lead to the formation of PTC/RET
fusion genes (16,17). Moreover, activated RET has
been reported in pancreatic cancer (18), prostate cancer (19) and melanoma (20). Heterodimers of MET with RET have
previously been reported. These heterodimers have differential
roles in tumor development and they provide insight into the
function of transphosphorylated RET as partners of MET in
MET-amplified lung cancers (21).
Thyroid cancers and cell lines harboring PTC/RET
translocations are sensitive to the multikinase inhibitor,
sorafenib, or sunitinib that inhibits RET (22–25),
suggesting that the KIF5B/RET gene fusion may identify a
drug-sensitive subset of NSCLCs. The full length KIF5B/RET
gene (variant 1) introduced into Ba/F3 cells has shown
IL-3-independent growth consistent with oncogenic transformation
(8). As the KIF5B/RET fusion
gene overexpresses the chimeric RET receptor tyrosine kinase,
subsequently leading to spontaneous cellular transformation, the
inhibition of RET receptor tyrosine kinase activity may suppress
tumor progression of KIF5B/RET fusion-positive lung cancer.
These KIF5B-RET Ba/F3 cells were sensitive to sunitinib,
sorafenib and vandetanib, multi-targeted kinase inhibitors that
inhibit RET, but not gefitinib, an EGFR kinase inhibitor
(8). Sunitinib, but not gefitinib,
inhibited RET phosphorylation in the KIF5B/RET Ba/F3 cells.
As regards the chromosomal translocations, the KIF5B/ALK
fusion gene has also been detected in lung cancer (26). KIF5B fusions were originally
found in hypereosinophilia (27),
and all these fusions contained a dimerization unit (coiled-coil
domain) which induces homodimerization (26,27).
In our study, all 3 KIF5B/RET cases were
female, never smokers, with mixed subtype adenocarcinomas, similar
to EGFR (2,8,12) or
EML4/ALK (3,4,8) or
erbB2 (12) mutations.
Takahashi et al reported that EML4/ALK fusion was
present in 1.6% of patients with NSCLC and occurred more frequently
in females and non-smokers (28).
Inamura et al reported that EML4/ALK fusion was
present in 3.4% of patients with NSCLC (29) and correlated with acinar-predominant
(P<0.0001) and non- or light smokers (P=0.04) (30). Yoshida et al reported that
solid or acinar growth patterns were more common in ALK-rearranged
lung carcinomas (31). In the
present study, KIF5B/RET fusions were present in patients
with similar clinicopathological backgrounds compared to ALK
fusions in Japanese NSCLC patients. In addition, a papillary
pattern was frequently similar to the PTC/RET translocation
in thyroid cancers.
As regards prognosis, the overall survival in the
KIF5B/RET fusion-positive patients was 100% at 5 years,
whereas that in the fusion-negative patients was 61.3% at 5 years
and 46.0% at 10 years. Although the 3 cases are insufficient to
discuss, the patients with KIF5B/RET fusion-positive lung
cancer may have better prognosis due to the sensitivity to
pemetrexed treatment for recurrence 39 months after surgery.
Camidge et al reported that, in comparison with
EML4/ALK-negative patients, EML4/ALK-positive
patients had a significantly longer progression-free survival on
pemetrexed (32). However, there
are conflicting published data regarding the natural history and
clinical outcomes of EML4/ALK-positive NSCLC (33,34).
In our case, a less advanced stage, gender and smoking status may
influence the survival data, and additional samples and data are
required to conclude the correlation between the fusion status and
prognosis. To identify the clinicopathological characteristics of
KIF5B/RET fusion-positive lung cancer, further studies are
warranted.
Acknowledgements
The authors would like to thank Mrs. Miki Mochizuki
and Yuka Toda for their excellent technical assistance. This study
was supported by Grants-in-Aid for Scientific Research, Japan
Society for the Promotion of Science (JSPS) (nos. 23659674,
24592097 and 21591820).
References
|
1
|
Lee YJ, Kim JH, Kim SK, et al: Lung cancer
in never smokers: change of a mindset in the molecular era. Lung
Cancer. 72:9–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paez JG, Jänne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Soda M, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wong DW, Leung EL, So KK, et al: The
EML4-ALK fusion gene is involved in various histologic types of
lung cancers from non smokers with wild-type EGFR and KRAS. Cancer.
115:1723–1733. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shaw AT, Yeap BY, Solomon BJ, et al:
Effect of crizotinib on overall survival in patients with advanced
non-small-cell lung cancer harbouring ALK gene rearrangement: a
retrospective analysis. Lancet Oncol. 12:1004–1012. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ju YS, Lee WC, Shin JY, et al: Fusion of
KIF5B and RET transforming gene in lung adenocarcinoma revealed
from whole-genome and transcriptome sequencing. Genome Res.
22:436–445. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lipson D, Capelletti M, Yelensky R, et al:
Identification of new ALK and RET gene fusions from colorectal and
lung cancer biopsies. Nat Med. 18:382–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kohno T, Ichikawa H, Totoki Y, et al:
KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 18:375–377.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeuchi K, Soda M, Togashi Y, et al: RET,
ROS1 and ALK fusions in lung cancer. Nat Med. 18:378–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sasaki H, Endo H, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sasaki H, Shimizu S, Endo K, et al: EGFR
and erbB2 mutation status in Japanese lung cancer patients. Int J
Cancer. 118:180–184. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sasaki H, Hikosaka Y, Kawano O, Moriyama
S, Yano M and Fujii Y: Evaluation of Kras mutation and copy number
gain in non-small cell lung cancer. J Thorac Oncol. 6:15–20. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sasaki H, Okuda K, Kawano O, et al: Nras
and Kras mutation in Japanese lung cancer patients: genotyping
analysis using LightCycler. Oncol Rep. 18:623–628. 2007.PubMed/NCBI
|
|
15
|
Sasaki H, Kawano O, Endo K, et al:
Uncommon V599E Braf mutations in Japanese patients with lung
cancer. J Surg Res. 133:203–206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grieco M, Santoro M, Berlingieri MT, et
al: PTC is a novel rearranged form of the ret proto-oncogene and is
frequently detected in vivo in human thyroid papillary carcinomas.
Cell. 60:557–563. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alberti L, Carniti C, Miranda C, Roccato E
and Pierotti MA: RET and NTRK1 proto-oncogenes in human diseases. J
Cell Physiol. 195:168–186. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng Q, Cheng Y, Zhu Q, et al: The
relationship between overexpression of glial cell-derived
neurotrophic factor and its RET receptor with progression and
prognosis of human pancreatic cancer. J Int Med Res. 36:656–664.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dawson DM, Lawrence EG, MacLennan GT, et
al: Altered expression of RET proto-oncogene product in prostatic
intraepithelial neoplasia and prostate cancer. J Natl Cancer Inst.
90:519–523. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ohshima Y, Yajima I, Takeda K, Kumasaka M,
Matsumoto M and Kato M: c-RET molecule in malignant melanoma from
oncogenic RET-carrying transgenic mice and human cell lines. PLoS
One. 5:e102792010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tanizaki J, Okamoto I, Sakai K and
Nakagawa K: Differential roles of trans-phosphorylated EGFR, HER2,
HER3, and RET as heterodimerisation partners of MET in lung cancer
with MET amplification. Br J Cancer. 105:807–813. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Henderson YC, Ahn SH, Kang Y and Clayman
GL: Sorafenib potently inhibits papillary thyroid carcinomas
harboring RET/PTC1 rearrangement. Clin Cancer Res. 14:4908–4914.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carlomagno F, Anaganti S, Guida T, et al:
BAY 43–9006 inhibition of oncogenic RET mutants. J Natl Cancer
Inst. 98:326–334. 2006.
|
|
24
|
Kim DW, Jo YS, Jung HS, et al: An orally
administered multitarget tyrosine kinase inhibitor, SU11248, is a
novel potent inhibitor of thyroid oncogenic RET/papillary thyroid
cancer kinases. J Clin Endocrinol Metab. 91:4070–4076. 2006.
View Article : Google Scholar
|
|
25
|
Dawson S, Conus NM, Toner GC, Raleigh JM,
Hicks RJ, McArthur G and Rischin D: Sustained clinical responses to
tyrosine kinase inhibitor sunitinib in thyroid carcinoma.
Anticancer Drugs. 19:547–552. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takeuchi K, Choi YL, Togashi Y, et al:
KIF5B-ALK, a novel fusion oncokinase identified by an
immunohistochemistry-based diagnostic system for ALK-positive lung
cancer. Clin Cancer Res. 15:3143–3149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Score J, Curtis C, Waghorn K, et al:
Identification of a novel imatinib responsive KIF5B-PDGFRA fusion
gene following screening for PDGFRA overexpression in patients with
hypereosinophilia. Leukemia. 20:827–832. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takahashi T, Sonobe M, Kobayashi M, et al:
Clinicopathologic features of non-small-cell lung cancer with
EML4-ALK fusion gene. Ann Surg Oncol. 17:889–897. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Inamura K, Takeuchi K, Togashi Y, et al:
EML4-ALK fusion is linked to histological characteristics in a
subset of lung cancers. J Thorac Oncol. 3:13–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Inamura K, Takeuchi K, Togashi Y, et al:
EML4-ALK lung cancers are characterized by rare other mutations, a
TTF-1 cell lineage, an acinar histology, and young onset. Mod
Pathol. 22:508–515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoshida A, Tsuta K, Nakamura H, et al:
Comprehensive histologic analysis of ALK-rearranged lung
carcinomas. Am J Surg Pathol. 35:1226–1234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Camidge DR, Kono SA, Lu X, et al:
Anaplastic lymphoma kinase gene rearrangements in non-small cell
lung cancer are associated with prolonged progression-free survival
on pemetrexed. J Thorac Oncol. 6:774–780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang P, Kulig K, Boland JM, et al: Worse
disease-free survival in never-smokers with ALK+ lung
adenocarcinoma. J Thorac Oncol. 7:90–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu SG, Kuo YW, Chang YL, et al: EML4-ALK
translocation predicts better outcome in lung adenocarcinoma
patients with wild-type EGFR. J Thorac Oncol. 7:98–104. 2012.
View Article : Google Scholar : PubMed/NCBI
|