Introduction
Intrahepatic cholangiocarcinoma (ICC) is the second
most common type of primary liver cancer. It arises from the second
or more distal branches of intrahepatic bile ducts. Although ICC
accounts for only 5–15% of primary liver malignancies, its
incidence and cancer-related mortality continues to rise. The
causes for this increasing incidence remain unknown and may be
related to predisposing genetic and environmental factors (1). Surgical resection has been shown to
improve long-term survival of patients with ICC, but the
resectibility rate for this cancer is low and the outcome even
after resection still remains disappointing. The reasons are likely
that early diagnosis of ICC is difficult and currently no effective
treatment in the advanced stages of the disease is available
(2,3). Clinicopathologic factors such as tumor
size, differentiation, gross type, cancer-free margin, vascular
invasion, and lymph node metastasis have been reported to be
significant prognositic factors (3,4).
Despite the recent advances in disease detection and treatment, the
predicted outcome for ICC remains dismal, partly because of limited
knowledge of the molecular processes involved in pathogenesis and
failure to detect the disease in the early stages. Therefore, it is
critical to identify biomarkers that enable early diagnosis,
provide insight into pathogenesis of the disease, and develop more
effective treatment options.
Fatty acid-binding proteins (FABPs) are members of
the intracellular lipid-binding protein family and are abundantly
expressed 14–15-kDa proteins that reversibly bind intracellular
hydrophobic ligands (5,6). These proteins are thought to be
involved in the uptake and intracellular transport of fatty acids.
In addition, they play many roles in gene regulation, cell
signaling, cell growth and differentiation (7). FABP5, also known as
psoriasis-associated fatty acid-binding protein (PA-FABP),
epidermal or cutaneous fatty acid-binding protein (E- or C-FABP)
was originally isolated from psoriatic skin samples (8). It was later found to be expressed in
many other tissues, including mammary gland (9). Several recent studies have reported
that FABP5 was very strongly expressed in pancreas, bladder and
prostate cancers (10–12), indicating that FABP5 may also play a
role in malignant progression in these tissues. Although the
precise role of FABP5 in these tissues is not clear, it is likely
to be involved in binding and transporting intracellular fatty
acids, some of which are signaling molecules reported to be
involved in the regulation of gene expression (13,14).
In the current study, we analyzed differential
expression profiles of human ICC and normal tissue specimens,
examined the expression of FABP5 protein in clinical ICC specimens,
and assessed the relationship between FABP5 expression pattern and
known clinicopathological variables. In addition, we investigated
the effect of stable overexpression and knockdown of FABP5 on
cancer cell proliferation and invasion to determine whether FABP5
contributes to carcinogenesis of cholangiocarcinoma. We report that
FABP5 is significantly overexpressed in ICC, and associated with
tumor size, lymph node metastasis, angio-invasion and TNM (tumor,
node and metastasis) staging. Moreover, FABP5 expression promotes
cancer cell proliferation and invasion.
Materials and methods
Patients and tissue samples
Pairs of cancerous and normal tissues diagnosed by a
pathologist were collected from sixteen patients with MF type ICC
who underwent radical or palliative surgical procedures at
Gyeongsang National University Hospital (GNUH) during the period
2006–2009. The clinical data collected included age, gender,
differentiation, operation type and TNM stage. None of the patients
received preoperative chemotherapy or radiotherapy. Informed
patient consent was obtained for all of the cases. The use of the
tissue samples for this project was approved by the GNUH
Institutional Review Board. Cancerous lesions and the corresponding
normal bile duct tissues were excised during surgery and stored at
−70°C for proteomic analysis and further study.
Protein extraction and two-dimensional
gel electrophoresis (2-DE)
Tissue samples (150–200 mg) were homogenized on ice
in 1 ml homogenization buffer (50 mM Tris-HCl, pH 7.2) containing a
protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA).
The homogenate was centrifuged, followed by TCA precipitation. The
protein precipitate was resuspended in lysis buffer (8 M urea, 4%
CHAPS, 40 mM Tris-base, 100 mM DTT and 2% (w/v) ampholyte) and the
protein concentration was measured using Bradford method with
Protein assay kit (Bio-Rad Laboratories, Hercules, CA). Equal
amounts of protein extracts of each tissue samples (50 μg for
silver-stained gels or up to 0.5 mg for Coomassie-stained gels)
were subjected to isoelectric focusing (pH 4.0 to 7.0) and
sequentially a gradient SDS-polyacrylamide gel (7.5–17.5%)
electrophoresis as previously described (15).
Protein visualization and image
analysis
Separated two-dimensional gels were fixed and then
pretreated in a solution of 0.02% sodium thiosulfate pentahydrate.
After washing twice with deionized water, the gels were impregnated
in a solution of 0.2% silver nitrate and 0.075% (v/v) formaldehyde,
transferred to a developing solution [0.06% (v/v) formaldehyde, 2%
sodium bicarbonate, and 0.0004% sodium sulfoxide]. For Coomassie
staining, the gels were fixed and rinsed twice in 2% phosphoric
acid. The gels were placed in equilibration (18% ethanol, 2%
phosphoric acid and 15% ammonium sulfate) followed by overnight
staining with Coomassie Brilliant Blue G-250 solution. The gels
were scanned using a high resolution scanner (GS-800 Calibrated
Imaging Densitometer, Bio-Rad Laboratories) and analyzed using
PDQuest™ version 8.0. software (Bio-Rad Laboratories). The
intensity of each spot was quantified by calculating the spot
volume after normalization of the gel image.
In-gel digestion
Selected protein spots were excised from 2-DE gels
that had been stained with Coomassie blue G-250. Gel pieces first
were destained with water/acetonitrile (1:1 v/v) then dehydrated in
acetonitrile followed by rehydration in 10 mM DTT/0.1 M ammonium
bicarbonate for 45 min at 56°C. After cooling the tubes to room
temperature and removing the liquid, 55 mM iodoacetamide in 0.1 M
ammonium bicarbonate was added and the tubes were incubated for 30
min at room temperature in the dark. The iodoacetamide solution was
removed and the gel particles were washed with 0.1 M ammonium
bicarbonate and acetonitrile and then dried in a vacuum centrifuge.
The dried gel pieces were incubated in freshly prepared digestion
buffer containing 50 mM ammonium bicarbonate, 5 mM
CaCl2, and 12.5 ng/μl trypsin overnight at 37°C. The
supernatant was then collected and the digested peptides extracted
three times in 5% formic acid/acetonitrile (1:1 v/v).
Matrix assisted laser
desorption/ionization-time of flight-mass spectrometry
(MALDI-TOF-MS) and database search
The digested peptides were redissolved in a solution
containing water, acetonitrile, and trifluoroacetic acid (93:5:2 by
volume) and sonicated for 5 min in a bath sonicator. Target
preparation was carried out by the ‘solution phase nitrocellulose
method’ described by Landry et al (16). Briefly, a saturated solution of
α-cyano-4-hydroxycinnamic acid (CHCA; ~40 mg/ml) and nitrocellulose
solution (20 mg/ml) were prepared separately in acetone and mixed
with 2-propanol at a ratio of 2:1:1, respectively. Internal
calibrants (50–200 fmole each), des-Arg-bradykinin (monoisotopic
mass = 904.4681), and neurotensin (monoisotopic mass = 1672.9715)
were added to the mixture. The matrix solution was mixed with the
sample at a 1:1 ratio, and 1 μl of mixed sample was spotted onto a
MALDI plate. The dried spots were analyzed with a Voyager-DE STR
MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA).
The identity of the proteins was assigned by comparing the observed
mass fingerprint with the predicted mass fingerprint of proteins in
the SWISS-Prot and NCBI databases using the MS-Fit search program
(17).
Western blot analysis
Lysates (40 μg) from ICC and its corresponding
normal bile duct tissues from 16 patients were separated by on a
15% SDS-polyacrylamide gel. The separated proteins were transferred
to a PVDF membrane by electroblotting. The membrane was blocked for
1 h in TBST solution (Tris-buffed saline, 20 mM Tris-HCl, pH 7.6,
137 mM NaCl, and 0.1% Tween-20) containing 5% skim milk. After
blocking, the membranes were washed in TBST twice for 10 min each
and incubated with anti-FABP5 rabbit polyclonal antibody for 1 h at
room temperature. After additional washes with TBST, the membranes
were incubated with secondary anti-rabbit IgG (1:10,000) for 1 h.
After final washes, proteins were visualized by the enhanced
chemiluminescence method (Pierce, Rockford, IL). β-actin was used
as a loading control in the stripped blot.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue blocks from
43 patients with MF-ICC were collected, sectioned at 4 μm and
mounted on slides. The slides were deparaffinized and rehydrated in
a graded alcohol series, and pretreated with 10 mM sodium citrate.
Immunostaining was performed with a rabbit polyclonal FABP5
antibody (1:500) using an LSAB kit (Dako, Glostrup, Denmark).
Antigen retrieval was facilitated by microwaving the tissue for 15
min, and the staining procedure was carried out according to the
standard avidin-biotin-peroxidase complex system previously
described (18).
Immunohistochemical staining was interpretated and categorized by
an arbitrary semi-quantitative scale as 0 (negative staining), +1
(0~20% positive), +2 (20~40% positive), or +3 (>40% positive) by
pathologists. The staining intensity was also simply categorized
into ‘weak’ (0 to +1) and ‘strong’ (+2 to +3) groups. Tumor size,
degree of differentiation, lymph node metastasis, vascular
invasion, perineural invasion and multiplicity were included in the
clinicopathological information (Table III).
 | Table IIICorrelation between FABP5 expression
and clinicopathological characteristics. |
Table III
Correlation between FABP5 expression
and clinicopathological characteristics.
| Level of expression
FABP5 |
|---|
|
|
|---|
| Pathological
variables | Weaka (n=18) (%) | Strongb (n=25) (%) | P-value |
|---|
| Size (cm) |
| <5 | 12 (57.1) | 9 (42.9) | 0.047 |
| >5 | 6 (27.3) | 16 (72.7) | |
| LN metastasis |
| Negative | 18 (51.4) | 17 (48.6) | 0.013 |
| Positive | 0 (0) | 8 (100) | |
| Angioinvasion |
| Negative | 18 (48.6) | 19 (51.4) | 0.032 |
| Positive | 0 (0) | 6 (100) | |
| Perineural
invasion |
| Negative | 16 (45.7) | 19 (54.3) | 0.434 |
| Positive | 2 (25.0) | 6 (75.0) | |
| Multifocality |
| Single | 16 (45.7) | 19 (54.3) | 0.434 |
| Multiple | 2 (25.0) | 6 (75.0) | |
|
Differentiation |
| Well to
moderate | 17 (50.0) | 17 (50.0) | 0.339 |
| Poor | 1 (11.1) | 8 (88.9) | |
| Stage |
| I | 15 (62.5) | 9 (37.5) | 0.007 |
| II | 2 (22.2) | 7 (77.8) | |
| III and IV | 1 (10.0) | 9 (90.0) | |
Cell culture and transfection
HuCCT1 cell line was purchased from the Health
Science Research Resources Bank (Osaka, Japan). HuCCT1 cells were
cultured with RPMI-1640, 10% fetal bovine serum (FBS) and 1×
penicillin/streptomycin at 37°C in 5% CO2 incubator.
Human hepatoma cell line, Hep3B were maintained in DMEM
supplemented with 10% FBS and 1× antibiotics in a humidified
atmosphere containing 5% CO2 and 95% air at 37°C. Human
FABP5 cDNA was cloned into pcDNA3.1 using standard RT-PCR
procedures. The empty vector was used as a control for possible
effects caused by the transfected plasmid. The constructs were
transfected into Hep3B cells using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA) according to the manufacturer’s protocol and stable
FABP5-expressing cell lines were selected by G418 (2 mg/ml).
Short-hairpin RNA-mediated FABP5-gene
silencing
The MISSION shRNA bacterial glycerol stock
containing 5 anti-FABP5 shRNA sequences and the SHC002 shRNA
Control were purchased from Sigma-Aldrich. The shRNA constructs
were transfected into HuCCT1 cells using a reagent (Lipofectamine
2000; Invitrogen) according to the manufacturer’s instructions.
Stable shRNA-expressing cell lines were established and selected by
puromycin (2 μg/ml). Results from shRNA TRCN0000059720 are
presented in this report.
Proliferation assay and invasion
assay
For proliferation assay, cells were placed in a
6-well plate at a concentration of 1×105 cells/well.
After incubation for 1–3 days, the viable cells were counted with a
hemocytometer after trypan blue staining. Invasion assays were
performed on 24-well transwells (Costar, Cambridge, MA, USA) with
polycarbonate filters (8 μm pore size). The transwells for invation
assays were coated with a uniform layer of BD Matrigel™ Basement
Membrane Matrix (BD Biosciences, Bedford, MA, USA). The stable cell
lines were resuspended in RPMI-1640 or DMEM containing 10% FBS and
seeded into the upper wells (1×105 cells/well) and
incubated at 37°C for 24 h. Invaded cells were fixed in 4% PFA,
stained with DAPI, and counted under the fluorescent microscope at
×100 magnification for 5 random fields.
Statistical analysis
Statistical analysis was performed by the
Chi-squared test using the SPSS program (version 11.0). All
P-values were based on a two-tailed statistical analysis, and
P-values of <0.05 were considered statistically significant.
Results
Proteomic analysis of ICC
We analyzed the proteomic profiles of the ICC tissue
and corresponding normal bile duct tissues from 16 patients to
establish a tumor specific protein expression profile. The
upregulated or downregulated protein expressions between the ICC
tissue samples and their healthy counterparts were evaluated using
PDQuest™ software. By making comparison with PDQuest software
quantification, the 151 protein spots that showed a statistically
meaningful expression difference (P<0.05) were selected. The
spots that showed more than a 3-fold increase or decrease of
intensity were defined as being the up- or downregulated proteins,
respectively. According to this definition, among the 67 candidate
protein spots, 50 proteins were upregulated and 17 proteins were
downregulated in ICC tissue compared to the non-cancerous bile duct
tissue. The protein spots were identified using MALDI-TOF-MS and
the MS-FIT search program. The identities of the proteins were
confirmed by comprehensively comparing the corresponding
experimental value of isoelectric point (pI), molecular weight
(MW), the number of matched peptides, and the sequence coverage to
known proteins in the SWISS-PROT and NCBI databases. The lists of
proteins that were found to be up- or downregulated in ICC tissue
are shown in Tables I and II, respectively.
 | Table IThe upregulated proteins identified in
ICC. |
Table I
The upregulated proteins identified in
ICC.
| SSP no. | Accession no. | Protein name | MW (kDa)/pI | Matched peptides
(%) | Sequence coverage
(%) | P-value |
|---|
| 0013 | P60660 | Myosin light
polypeptide 6 | 16.9/4.6 | 26 | 30.5 | 0.0126 |
| 0111 | P28065 | Proteasome subunit β
type-9 precursor | 23.3/4.9 | 40 | 23.7 | 0.0190 |
| 0301 | P07951 | Tropomyosin β
chain | 32.9/4.7 | 65 | 35.9 | 0.0031 |
| 0302 | Q9BSJ2 | Proliferating cell
nuclear antigen | 28.8/4.6 | 44 | 31.0 | 0.0059 |
| 0305 | P09493 | Tropomyosin α-1
chain | 32.7/4.7 | 33 | 21.1 | 0.0027 |
| 0306 | P09493 | Tropomyosin α-1
chain | 32.7/4.7 | 56 | 25.0 | 0.0191 |
| 0312 | P61978 | Heterogeneous
nuclear ribonucleoprotein K | 50.9/5.4 | 33 | 18.1 | 0.0063 |
| 0603 | P49585 | Choline-phosphate
cytidylyltransferase A | 41.7/6.8 | 28 | 13.4 | 0.0092 |
| 0606 | Q12765 | Secernin-1 | 46.4/4.7 | 22 | 13.0 | 0.0073 |
| 1006 | gi|37014297 | Immunoglobulin E
heavy chain variable region | 14.6/4.86 | 8 | 17.0 |
6.2×10−5 |
| 1008 | Q15814 | Tubulin-specific
chaperone C | 39.2/5.6 | 33 | 13.9 | 0.0092 |
| 1018 | CPLX4_Human | Complexin-4 | 18.4/4.5 | 8 | 17.0 | 0.0336 |
| 1101 | P04632 | Calpain small
subunit 1 | 28.3/5.0 | 46 | 29.5 | 0.0093 |
| 1105 | P52565 | Rho
GDP-dissociation inhibitor 1 | 23.2/5.0 | 22 | 21.1 | 0.0114 |
| 1108 | P63104 | 14-3-3 protein
zeta/δ | 27.7/4.7 | 47 | 48.6 | 0.0492 |
| 1210 | Q99426 | Tubulin folding
cofactor B | 27.3/5.1 | 25 | 23.0 | 0.0036 |
| 1211 | O00299 | Chloride
intracellular channel protein 1 | 26.9/5.1 | 35 | 23.7 | 0.0008 |
| 1214 | gi|119627265 | hCG1643319 | 28.1/5.6 | 14 | 18.0 | 0.0354 |
| 1301 | P35237 | Serpin B6,
Placental thrombin inhibitor | 42.6/5.2 | 45 | 30.1 | 0.0202 |
| 1415 | Q14320 | Protein FAM50A | 40.2/6.4 | 30 | 13.3 | 0.0108 |
| 1421 | P60709 | Actin, cytoplasmic
1 | 41.7/5.3 | 30 | 23.2 | 0.0243 |
| 1514 | P50452 | Serpin B8 | 42.8/5.4 | 29 | 15.0 | 0.0414 |
| 1607 | P01009 | α-1-antitrypsin
precursor | 46.7/5.4 | 38 | 24.2 | 0.0227 |
| 2005 | O75368 | SH3 domain-binding
glutamic acid-rich-like protein | 12.8/5.2 | 27 | 28.9 | 0.0015 |
| 2010 | Q8WU39 | Proapoptotic
caspase adapter protein | 20.7/5.4 | 37 | 34.9 | 0.0002 |
| 2208 | P19623 | Spermidine
synthase | 33.8/5.3 | 23 | 16.2 | 0.0459 |
| 2302 | Q04323 | SAPK substrate
protein 1 | 33.3/5.2 | 33 | 21.5 | 0.0068 |
| 2305 | O76003 | Glutaredoxin-3 | 37.4/5.3 | 31 | 23.9 | 0.0153 |
| 3001 | Q14019 | Coactosin-like
protein | 15.9/5.5 | 53 | 50.7 | 0.0004 |
| 3113 | Q9BY32 | Inosine
triphosphate pyrophosphatase | 21.4/5.5 | 36 | 29.9 | 0.0013 |
| 3301 | P60709 | Actin, cytoplasmic
1 | 41.7/5.3 | 43 | 22.7 | 0.0033 |
| 3303 | P60709 | Actin, cytoplasmic
1 | 41.7/5.3 | 35 | 17.1 | 0.0129 |
| 3704 | P10809 | Hsp60 | 61.1/5.7 | 52 | 21.3 | 0.0133 |
| 4008 | P26447 | Metastasin | 11.7/5.9 | 33 | 36.6 | 0.0036 |
| 4408 | P36952 | Maspin | 42.1/5.7 | 46 | 24.8 | 0.0115 |
| 4607 | P48637 | Glutathione
synthetase | 52.4/5.7 | 47 | 16.9 | 0.0008 |
| 4703 | Q99829 | Copine-1 (Copine
I) | 59.1/5.5 | 26 | 5.8 | 0.0063 |
| 4826 | Q02156 | nPKC-epsilon | 83.7/6.7 | 31 | 10.9 | 0.0173 |
| 5008 | P31949 | Calgizzarin | 11.7/6.6 | 41 | 49.5 | 0.0004 |
| 5104 | P09211 | Glutathione
S-transferase P | 23.4/5.4 | 30 | 40.5 | 0.0188 |
| 5405 | P36952 | Serpin B5
precursor | 42.1/5.7 | 50 | 54.7 | 0.0187 |
| 5411 | Q03154 | Aminoacylase-1 | 45.9/5.8 | 50 | 37.3 | 0.0031 |
| 5414 | P40121 | Macrophage-capping
protein | 38.5/5.9 | 53 | 29.6 | 0.0171 |
| 5502 | P35998 | 26S protease
regulatory subunit 7 | 48.6/5.7 | 30 | 18.7 | 0.0122 |
| 5615 | Q96KP4 | Cytosolic
non-specific dipeptidase | 52.9/5.7 | 46 | 17.7 | 0.0389 |
| 5802 | P06396 | Gelsolin
precursor | 85.7/5.9 | 54 | 19.8 | 0.0010 |
| 6001 | P80511 | Calgranulin-C | 10.6/5.8 | 25 | 45.7 | 0.0373 |
| 6010 | Q01469 | Fatty acid-binding
protein 5 | 15.2/6.6 | 31 | 43.7 | 0.0125 |
| 6112 | Q9ULZ3 | Target of
methylation induced silencing 1 | 21.6/6.0 | 36 | 19.5 | 0.0081 |
| 6602 | Q8N1M1 | Bestrophin-3 | 76.1/6.1 | 26 | 6.0 | 0.0015 |
 | Table IIThe downregulated proteins identified
in ICC. |
Table II
The downregulated proteins identified
in ICC.
| SSP no. | Accession no. | Protein name | MW (kDa)/pI | Matched peptides
(%) | coverage (%) | Sequence
P-value |
|---|
| 1104 | P21964 | Catechol
O-methyltransferase | 30.0/5.3 | 38 | 32.8 | 0.0128 |
| 2308 | Q15417 | Calponin-3 | 36.4/5.7 | 26 | 17.9 | 0.0031 |
| 2702 | Q9BQE3 | Tubulin α-1C
chain | 49.9/5.0 | 40 | 39.2 | 0.0061 |
| 2720 | P08670 | Vimentin | 53.7/5.1 | 23 | 12.2 | 0.0185 |
| 3215 | Q9HC38 | Glyoxalase
domain-containing protein 4 | 34.8/5.4 | 29 | 18.2 | 0.0126 |
| 4011 | Q12988 | Heat shock protein
β-3 | 17.0/5.7 | 33 | 18.0 | 0.0001 |
| 4013 | Q99584 | Protein
S100-A13 | 11.5/5.9 | 28 | 39.8 | 0.0198 |
| 4017 | Q9BV57 |
1,2-dihydroxy-3-keto-5-methylthiopentene
dioxygenase | 21.5/5.4 | 35 | 40.2 | 0.0008 |
| 4121 | P07203 | Glutathione
peroxidase 1 | 21.9/6.1 | 25 | 23.9 | 0.0155 |
| 4317 | P47755 | F-actin-capping
protein subunit α-2 | 32.9/5.6 | 35 | 33.2 | 0.0388 |
| 4507 | Q9Y2T3 | Guanine
deaminase | 51.0/5.4 | 52 | 33.7 | 0.0406 |
| 4708 | P48643 | TCP-1-epsilon | 59.7/5.5 | 50 | 29.9 | 0.0023 |
| 5107 | P62993 | Growth factor
receptor-bound protein 2 | 25.2/5.9 | 40 | 32.7 | 0.0133 |
| 5219 | P50053 | Ketohexokinase | 32.7/5.6 | 25 | 20.1 | 0.0331 |
| 5610 | P37837 | Transaldolase | 37.5/6.4 | 46 | 23.7 | 0.0022 |
| 6411 | P30740 | Leukocyte elastase
inhibitor (LEI) | 42.7/5.9 | 55 | 32.7 | 0.0091 |
| 8015 | P68871 | Hemoglobin subunit
β | 16.0/6.7 | 27 | 46.9 | 0.0189 |
Verification of FABP5 expression in ICC
tissue specimens by western blot analysis and
immunohistochemistry
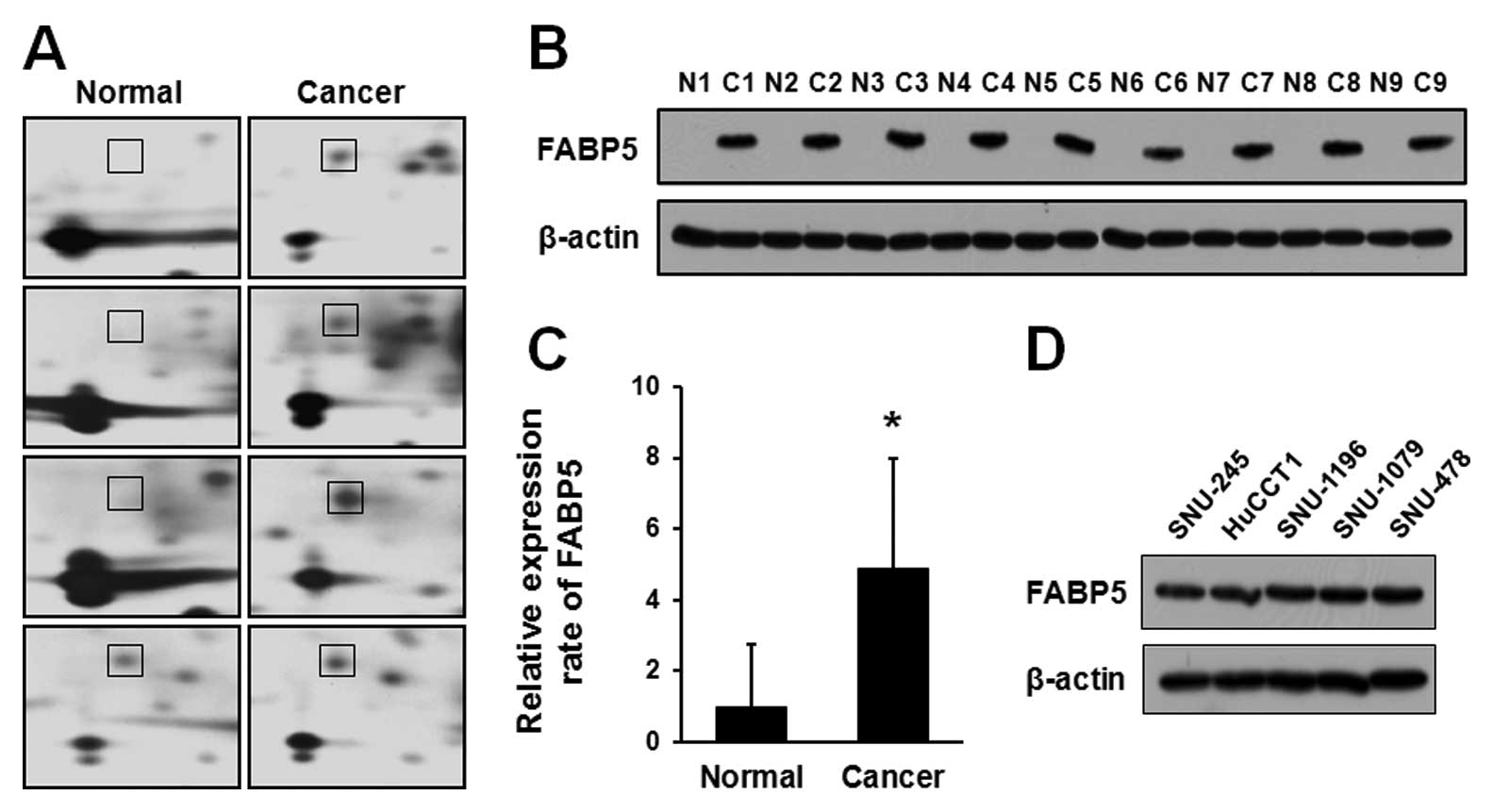
Among the identified proteins, the spot 6010 showed
markedly increased intensity in all of investigated patients with
an average increase of 4.49-fold (Fig.
1A). Statistical analysis of the spots confirmed that spot 6010
was significantly upregulated (P=0.0125) and subsequently
identified as FABP5 by MALDI-TOF-MS, with a molecular mass of 15.2
kDa and a pI 6.6. The sequence coverage of protein isolated from
peptide mass matching in the program was 43.7% (Table I).
Expression levels of FABP5 were also examined by
western blot analysis in paired ICC and normal bile duct tissues
obtained from 16 patients. Nine representative samples of western
blotting for FABP5 expression are shown in Fig. 1B. Western blot analysis of FABP5
expression in the same sample as those used for 2-DE confirmed that
FABP5 was highly expressed in ICC samples, while the normal bile
duct from the same patients showed undetectable FABP5 levels. As
shown in Fig. 1C, the upregulation
ratio of FABP5 in ICC to normal bile duct tissues was about 5-fold.
We next compared expression of FABP5 in cholangiocarcinoma cell
lines (Fig. 1D). Western blot
analysis revealed that all five cholangiocarcinoma cell lines
highly expressed FABP5.
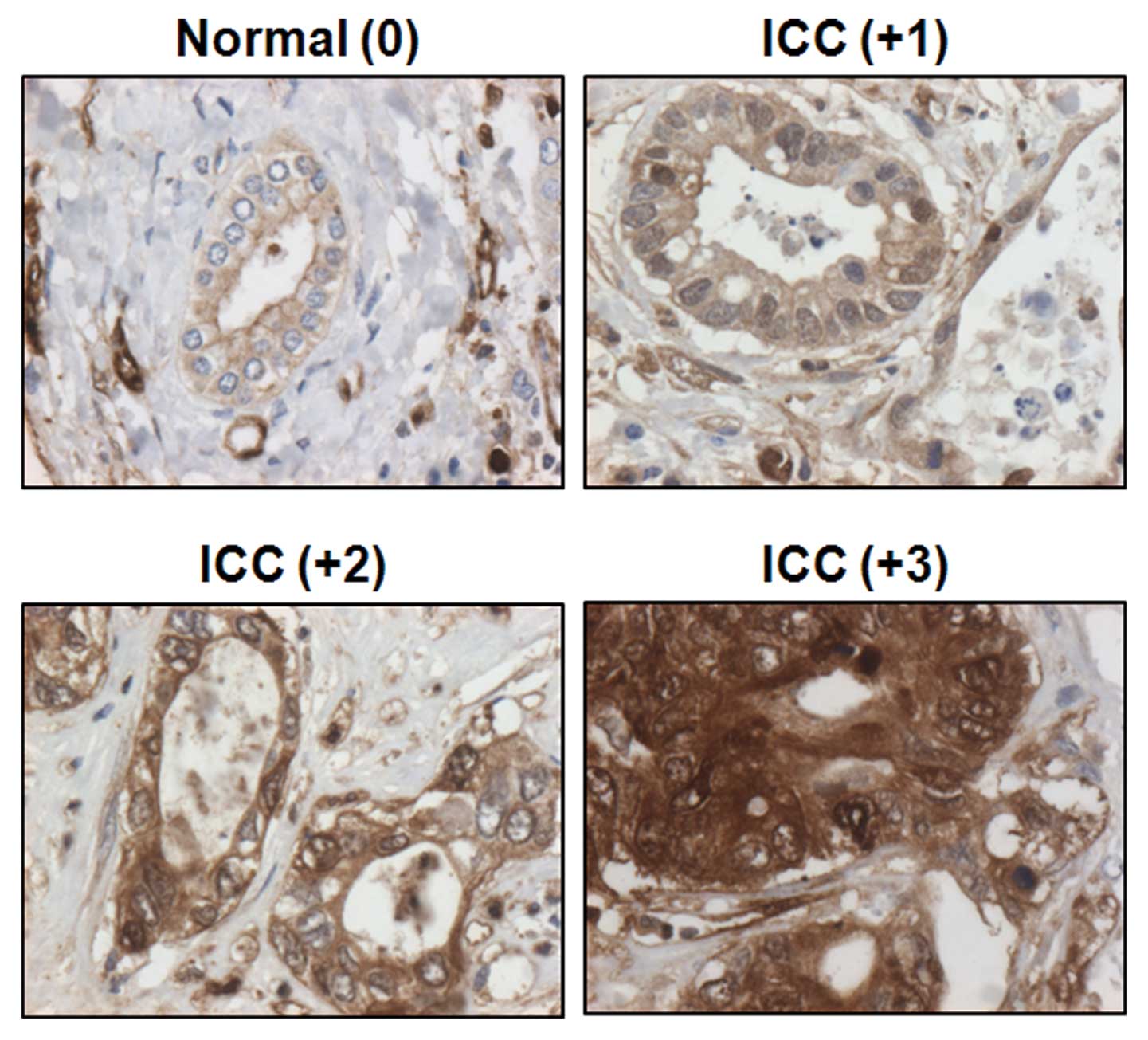
FABP5 was primarily observed in the cytoplasm of
epithelial cells in both cancerous and non-cancerous tissues
(Fig. 2). Immunohistochemical
staining was interpretated and categorized by an arbitrary
semi-quantitative scale as 0 (negative staining), +1 (0~20%
positive), +2 (20~40% positive), and +3 (>40% positive) by
pathologists. All normal bile ducts stained weakly for FABP5 (0 or
+1), while the strong staining intensity was verified in ICC (+1 to
+3).
Correlation between FABP5 expression and
clinicopathological variables
In order to investigate the clinical significance of
FABP5 expression, we performed immunohistochemical analysis on
tissue sections from a larger population of ICC patients. A total
of 43 patients with MF type ICC who underwent several types of
major hepatectomy and lymph node dissection, either alone or in
conjunction with extrahepatic bile duct resection were enrolled in
this study. There were 33 men and 10 women with a mean age of 63.21
years (range, 47–80 years). If the tumor seemed to invade adjacent
organs grossly in the operative field, combined resection was
performed to achieve negative resection margins. Clinical data and
outcome of the patients were collected from medical records
retrospectively. The results of the study revealed weak FABP5
staining (0 or +1) in 18 of the 43 cases, which were classified as
the weak positive group. The remaining 25 cases exhibited strong
FABP5 staining and were classified as the strong positive group (+2
or +3). The expression levels of FABP5 were compared with the
clinical and pathological characteristics of each case of MF type
ICC, including size, differentiation, lymph node metastasis,
vascular invasion, perineural invasion, multiplicity and stage
(Table III). All eight patients
with lymph node metastasis showed strong staining intensity against
FABP5. Univariate analysis revealed that tumor size (P=0.047),
lymph node metastasis (P=0.013), angio-invasion (P=0.032), and
staging (P=0.007) significantly correlated with expression levels
of FABP5.
Effect of FABP5 on the proliferation and
invasion of ICC cells
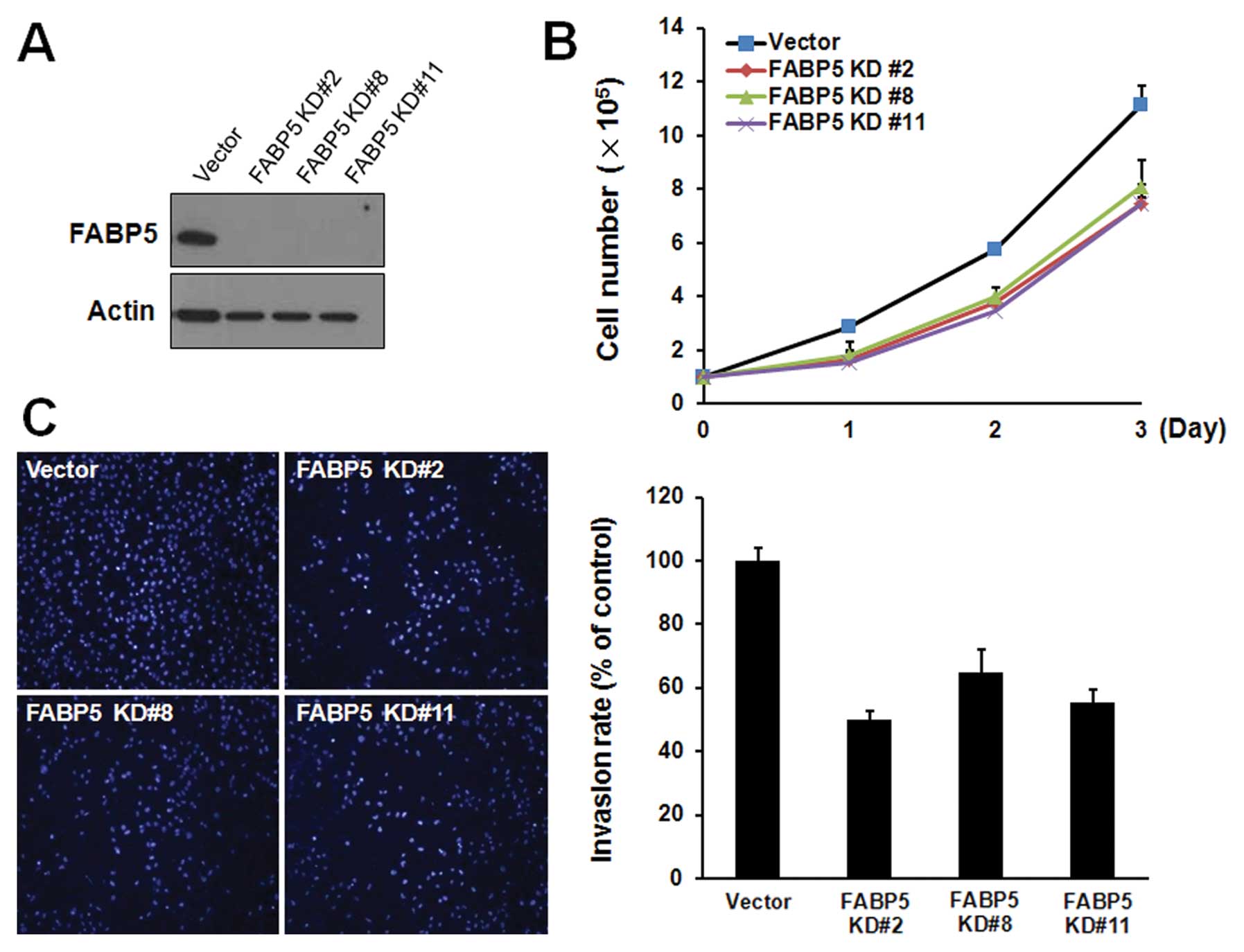
Suppression of FABP5 expression in oral squamous
cell carcinoma has been reported to inhibit proliferation and
invasion (19). Therefore, we first
investigated whether FABP5 was critical for the proliferation of
cholangiocarcinoma cells. We used a short hairpin RNA (shRNA) to
silence the expression of FABP5. HuCCT1 cells were transfected with
control shRNA or FABP5 specific shRNA plasmid. After puromycin (2
μg/ml) selection, western blot analysis confirmed that FABP5-shRNA
clones (named FABP5-KD #2, FABP5-KD #8, FABP5-KD #11) showed an
almost complete silence of FABP5 expression compared to control
shRNA clones (Fig. 3A). Depletion
of FABP5 expression significantly (P<0.01) inhibited the cell
proliferation rate of HuCCT1 cells without significant increase of
dead cells (Fig. 3B). To examine
the effects of FABP5 depletion on tumor cell invasion in
FABP5-depleting HuCCT1 clones, we then performed in vitro
invasion assay. FABP5 depletion caused a significant reduction in
the invasiveness of FABP5-depleted HuCCT1 cells (Fig. 3C).
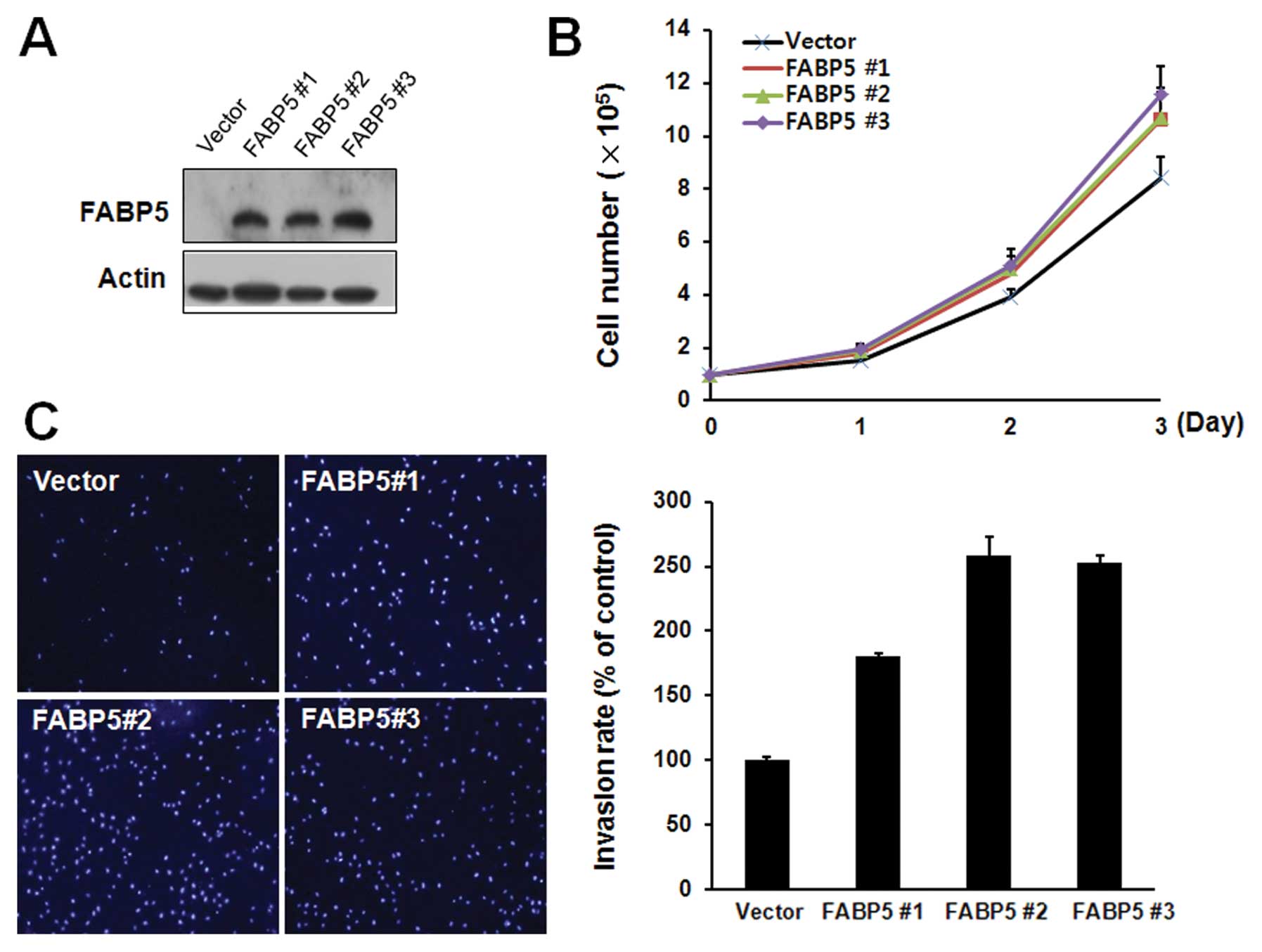
To confirm the enhancing effects of FABP5 on cell
proliferation and invasion, we also examined the effects of the
FABP5 overexpression on the FABP5-negative hepatoma cell line Hep3B
by stable transfection with a plasmid encoding the FABP5 protein.
After G418 (2 mg/ml) selection, the expression of FABP5 in selected
clones (named FABP5 #1, FABP5 #2, FABP5 #3) was determined by
western blotting (Fig. 4A). Cell
proliferation rate, counted by trypan blue assay (Fig. 4B) and Matrigel invasion assays
(Fig. 4C) of FABP5 overexpressing
cell clones were significantly increased compared to the control
vector expressing clones. Taking these results together, the
expression of FABP5 seems to be involved in the increased cell
proliferation and invasion of ICC cells.
Discussion
Carcinogenesis is a multistep process involving
cumulative genetic and epigenetic alterations such as the
activation of oncogenes or the inactivation of tumor suppressor
genes (1). Recent extensive
molecular studies of cholangiocarcinoma demonstrated that numerous
molecular modifications, including dysregulation of cell growth and
survival pathways, aberrant gene expression, invasion and
metastasis, and tumor microenvironment play important roles in
multistep cholangiocarcinogenesis (20). ICC is a good model for the study of
multistep carcinogenesis because of the progress from hyperplasia
to dysplasia, and finally to adenocarcinoma (21).
To gain a comprehensive understanding of cellular
function, the logical method of analysis is proteomics, in which
the global protein expression patterns in a cell, tissue, or
organism can be analyzed. In general, proteomics deal with the
large-scale determination of gene and cell function directly at the
protein level. Many proteomic studies investigating HCC have been
done already, but only a few studies using ICC tissues or cell
lines have been published (22,23).
In this study, we obtained the protein profiles of the normal bile
duct and ICC tissues. By comparing these profiles, we obtained 67
proteins that showed differential expressions (more than a 3-fold
increased or decreased, P<0.05) in the ICC tissues (Tables I and II). Fifty proteins, including FABP5,
proapoptotic caspase adapter protein (PACAP), metastasin, protein
kinase C epsilon (PKCɛ), calgizzarin, target of methylation-induced
silencing 1 (TMS1), calgranulin C and maspin, were upregulated in
the ICC tissues, while 17 proteins, including heat shock protein
β-3 (HspB3), glyoxalase domain-containing protein 4 (GLOD4),
glutathione peroxidase 1, T-complex protein 1 subunit epsilon
(TCP-1ɛ), ketohexokinase and transaldolase, were downregulated.
Among the set of differentially expressed proteins, we identified
several proteins previously described as proteins that are involved
in ICC progression or differentially expressed by ICC. Three
proteins, FABP5, TMS1 and ketohexokinase in ICC were newly
identified in this study. Subsequently, we focused on FABP5, which
have previously been reported as upregulated protein in other
cancers. FABP5 was barely detectable in normal bile duct tissues.
However, the expression was increased in ICC when the rate of
increase in ICC was more evident in western blotting (Fig. 1). Furthermore, our results were
validated by immunohistochemical staining of 43 tumor sections.
These results showed that the expression of FABP5 was suitably
visible in the inflammatory bile duct, and was greatly increased in
many ICC tissues (Fig. 2).
Comparative analysis of the clinicopathological
characteristics of carcinomas with weak FABP5 staining (0 or +1)
and those with strong FABP5 staining (2+ or +3) indicated that
tumors with strong FABP5 expression were significantly associated
with tumor size (P=0.047), lymph node metastasis (P=0.013),
angioinvasion (P=0.032) and staging (P=0.007) (Table 3). Lymph node metastasis is a
negative prognostic factor in patients with ICC (3). Analysis of prognostic factors after
surgical resection demonstrated that mixed type of mass-forming
(MF) and periductal-infiltrating (PI) ICC, lymph node metastasis,
and vascular invasion were important predictive factors related to
poor survival (3). Another study
revealed that lymph node metastasis was closely associated with
negative prognostic factors including MF or PI type, poorly or
undifferentiated tumors, vascular invasion, and perineural invasion
(4). In the current study, all of
the 43 enrolled patients had MF type ICC and underwent standard
lymph node dissection, and lymph node metastasis was found in eight
cases (18.6%). Cancerous tissues from all patients with lymph node
metastasis exhibited strong staining for FABP5. Our results
demonstrated that increased expression of FABP5 is significantly
correlated with known factors of poor prognosis and poor
outcome.
FABPs are 14-to-15 kDa proteins of 126–134 amino
acids in length, and are designated according to the tissue from
which they were first isolated or identified, such as liver FABP
(L-FABP), heart FABP (H-FABP), adipose FABP (A-FABP) and epidermal
FABP (E-FABP or FABP5). Among them, E-FABP, also known as
psoriasis-associated (PA)-FABP, keratinocyte FABP, was originally
isolated from psoriatic skin samples (8). It is a small (15.2 kDa) cytoplasmic
protein and has binding activity typical of the FABP family of
proteins in that it has a high affinity for long-chain fatty acids.
Several recent studies have implicated FABP5 in tumorigenesis.
Adamson et al demonstrated that FABP5 was increased in
prostatic carcinoma cells, and that suppression of FABP5 expression
inhibited the expression of vascular endothelial growth factor
(VEGF) (12). FABP5 contributes to
metastasis by upregulating the expression of the VEGF gene, which
is one of the most potent stimulators of angiogenesis (24). In addition to prostatic carcinoma, a
recent study reported that FABP5 may also play a role in the
malignant progression of pancreatic carcinoma, and that
overexpression of FABP5 in chemo-resistant pancreatic cancer cell
lines enhances intracellular compartmentation or removal of
cytotoxic drugs (10). Another
proteomic study using human HCC cell lines revealed increased
expression of FABP5 in HCC tissues compared with normal hepatocytes
(25). A more recent study reported
that suppression of FABP5 expression in oral squamous cell
carcinoma inhibited cell proliferation and invasion (19). To elucidate the biological
significance of elevated levels of FABP5, studies utilizing
siRNA-mediated silencing of FABP5 were performed. We showed that
FABP5 is highly expressed in ICC cells and is involved in the
enhanced cell proliferation and invasion through experiments that
used both FABP5 downregulation and upregulation. The shRNA-mediated
silencing of FABP5 in HuCCT1 cells significantly inhibited cell
proliferation and invasion (Fig.
3). On the contrary, Hep3B cells stably expressing FABP5 showed
increased invasion and proliferation abilities of cancer cell
(Fig. 4). Although the precise role
of FABP5 in carcinogenesis is not clear, it is likely to be
involved in the binding and transport of intracellular fatty acids,
some of which may be signaling molecules involved in carcinogenesis
(26). Intracellular fatty acids
(FAs) are bound by FABPs, which are important carriers of the
intracellular FAs. Elevated expression of FABPs may result in the
increased mobilization of FAs, and thus enhance FA signaling
activity.
FAs, especially polyunsaturated FAs (PUFAs), such as
arachidonic acid (AA) and linoleic acid, can act as signaling
molecules recognized by the nuclear peroxisome
proliferator-activated receptor (PPAR) (27). PPAR family members are nuclear
receptors known to regulate the transcription of many genes
involved in lipid metabolism (28).
Of the three PPAR isotypes (PPARα, PPAR β/δ and PPARγ), PPARβ/δ is
known to be a downstream gene of Wnt/β-catenin signaling pathway
and it has been shown to promote human cholangiocarcioma cell
growth (29). It was demonstrated
that the positive feedback loop between PPARβ/δ and prostaglandin
E2 (PGE2) played an important role in cholangiocarcinoma cell
growth in human cholangiocarcinoma cell lines. Activation of
PPARβ/δ has also been shown to increase the expression of
cyclooxygenase-2 (COX-2) and the production of COX-2-derived PGE2,
resulting in subsequent activation of PPARβ/δ through cPLA2a
phosphorylation-induced AA release (30,31).
In turn, cPLA2a-derived AA activates PPARβ/δ in the nucleus. This
positive feedback loop is likely important for increased E-FABP
expression in cholangiocarcinoma.
In conclusion, the current study showed that the
expression of FABP5 was significantly increased in ICC with lymph
node metastasis, vascular invasion, and large tumor size and it can
regulate proliferation and invasion in cholangiocarcinoma cells
in vitro. This finding suggests a potential role of FABP5 in
tumor progression. We propose that FABP5 may serve as a prognostic
biomarker and potential therapeutic target for ICC.
Acknowledgements
This study was supported by a grant from the
National R&D Program for Cancer Control, Ministry for Health,
Welfare and Family Affairs, Republic of Korea (0820050) and a
clinical research grant from the Gyeongsang National University
Hospital (2007 and GNUHCRF 2010-001).
References
|
1
|
Blechacz B and Gores GJ:
Cholangiocarcinoma: advances in pathogenesis, diagnosis and
treatment. Hepatology. 48:308–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Okabayashi T, Yamamoto J, Kosuge T,
Shimada K, Yamasaki S, Takayama T and Makuuchi M: A new staging
system for mass-forming intrahepatic cholangiocarcinoma. Cancer.
92:2374–2383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guglielmi A, Ruzzenente A, Campagnaro T,
Pachera S, Valdegamberi A, Nicoli P, Cappellani A, Malfermoni G and
Iacono C: Intrahepatic cholangiocarcinoma: prognostic factors after
surgical resection. World J Surg. 33:1247–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi SB, Kim KS, Choi JY, Park SW, Choi
JS, Lee WJ and Chung JB: The prognosis and survival outcome of
intrahepatic cholangiocarcinoma following surgical resection:
association of lymph node metastasis and lymph node dissection with
survival. Ann Surg Oncol. 16:3048–3056. 2009. View Article : Google Scholar
|
|
5
|
Zimmerman AW and Veerkamp JH: New insights
into the structure and function of fatty acid-binding proteins.
Cell Mol Life Sci. 59:1096–1116. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coe NR and Bernlohr DA: Physiological
properties and functions of intracellular fatty acid-binding
proteins. Biochim Biophys Acta. 1391:287–306. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Glatz JF and Storch J: Unravelling the
significance of cellular fatty acid-binding proteins. Curr Opin
Lipidol. 12:267–274. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Madsen P, Rasmussen HH, Leffers H, Honoré
B and Celis JE: Molecular cloning and expression of a novel
keratinocyte protein (psoriasis-associated fatty acid-binding
protein [PA-FABP]) that is highly up-regulated in psoriatic skin
and that shares similarity to fatty acid-binding proteins. J Invest
Dermatol. 99:299–305. 1992.PubMed/NCBI
|
|
9
|
Haunerland NH and Spener F: Fatty
acid-binding proteins - insights from genetic manipulations. Prog
Lipid Res. 43:328–349. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sinha P, Hütter G, Köttgen E, Dietel M,
Schadendorf D and Lage H: Increased expression of epidermal fatty
acid binding protein, cofilin and 14-3-3-sigma (stratifin) detected
by two-dimensional gel electrophoresis, mass spectrometry and
microsequencing of drug-resistant human adenocarcinoma of the
pancreas. Electrophoresis. 20:2952–2960. 1999. View Article : Google Scholar
|
|
11
|
Ostergaard M, Rasmussen HH, Nielsen HV,
Vorum H, Orntoft TF, Wolf H and Celis JE: Proteome profiling of
bladder squamous cell carcinomas: identification of markers that
define their degree of differentiation. Cancer Res. 57:4111–4117.
1997.PubMed/NCBI
|
|
12
|
Adamson J, Morgan EA, Beesley C, Mei Y,
Foster CS, Fujii H, Rudland PS, Smith PH and Ke Y: High-level
expression of cutaneous fatty acid-binding protein in prostatic
carcinomas and its effect on tumorigenicity. Oncogene.
22:2739–2749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu HE, Lambert MH and Montana VG:
Molecular recognition of fatty acids by peroxisome
proliferator-activated receptors. Mol Cell. 3:397–403. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morgan E, Kannan-Thulasiraman P and Noy N:
Involvement of fatty acid binding protein 5 and PPARβ/δ in prostate
cancer cell growth. PPAR Res. 2010:2346292010.
|
|
15
|
Jung EJ, Moon HG and Cho BI: Galectin-1
expression in cancer-associated stromal cells correlates tumor
invasiveness and tumor progression in breast cancer. Int J Cancer.
120:2331–2338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landry F, Lombardo CR and Smith JW: A
method for application of samples to matrix-assisted laser
desorption ionization time-of-flight targets that enhances peptide
detection. Anal Biochem. 279:1–8. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yates JR: Database searching using mass
spectrometry data. Electrophoresis. 19:893–900. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu SM, Raine L and Fanger H: Use of
avidin-biotin-peroxidase complex (ABC) in immunoperoxidase
techniques: a comparison between ABC and unlabeled antibody (PAP)
procedures. J Histochem Cytochem. 29:577–580. 1981. View Article : Google Scholar
|
|
19
|
Fang LY, Wong TY, Chiang WF and Chen YL:
Fatty-acid-binding protein 5 promotes cell proliferation and
invasion in oral squamous cell carcinoma. J Oral Pathol Med.
39:342–348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sirica AE: Cholangiocarcinoma: molecular
targeting strategies for chemoprevention and therapy. Hepatology.
41:5–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shimonishi T, Sasaki M and Nakanuma Y:
Precancerous lesions of intrahepatic cholangiocarcinoma. J
Hepatobiliary Pancreat Surg. 7:542–550. 2000. View Article : Google Scholar
|
|
22
|
Kawase H, Fujii K, Miyamoto M, Kubota KC,
Hirano S, Kondo S and Inagaki F: Differential LC-MS-based
proteomics of surgical human cholangiocarcinoma tissues. J Proteome
Res. 8:4092–4103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol
K, Chokchaichamnankit D, Sirisinha S and Svasti J: Proteomic
analysis of cholangiocarcinoma cell line. Proteomics. 4:1135–1144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jing C, Beesley C, Foster CS, Chen H,
Rudland PS, West DC, Fujii H, Smith PH and Ke Y: Human cutaneous
fatty acid-binding protein induces metastasis by up-regulating the
expression of vascular endothelial growth factor gene in rat Rama
37 model cells. Cancer Res. 61:4357–4364. 2001.
|
|
25
|
Fujii K, Kondo T, Yokoo H, Yamada T,
Iwatsuki K and Hirohashi S: Proteomic study of human hepatocellular
carcinoma using two-dimensional difference gel electrophoresis with
saturation cysteine dye. Proteomics. 5:1411–1422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Glatz JFC, Schaap FG, Binas B, Bonen A,
van der Vusse GJ and Luiken J: Cytoplasmic fatty acid-binding
protein facilitates fatty acid utilization by skeletal muscle. Acta
Physiol Scand. 178:367–371. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schroeder F, Petrescu AD, Huang H,
Atshaves BP, McIntosh AL, Martin GG, Hostetler HA, Vespa A,
Landrock D and Landrock KK: Role of fatty acid binding proteins and
long chain fatty acids in modulating nuclear receptors and gene
transcription. Lipids. 43:1–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Desvergne B and Wahli W: Peroxisome
proliferator-activated receptors: nuclear control of metabolism.
Endocr Rev. 20:649–688. 1999.PubMed/NCBI
|
|
29
|
Lim K, Han C, Xu L, Isse K, Demetris AJ
and Wu T: Cyclooxygenase-2-derived prostaglandin E2 activates
beta-catenin in human cholangiocarcinoma cells: evidence for
inhibition of these signaling pathways by omega 3 polyunsaturated
fatty acids. Cancer Res. 68:553–560. 2008. View Article : Google Scholar
|
|
30
|
Wu T: Cyclooxygenase-2 and prostaglandin
signaling in cholangiocarcinoma. Biochim Biophys Acta.
1755:135–150. 2005.PubMed/NCBI
|
|
31
|
Xu L, Han C and Wu T: A novel positive
feedback loop between peroxisome proliferator-activated
receptor-delta and prostaglandin E2 signaling pathways for human
cholangiocarcinoma cell growth. J Biol Chem. 281:33982–33996. 2006.
View Article : Google Scholar
|