Introduction
Lung cancer is a leading cause of cancer-related
death worldwide (1). The 5-year
survival rate for patients with non-small cell lung cancer (NSCLC)
is less than 50% (2). The
development of widespread distant metastases after initial surgery
is the main reason for cancer-related deaths in operable NSCLC.
Overcoming widespread metastases might contribute to a marked
improvement in the treatment outcome of lung cancer. Understanding
the mechanism(s) of metastasis of lung cancer is indispensable for
the development of novel treatment strategies.
It is thought that development of distant metastases
requires a multi-step process, such as invading the surrounding
tissue, entering the lymphatic or blood vessels, surviving in the
circulation, extravasating into a new tissue, and growing in the
new organ (3). Lung cancers can
metastasize to a variety of organs, but like other cancers, there
is an organ preference for metastatic spread in lung cancer. The
organ-preference patterns of tumor metastasis have been proposed to
be the product of favorable interactions between metastatic tumor
cells the ‘seed’ and the organ microenvironment the ‘soil’
(4).
To date, part of the physics and biology of cancer
metastasis has been examined and several molecules associated with
metastasis have been identified and shown to play key roles in
metastasis (5). However, the
molecular mechanisms of metastasis in lung cancer remain
unresolved.
Precise analyses of the pathological, biological,
molecular, and genetic aspects of tumor metastasis using tissue
specimens from metastatic organs might contribute to further
understanding of the mechanism of metastasis and to the development
of new treatment modalities. However, it is much more difficult to
obtain tissue specimens from metastatic organs than from primary
tumors, not least because surgery is not usually the standard
treatment for metastasis of various cancers. Furthermore,
preclinical therapeutic studies on new treatment modalities are
needed before clinical trials in patients with advanced metastatic
disease.
Several mouse models of metastasis have been
developed for various cancers (6–9).
However, the majority of human lung cancer cell lines do not have
the ability to metastasize to multiple organs beyond the lung in
conventional athymic nude mice, even though the models were
developed by directly injecting human cancer cells into the vessels
of these nude mice (10). One way
to address this is to establish human cancer cell lines with a
robust ability to metastasize to multiple organs in conventional
nude mice (7,8,11).
Such highly metastatic cell lines might provide mouse metastasis
models without troublesome manipulation and would be particularly
suitable for comparative studies on biological and molecular
mechanisms of metastasis using subclones with differing metastatic
potential.
Important data on the molecular mechanisms of
metastasis in cancers have been produced from comparative studies
of biological behavior and genetic status among clonally related
cell lines with different metastatic potentials (11–15).
Several genes associated with metastasis have been identified by
comparing the gene expression in cells with different metastatic
potentials, indicating that this strategy is useful for determining
the molecular mechanisms of metastasis (12,16,17).
However, the metastatic potential of these cell lines is
practically restricted to the lung in mouse models. Cell lines with
high metastatic potential to multiple organs would be more
appropriate for comparative studies, because such cell lines would
more closely reflect the features of human cancer metastasis.
In this study, to develop a convenient and reliable
multiple organ metastasis model system, we established a highly
metastatic subline, PC14HM, from the PC14 human lung adenocarcinoma
cell line using an in vivo method. The highly metastatic
subline had the ability to metastasize to multiple organs. To
search for genetic determinants of metastasis in human lung cancer,
we compared gene expression profiles between the new subline and
the parent cells using microarray and real-time reverse
transcription polymerase chain reaction (RT-PCR) analyses.
Materials and methods
Human lung cancer cell lines
Human lung cancer PC14 cells were maintained in
RPMI-1640 medium (Sigma, St. Louis, MO), supplemented with 10%
heat-inactivated fetal bovine serum (Life Technologies, Rockville,
MD), penicillin, and streptomycin in a humidified atmosphere of 5%
CO2 and 95% air at 37°C.
Mice
Female BALB/c nude mice (5 weeks old) were obtained
from Charles River Laboratories Japan, Inc. (Tokyo, Japan). Sterile
food and water were available to mice ad libitum. The mice
were maintained in sterile cages on sterile bedding and housed in
rooms at constant temperature and humidity.
Experimental metastasis
Human lung cancer cell lines were injected
intravenously (i.v.) into the lateral tail vein of nude mice at a
density of 2×106 cells/0.2 ml. Mice were sacrificed when
they became moribund, and an autopsy was performed. Visceral organs
were removed and inspected for the presence of metastases.
Selection of the highly metastatic
subline
A human lung cancer cell line with high metastatic
potential to multiple organs was established by a method previously
described (7). Briefly, mice were
injected i.v. with the parental PC14 cells and sacrificed when they
became moribund. Under sterile conditions, organs with macroscopic
metastasis were removed and a metastasized nodule was dissected
free of connective tissue and blood clots, and rinsed twice with
medium. The nodule was finely minced using sterile scalpel blades
in medium, and the cells were seeded in tissue culture dishes.
Then, 4–6 weeks later, the cells were reinjected i.v. into nude
mice. This process was repeated three times to establish the highly
metastatic subline, PC14HM. PC14HM was derived from an adrenal
metastasis, which is uncommon, after i.v. injection of parental
PC14 cells. The metastasized cells in the bone after reinjection of
the first metastatic subline were reinjected again and cancer cells
in the metastatic nodule in the bone were selected as the highly
metastatic subline, PC14HM.
Tumorigenicity in nude mice
Tumorigenicity was examined by injecting
1×107 cells/0.2 ml into the subcutis of the dorsal
flanks of nude mice. The tumor size was measured twice a week.
Tumor volume (V) was calculated using the formula V = 1/2 × length
× (width)2.
Cell proliferation assay
Cells were seeded in a 96-well plate at a density of
1×104 cells/well, and the number of cells at each time
point was estimated using the TetraColor One Cell Proliferation
Assay System (Seikagaku Corp., Tokyo, Japan) according to the
manufacturer’s protocol. Doubling times of the cells were
calculated from the logarithmic phase of the growth curve.
cDNA microarray analysis
Total RNA was extracted using the RNeasy mini-kit
(Qiagen, Tokyo, Japan) according to the supplier’s protocol.
Microarray analysis was performed using a Filgen Array Human 35K
(Filgen, Inc., Nagoya, Japan), consisting of 34,580 cDNA fragments
of human genes, 8 cDNA fragments of control human housekeeping
genes, and 6 cDNA fragments derived from other species arrayed and
immobilized. The 34,580 70-mer probes represented 24,650 genes and
37,123 gene transcripts. Fluorescent probes were synthesized by
incorporating Cy3- or Cy5-dUTP (Amersham Biosciences, Arlington
Heights, IL) using 2 μg total RNA as a template and the RNA
Transcript SureLABEL Core kit (Takara Bio, Tokyo, Japan), and
hybridized to the chip according to the manufacturer’s protocol.
Imaging and quantifying analyses were performed using the GenePix
4000B scanner (Molecular Devices Japan, Tokyo, Japan) and the
Array-Pro Analyzer software (ver. 4.5; Media Cybernetics, Inc.,
Bethesda, MD). Gene expression was quantified as the ratio of
PC14HM to PC14. For a cDNA fragment to be selected as being
differentially expressed, it had to be expressed in PC14HM cells at
a level at least three times higher or lower than in PC14 cells.
These genes were then classified based on the described function.
cDNA fragments with >10-fold difference in expression between
the two cell lines are listed.
Clinical samples
Surgically resected specimens of lung cancer and
matched normal lung tissue were obtained from 36 patients who
underwent surgery at Gunma University Hospital between October 2002
and March 2006. The specimens consisted of 28 adenocarcinomas and 8
squamous cell carcinomas. After surgical removal, a portion of each
sample was immediately frozen and stored at −80°C until total RNA
extraction. Institutional approval was obtained from the
institutional review board of Gunma University Hospital. Written
informed consent was obtained from the patients.
Quantitative real-time PCR (qPCR)
analysis
Four genes, HRB-2, HS3ST3A1,
RAB7 and EDG1, were randomly selected for this
experiment from the list of differentially expressed genes.
HRB-2, HS3ST3A1 and RAB7 were upregulated in
PC14HM cells. Expression of EDG1 in PC14HM was approximately
200 times lower than in the parental cells in the microarray
analysis. Total RNA was extracted using the RNeasy mini-kit
(Qiagen) according to the supplier’s protocol. Randomly primed
cDNAs were reverse-transcribed from 5 μg total RNAs using the
SuperScript first-strand synthesis system for RT-PCR (Invitrogen,
Carlsbad, CA) according to the supplier’s protocol. Quantitative
analysis of gene expression was performed using the TaqMan PCR
method. PCR primers and TaqMan probes were purchased from Applied
Biosystems (Foster City, CA). A cDNA conversion mixture (1 μl) was
used for PCR amplification. The PCR mixture was prepared using a
Quantitect Probe PCR kit (Qiagen) according to the manufacturer’s
protocol. Amplification was performed on an ABI 7300 Fast Real-Time
PCR System (Applied Biosystems) programmed to hold at 50°C for 2
min, hold at 95°C for 15 min, and complete 45 cycles of 94°C for 15
sec and 60°C for 30 sec. The Sequence Detection software (ver.
1.3.1; Applied Biosystems) was used to measure the quantity of
template molecule from the standard curve of the Ct value for each
gene generated from cDNA from human brain poly(A)+ RNA
(Clontech, Palo Alto, CA). The level of mRNA expression in human
lung cancer specimens for each gene was expressed as the value of
the quantity for each gene relative to that for ACTB and
compared to that of adjacent normal lung tissues. Each assay was
performed in duplicate.
Statistical analysis
Differences in the number of metastatic nodules
among each group of mice were analyzed for statistical significance
using a Student’s t-test. Differences in survival among each group
of mice were analyzed for statistical significance using the
log-rank test. Differences in the expression of the four selected
genes between human NSCLC and normal lung were assessed using a
Student’s t-test. Differences were considered significant at the
95% confidence limit. All statistical analyses were conducted using
the Statview software (Abacus Concepts Inc., Berkeley, CA).
Results
Metastatic potential of the highly
metastatic subline PC14HM from PC14 cells
To evaluate the metastatic potential of the PC14HM
cells, PC14 and PC14HM cells were injected simultaneously into the
tail veins of five mice. In the five mice injected with PC14 cells,
only one developed macroscopic metastases in the lung and bone 69
days after injection (Table I). The
other four mice were sacrificed between 96 and 130 days after
injection. No obvious metastases were observed in these mice. On
the other hand, mice injected with PC14HM cells became moribund
between 19 and 39 days after injection and autopsies revealed that
all five mice developed macroscopic metastatic nodules in the
multiple organs, such as the lungs, lymph nodes, bones, and adrenal
glands (Table I). The distribution
of organs with metastasized cancer was similar to that seen in
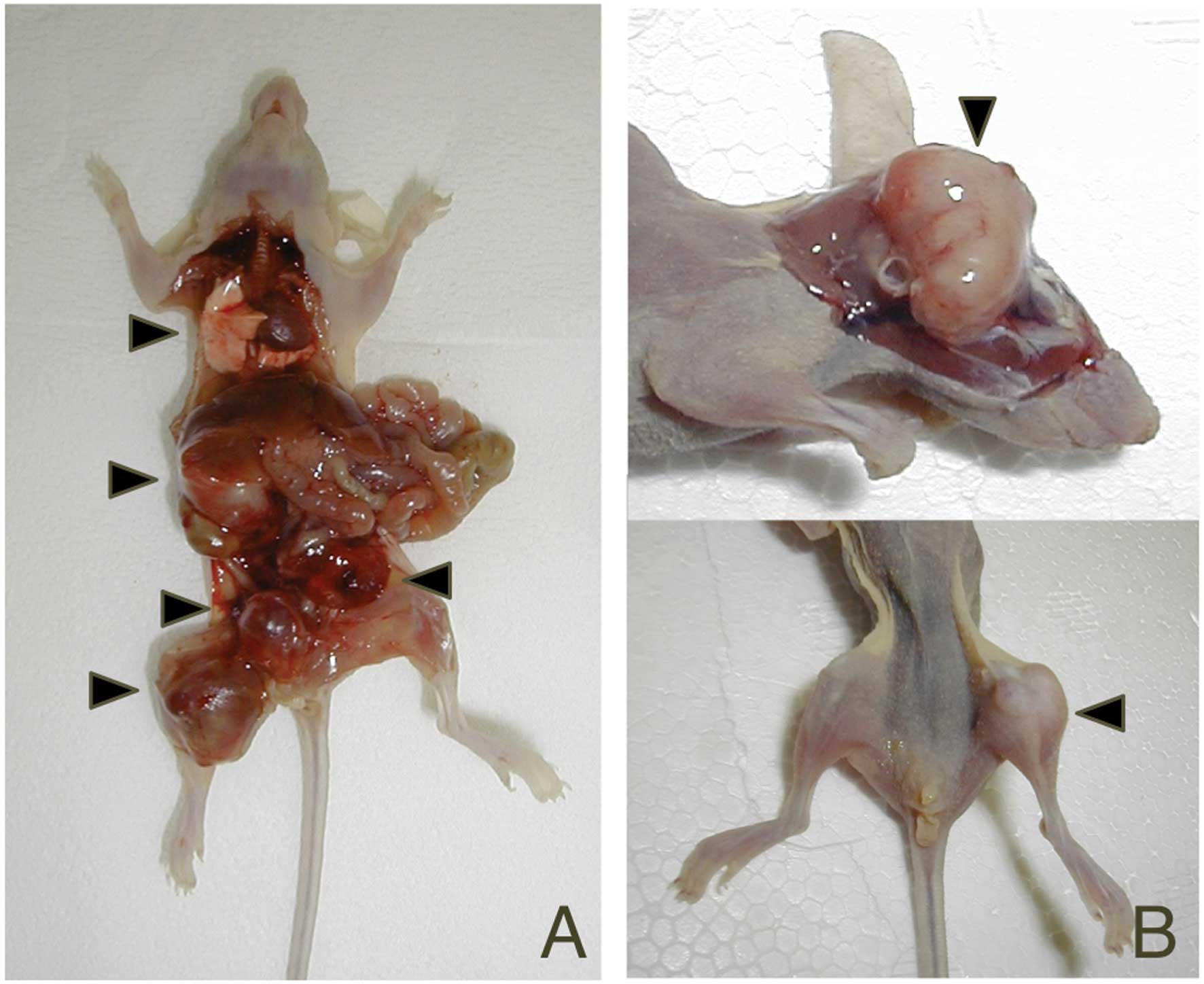
human lung cancer (Fig. 1). In
particular, bone metastases developed in all five mice injected
with PC14HM cells. Differences in the number of metastatic nodules
and the survival periods were statistically significant between the
highly metastatic subline and the parental cells (P=0.0101,
P=0.0027, respectively; Table
II).
 | Table IExperimental metastasis of PC14 cells
and the subline in BALB/c nu/nu mice. |
Table I
Experimental metastasis of PC14 cells
and the subline in BALB/c nu/nu mice.
| Cell line | Survival
(days) | Experimental
metastasis |
|---|
| PC14 (parent) | 69 | Lung, bone
(cranium) |
| 96 | ND |
| 126 | ND |
| 128 | ND |
| 130 | ND |
| PC14HM | 19 | Bone |
| 32 | Lung, bone
(pelvis), LN (neck, Md) |
| 37 | Lung, bone
(scapula), Lt. Ad, LN (axilla) |
| 39 | Bone (pelvis), Lt.
Ad |
| 39 | Lung, bone (thighs,
leg), LN (neck) |
 | Table IICharacteristics of PC14 cells and the
highly metastatic subline. |
Table II
Characteristics of PC14 cells and the
highly metastatic subline.
| | | Experimental
metastasis |
|---|
| | |
|
|---|
| Cell line | Doubling time in
vitro (h) | In vivo
tumor growth (days)a | Incidenceb | Metastases/mouse
(n) | Survival (days)
(mean ± SD) |
|---|
| PC14 (parent) | 41 | 18.9 | 1/5 | 0.4 | 109.8±26.7 |
| PC14HM | 42 | 17.6 | 5/6 | 3.2c | 33.2±8.4d |
Effect of growth rate and tumorigenicity
on metastatic ability
The in vitro population doubling times of
PC14 and PC14HM were 41 and 42 h, respectively. No statistically
significant difference was observed between the cell lines
(Table II). To compare
tumorigenicity between the two cell lines, the average number of
days for tumors to reach 1000 mm3 was compared. The PC14
tumors took 18.9 days to reach 1000 mm3 after
subcutaneous injection, whereas the PC14HM tumors took 17.6 days
(Table II). There was no
statistically significant difference between the cell lines.
Differences in gene expression between
PC14 and PC14HM cells
According to the cDNA microarray analysis, 981 cDNAs
(among the 34,580 cDNAs) showed 3-fold differences in expression
between the PC14 and PC14HM cells (data not shown). The expression
of 537 cDNAs was more abundant in PC14HM than in PC14 cells,
whereas that of 444 cDNAs was lower in PC14HM than in PC14 cells.
Functional classification based on gene ontology biological
processes revealed that >30 genes associated with metabolism,
cell growth and/or maintenance, cell communication, transcription,
and development were contained in the differentially expressed
genes (Table III). More than
10-fold differences in expression were found in 28 cDNAs between
the two cell lines. Tables IV and
V list the genes expressed in
PC14HM cells at least 10-fold higher or lower than in PC14 cells,
respectively. In listing the genes, two cDNAs without gene
information were omitted from the 11 cDNAs that were expressed in
PC14HM more abundantly than in PC14 cells. Thus, 9 upregulated
genes and 17 downregulated genes are listed in Tables IV and V, respectively.
 | Table IIIFunctional classification of genes
differentially expressed between the PC14HM highly metastatic
subline and parental PC14 cells. |
Table III
Functional classification of genes
differentially expressed between the PC14HM highly metastatic
subline and parental PC14 cells.
| No. of genes |
|---|
|
|
|---|
| Function | Upregulated | Downregulated |
|---|
| Metabolism | 78 | 65 |
| Cell growth and/or
maintenance | 30 | 29 |
| Cell
communication | 23 | 24 |
| Transcription | 21 | 18 |
| Development | 22 | 13 |
| Response to
stress | 10 | 13 |
| Signal
transduction | 10 | 6 |
| Protein
transport | 7 | 4 |
| Antigen
processing | 1 | 7 |
| Translation | 5 | 1 |
| Morphogenesis | 4 | 2 |
| Apoptosis | 3 | 3 |
| Cell cycle | 3 | 1 |
| Cell-cell
adhesion | 0 | 4 |
 | Table IVGenes upregulated in the PC14HM
highly metastatic subline compared to parental PC14 cells. |
Table IV
Genes upregulated in the PC14HM
highly metastatic subline compared to parental PC14 cells.
| Gene symbol | Mean net intensity
ratio | GB accession | Description | Biological
process | Chromosome |
|---|
| HRB2 | 23.167992 | BC033887 | HIV-1 rev binding
protein 2 | - | 12q21.1 |
| PPA2 | 13.245953 | AF217187 | PPA2
pyrophosphatase (inorganic) 2 | Metabolism | 4q25 |
| PAEP | 12.798439 | M61886 |
Progestagen-associated endometrial
protein | Development | 9q34.3 |
| UBL5 | 12.568171 | BM703541
BP291575 | Ubiquitin-like
5 | - | 19p13.2 |
| GTPBP4 | 11.925421 | AK001548 | GTP binding protein
4 | - | 10p15.3 |
| HS3ST3A1 | 11.627976 | AF105376 | Heparin sulfate
(glucosamine) 3-O-sulfotransferase 3A1 | - | 17p12 |
| NP_055594 | 11.507741 | AB014569 | KIAA0669 gene
product | Transcription,
DNA-dependent | 3q25.1 |
| NUMB | 11.411299 | AF171938
AK023670 | Numb homolog
(Drosophila) | Development | 14q24.2 |
| RAB7 | 10.855008 | AK024417 | RAB7, member RAS
oncogene family | Protein
transport | 3q21.3 |
 | Table VGenes downregulated in the PC14HM
highly metastatic subline compared to parental PC14 cells. |
Table V
Genes downregulated in the PC14HM
highly metastatic subline compared to parental PC14 cells.
| Gene symbol | Mean net intensity
ratio | GB accession | Description | Biological
process | Chromosome |
|---|
| EDG1 | 0.004762 | M31210
BC018650 | Endothelial
differentiation sphingolipid G-protein-coupled receptor, 1 | Signal
transduction | 1 p21.2 |
| MGST1 | 0.020232 | J03746 | Microsomal
glutathione S-transferase 1 | - | 12p12.3 |
| G1P3 | 0.030553 | - | Interferon,
α-inducible protein (clone IFI-6–16) | Response to
stress | 1 p36.11 |
| IL6 | 0.041293 | BC069201
BC012337 | Interleukin 6
(interferon, β 2) | Negative regulation
of cell proliferation | 7 p15.3 |
| NP_079406 | 0.053462 | X04430 | Hypothetical
protein FLJ22761 | Energy
pathways | 10q22.1 |
| MX1 | 0.059693 | AK096355
M33882 | Myxovirus
(influenza virus) resistance 1, interferon-inducible protein p78
(mouse) | Cell
communication | 21q22.3 |
| LGALS3BP | 0.064397 | BC015761
L13210 | lectin,
galactoside-binding, soluble, 3 binding protein | Signal
transduction | 17q25.3 |
| FN1 | 0.069631 | BX647421
BC000055 | Fibronectin 1 | Cell motility | 2q35 |
| FSTL1 | 0.071251 | X02761
BX640803 | Follistatin-like
1 | - | 3q13.33 |
| NP_076933 | 0.071905 | AK090410 | Hypothetical
protein MGC3265 | - | 5q32 |
| IL8 | 0.072615 | M17017 | Interleukin 8 | - | 4q13 |
| SFN | 0.07789 | BC023552 | Stratifin | Cell
communication | 1p36.11 |
| UBE2L6 | 0.0827 | AK093462
AF061736 |
Ubiquitin-conjugating enzyme E2L 6 | Metabolism | 11 q12.1 |
| HSPA5BP1 | 0.08444 | AK000546 | Heat shock 70 kDa
protein 5 (glucose-regulated protein, 78 kDa) binding protein
1 | - | 11 q12.2 |
| TGFBI | 0.084707 | BC026352
M77349 | Transforming growth
factor, β-induced, 68 kDa | Cell growth and/or
maintenance | 5q31.1 |
| FOXF1 | 0.090622 | U13219 | Forkhead box
F1 | Transcription from
Pol II promoter | 16q24.1 |
| CEBPD | 0.095934 | BC094715.1 | CCAAT/enhancer
binding protein (C/EBP), δ | - | 8p11.2 |
Expression of genes differentially
expressed between PC14 and PC14HM cells in human NSCLC and normal
lung tissues
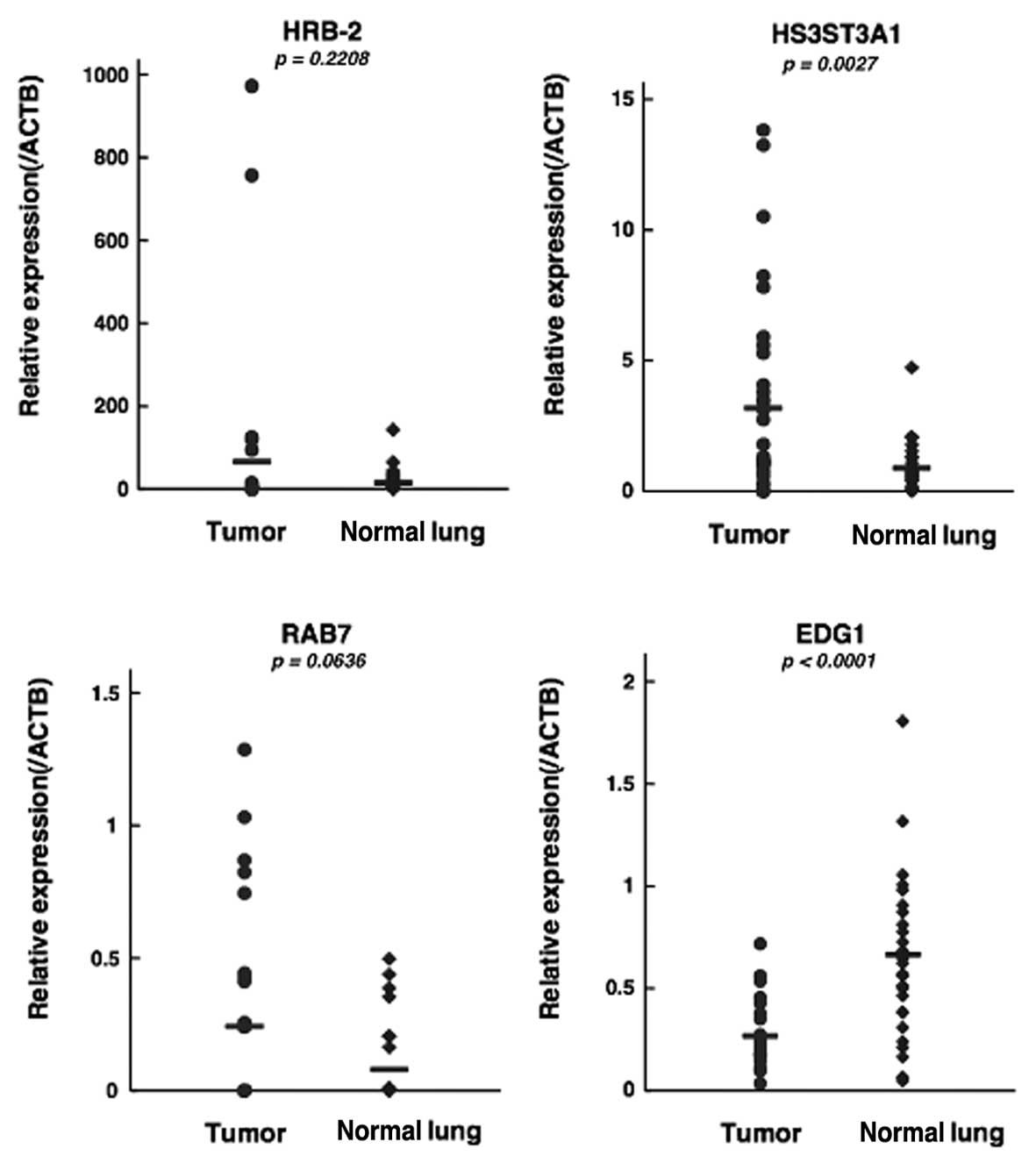
In the qPCR analysis, the mean relative expression
of the three genes that were upregulated in PC14HM cells,
HRB-2, HS3ST3A1 and RAB7, was also elevated in
human lung cancer compared to normal lung (Fig. 2). Expression of HS3ST3A1
between lung cancer and normal lung tissues showed a statistically
significant difference (P=0.0027; Fig.
2). Expression of EDG1, which was downregulated in
PC14HM cells, was also reduced in human lung cancer compared to
normal lung, and the difference was statistically significant
(P<0.0001; Fig. 2).
Discussion
Metastases of lung cancer often appear in multiple
organs. Various organs can be attacked by lung cancer cells, which
tend to ‘prefer’ certain organs over others (18). To understand the mechanisms of
metastasis of human lung cancer, it is important to identify the
metastatic potential of multiple organs. There is limited
information about the metastasis of human lung cancer cells for
several reasons, including the fact that it is difficult to obtain
tissue specimens from multiple metastatic organs and because there
is no relevant and simple animal model of the metastasis of human
lung cancer.
Until now, the development of animal models for
multiple organ metastasis of human lung cancer has required complex
immunocompromised host animals, such as severe combined
immunodeficient (SCID) mice depleted of natural killer cells with
anti-IL-2 receptor β chain antibody, or other genetically modified
rodents, such as SCID-beige mice or NOD/SCID mice (6,9,11,19,20).
To establish a more useful and convenient animal model system for
studying metastasis of human lung cancer, we isolated a highly
metastatic subline, PC14HM, from the parental PC14 lung
adenocarcinoma cell line. Tail vein injection of the highly
metastatic subline caused multiple organ metastases in all
inoculated nude mice, whereas few metastases were observed after
injection of the parental cells. Comparison of gene expression
between the highly metastatic subline and their parental cells
identified candidate genes that may be involved in regulating the
metastatic potential of human lung cancers.
We chose to implant cells via tail vein injection,
even though such models generally do not experience several steps
of the metastasis process (such as growth in primary organs and
invasion into the surrounding tissue and lymphatic or blood
vessels) because this method tends to produce a more stable
multi-organ metastasis model (8,9).
Orthotopically or subcutaneously implanted models show all steps in
the metastatic process, but are only rarely successfully produced
(8,9,21).
There may be several reasons for this; for one, the survival time
of mice injected orthotopically or subcutaneously is much shorter
than the period it takes tumor cells to metastasize to distant
organs, because of the typically rapid growth of primary tumors
(9). Subcutaneous inoculation of
PC14HM or the parental cells indeed did not cause metastasis when
the subcutaneous nodules grew completely.
Mice inoculated with PC14HM cells became moribund
within 40 days and developed multiple metastatic nodules (average,
2.8) in multiple organs, such as the lung, bone, adrenal gland and
lymph node. This stability in the survival period, incidence, and
number of metastases, and the wide variety of distribution of
organs with metastases are useful aspects of a multi-organ
metastasis model for molecular and genetic studies and for the
development of novel treatment strategies of cancer metastasis.
The distribution of metastasized organs in our
models was similar to the pattern of metastasis of human lung
adenocarcinomas, which is thought to reflect organ heterogeneity
and organ ‘preferences’. Lung, bone, adrenal gland and lymph nodes
are frequent metastasis targets with progression of the disease,
and our model system may be useful for studies on organ preferences
in the metastatic spread of lung cancer. Bone and brain metastasis
especially worsen the patient’s quality of life.
Stable development of bone metastases is a
distinctive feature of our model. A previous bone metastasis model
was reported, but it requires complex manipulations, such as
injection of human cancer cell lines into the left ventricle of
nude mice (22), while our model
system adopts conventional tail-vein injection, a much easier
procedure. In the course of developing PC14HM, the subline
experienced two cycles of experimental bone metastasis in nude
mice. It would be intriguing to investigate whether the distinct
bone metastatic ability of PC14HM is the result of a particular
organ preference for metastasis to bone.
Brain metastases were not detected macroscopically
in any mouse injected with PC14HM cells at the time of autopsy.
Conventional experimental metastasis models using severe combined
immunodeficient mice also do not develop brain metastases (6). Development of brain metastasis has
been reported to require injection of human lung cancer cells into
the carotid artery of mice and histopathological conformation of
metastatic nodules (23). Thus, to
date, research on brain metastasis using mouse models has only been
conducted using unique equipment and complex manipulations.
The process of cancer metastasis involves a series
of linked sequential steps, including local tumor growth,
angiogenesis, invasion, detachment, intravasation, circulation,
adhesion, extravasation, and growth in distant organs (3,8). In
this study, proliferative activity in vitro and
tumorigenicity in vivo showed no significant difference
between PC14HM and parental PC14 cells, indicating no obvious
correlation between proliferative activity and/or tumorigenicity
and metastatic potential of the cell lines. Based on these results,
proliferative activity and/or tumorigenicity may be less important
for the regulation of metastatic potential in PC14 cells.
Functional studies of clonally related cancer cell lines with
different metastatic potentials have demonstrated a relationship
between metastatic potential and biological properties other than
proliferative activity, such as invasiveness, motility, and
adherence (11,12). Further studies on other biological
properties of PC14HM and PC14 cells may provide information on
important biological determinants regulating the metastatic
potential of lung cancer cells.
Cell regulatory pathways that are deregulated in
lung cancer include the Rb/p16/cyclin D1, p53/MDM2/p19ARF, Wnt/APC,
EGFR/Ras, PP2a, and telomerase pathways (1). PC14 harbors a homozygous deletion of
p16 (24) and wild-type
K-ras and EGFR (25).
Activating mutations in the EGFR kinase domain is strongly
associated with responsiveness to EGFR kinase inhibitors (26). Also, the presence of these
EGFR kinase mutations is associated with improved
progression-free survival and overall survival after therapy with
EGFR kinase inhibitors, compared to patients with wild-type
EGFR (27). Thus, improving
the outcome of treatment in patients with metastatic lung cancer
harboring wild-type EGFR remains an essential clinical
problem to be resolved. Our model system using PC14 may be useful
for the assessment of novel therapies against metastatic lung
cancer with wild-type EGFR.
To search for genetic determinants for metastasis in
PC14 cells, we used a cDNA microarray analysis with the highly
metastatic PC14HM cells and their parent cells. Among 34,580 genes
examined, 537 upregulated and 444 downregulated genes with at least
a 3-fold difference in expression were identified. Functional
classification of these differentially expressed genes demonstrated
that more than 30 genes were associated with metabolism, cell
growth and/or maintenance, cell communication, transcription, and
development. More than 10-fold differences in expression were
observed in 26 cDNAs between the two cell lines. In this screen of
genes with highly differential expression, multiple genes were
associated with the biological processes listed above. It is
possible that these functional clusters contain genes that are
important for understanding and ultimately treating cancer
metastases.
To evaluate the expression of the differentially
expressed genes in human lung cancer, qPCR analysis using a human
lung cancer specimen and adjacent normal lung tissue was performed.
We randomly selected four differentially expressed genes,
HRB-2, HS3ST3A1, RAB7 and EDG1, for
qPCR analysis. The expression of three genes, HRB-2,
HS3ST3A1 and RAB7, upregulated in PC14HM cells, was
higher in a human lung cancer specimen than in adjacent normal lung
tissue, whereas expression of a downregulated gene, EDG1,
was markedly lower in the human lung cancer specimen. Differences
in the expression levels of two of the four selected genes,
HS3ST3A1 and EDG1, were statistically significant.
These findings may reflect the functions of these genes in human
lung cancer cells, indicating possible associations with not only
metastatic potential but also carcinogenesis and progression.
EDG1, also known as S1PR1, encodes a
member of the endothelial differentiation gene (Edg) family of
proteins, which are G protein-coupled receptors for
sphingosine-1-phosphate (S1P), a biologically active metabolite of
sphingolipid (28,29). S1P-S1PR1 signaling has been proposed
to contribute to cancer progression by regulating tumor
proliferation, invasion, and angiogenesis (28,30).
Inhibition of S1P-S1PR1 signaling has been considered a novel
target for cancer therapy (28,31).
However, recently, Metodieva et al reported reduced
expression of S1PR1 in human NSCLC specimens compared to
that in peripheral non-tumor tissues (32). On the other hand, receptor
subtype-specific positive and negative regulation of S1P-S1PR1
signaling has been proposed. Furthermore, the direction of
signaling is thought to depend on cell type (33,34).
In this study, expression of Edg family genes other than
EDG1 was not significantly different between PC14HM cells
and the parent cells (data not shown). Although further study is
clearly necessary to validate the functional significance of the
Edg family in lung cancer, it is possible that reduced expression
of EDG1 in lung cancer cells influences metastatic potential
in lung cancer.
HS3ST3A1 encodes the enzyme
3-O-sulfotransferase, which catalyzes the biosynthesis of a
specific subtype of heparan sulfate (HS), 3-O-sulfated heparan
sulfate. This HS subtype has specific functional significance for
herpes simplex virus-1 infection (35). To the best of our knowledge, no
function of this molecule in cancer cells has previously been
suggested. However, heparan sulfate cleavage by heparanase is
strongly implicated in cell dissemination associated with tumor
metastasis, angiogenesis, and inflammation (36). Upregulation of HS3ST3A1 in
PC14HM cells is thought to increase the biosynthesis of the
specific HS subtype and to contribute to elevated metastatic
potential. The function of HRB2 in human cells has not been
determined.
Rab7 is a late endosome/lysosome-associated small
GTPase, perhaps the only lysosomal Rab protein identified to date
(37). Rab7 plays critical roles in
the endocytic processes, participating in multiple regulatory
mechanisms in endosomal sorting, the biogenesis of lysosomes, and
phagocytosis. These processes are closely related to substrate
degradation, antigen presentation, cell signaling, cell survival,
and microbial pathogen infection. Consistently, mutations in or
dysfunction of Rab7 result in traffic disorders, which cause
various diseases, such as neuropathies, cancers, and lipid
metabolism diseases (37). Frasa
et al reported that Rab7 participated in the regulation of
E-cadherin turnover and stability of cell-cell contacts by
interacting with EGF and its downstream effector, Rac1 (38). In addition, Rab7-mediated lysosome
trafficking is implicated in hepatocyte growth factor-induced
invasion by prostate tumor cells (39). Elevated expression of RAB7 in
PC14HM cells and human cancer specimens may suggest a role of this
molecule in carcinogenesis, cell adhesion, and invasion. Overall,
the findings with these randomly selected differentially expressed
genes may suggest that the panel of differentially expressed genes
will contain functionally significant genes for metastasis in lung
cancer.
In summary, we established a human lung cancer cell
line with high metastatic potential to multiple organs from the
PC14 cell line using an in vivo selection method. Using
microarray analysis, we were able to identify a panel of genes that
were differentially expressed between the highly metastatic subline
and the parent cells. Although further studies are needed, as
described above, these genes may contain candidates for molecular
determinants of metastasis in lung cancer and may even provide
candidate molecular targets for the treatment of metastatic lung
cancer. This metastasis model system will be useful to further
understand the molecular mechanism(s) of, and discover and
characterize novel therapeutic strategies for, the metastasis of
human lung cancer.
Acknowledgements
This study was supported, in part, by a Grant-in-Aid
for Scientific Research (C) 17591457 from the Japan Society for the
Promotion of Science (JSPS).
References
|
1
|
Minna JD, Roth JA and Gazdar AF: Focus on
lung cancer. Cancer Cell. 1:49–52. 2002. View Article : Google Scholar
|
|
2
|
Sawabata N, Asamura H, Goya T, et al:
Japanese Lung Cancer Registry Study: first prospective enrollment
of a large number of surgical and nonsurgical cases in 2002. J
Thorac Oncol. 5:1369–1375. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fidler IJ and Poste G: The ‘seed and soil’
hypothesis revisited. Lancet Oncol. 9:8082008.
|
|
5
|
Wirtz D, Konstantopoulos K and Searson PC:
The physics of cancer: the role of physical interactions and
mechanical forces in metastasis. Nat Rev Cancer. 11:512–522. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yano S, Nishioka Y, Izumi K, et al: Novel
metastasis model of human lung cancer in SCID mice depleted of NK
cells. Int J Cancer. 67:211–217. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kimura K, Nakano T, Park YB, et al:
Establishment of human osteosarcoma cell lines with high metastatic
potential to lungs and their utilities for therapeutic studies on
metastatic osteosarcoma. Clin Exp Metastasis. 19:477–485. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Francia G, Cruz-Munoz W, Man S, Xu P and
Kerbel RS: Mouse models of advanced spontaneous metastasis for
experimental therapeutics. Nat Rev Cancer. 11:135–141. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khanna C and Hunter K: Modeling metastasis
in vivo. Carcinogenesis. 26:513–523. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagamachi Y, Tani M, Shimizu K, Tsuda H,
Niitsu Y and Yokota J: Orthotopic growth and metastasis of human
non-small cell lung carcinoma cell injected into the pleural cavity
of nude mice. Cancer Lett. 127:203–209. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jia D, Yan M, Wang X, et al: Development
of a highly metastatic model that reveals a crucial role of
fibronectin in lung cancer cell migration and invasion. BMC Cancer.
10:3642010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakano T, Tani M, Ishibashi Y, et al:
Biological properties and gene expression associated with
metastatic potential of human osteosarcoma. Clin Exp Metastasis.
20:665–674. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yokota J: Tumor progression and
metastasis. Carcinogenesis. 21:497–503. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bittner M, Meltzer P, Chen Y, et al:
Molecular classification of cutaneous malignant melanoma by gene
expression profiling. Nature. 406:536–540. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hashimoto Y, Shindo-Okada N, Tani M,
Takeuchi K, Toma H and Yokota J: Identification of genes
differentially expressed in association with metastatic potential
of K-1735 murine melanoma by messenger RNA differential display.
Cancer Res. 56:5266–5271. 1996.PubMed/NCBI
|
|
16
|
Clark EA, Golub TR, Lander ES and Hynes
RO: Genomic analysis of metastasis reveals an essential role for
RhoC. Nature. 406:532–535. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hashimoto Y, Shindo-Okada N, Tani M, et
al: Expression of the Elm1 gene, a novel gene of the CCN
(connective tissue growth factor, Cyr61/Cef10, and neuroblastoma
overexpressed gene) family, suppresses in vivo tumor growth and
metastasis of K-1735 murine melanoma cells. J Exp Med. 187:289–296.
1998. View Article : Google Scholar
|
|
18
|
Raynaud CM, Mercier O, Dartevelle P, et
al: Expression of chemokine receptor CCR6 as a molecular
determinant of adrenal metastatic relapse in patients with primary
lung cancer. Clin Lung Cancer. 11:187–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bjorge JD, Pang AS, Funnell M, et al:
Simultaneous siRNA targeting of Src and downstream signaling
molecules inhibit tumor formation and metastasis of a human model
breast cancer cell line. PLoS One. 6:e193092011. View Article : Google Scholar
|
|
20
|
Bulk E, Sargin B, Krug U, et al: S100A2
induces metastasis in non-small cell lung cancer. Clin Cancer Res.
15:22–29. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoffman RM: Orthotopic metastatic mouse
models for anticancer drug discovery and evaluation: a bridge to
the clinic. Invest New Drugs. 17:343–359. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sawabata N, Miyaoka E, Asamura H, et al:
Japanese lung cancer registry study of 11,663 surgical cases in
2004: demographic and prognosis changes over decade. J Thorac
Oncol. 6:1229–1235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yano S, Shinohara H, Herbst RS, et al:
Expression of vascular endothelial growth factor is necessary but
not sufficient for production and growth of brain metastasis.
Cancer Res. 60:4959–4967. 2000.PubMed/NCBI
|
|
24
|
Arifin M, Tanimoto K, Putra AC, Hiyama E,
Nishiyama M and Hiyama K: Carcinogenesis and cellular
immortalization without persistent inactivation of p16/Rb pathway
in lung cancer. Int J Oncol. 36:1217–1227. 2010.PubMed/NCBI
|
|
25
|
Noro R, Gemma A, Kosaihira S, et al:
Gefitinib (IRESSA) sensitive lung cancer cell lines show
phosphorylation of Akt without ligand stimulation. BMC Cancer.
6:2772006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yeh HH, Ogawa K, Balatoni J, et al:
Molecular imaging of active mutant L858R EGF receptor (EGFR)
kinase-expressing non-small cell lung carcinomas using PET/CT. Proc
Natl Acad Sci USA. 108:1603–1608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee H, Deng J, Kujawski M, et al:
STAT3-induced S1PR1 expression is crucial for persistent STAT3
activation in tumors. Nat Med. 16:1421–1428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spiegel S and Milstien S: The outs and the
ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol.
11:403–415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Visentin B, Vekich JA, Sibbald BJ, et al:
Validation of an anti-sphingosine-1-phosphate antibody as a
potential therapeutic in reducing growth, invasion, and
angiogenesis in multiple tumor lineages. Cancer Cell. 9:225–238.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chae SS, Paik JH, Furneaux H and Hla T:
Requirement for sphingosine 1-phosphate receptor-1 in tumor
angiogenesis demonstrated by in vivo RNA interference. J Clin
Invest. 114:1082–1089. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Metodieva SN, Nikolova DN, Cherneva RV,
Dimova II, Petrov DB and Toncheva DI: Expression analysis of
angiogenesis-related genes in Bulgarian patients with early-stage
non-small cell lung cancer. Tumori. 97:86–94. 2011.PubMed/NCBI
|
|
33
|
Yamaguchi H, Kitayama J, Takuwa N, et al:
Sphingosine-1-phosphate receptor subtype-specific positive and
negative regulation of Rac and haematogenous metastasis of melanoma
cells. Biochem J. 374:715–722. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takuwa Y: Subtype-specific differential
regulation of Rho family G proteins and cell migration by the Edg
family sphingosine-1-phosphate receptors. Biochim Biophys Acta.
1582:112–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shukla D, Liu J, Blaiklock P, et al: A
novel role for 3-O-sulfated heparan sulfate in herpes simplex virus
1 entry. Cell. 99:13–22. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arvatz G, Shafat I, Levy-Adam F, Ilan N
and Vlodavsky I: The heparanase system and tumor metastasis: is
heparanase the seed and soil? Cancer Metastasis Rev. 30:253–268.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang M, Chen L, Wang S and Wang T: Rab7:
roles in membrane trafficking and disease. Biosci Rep. 29:193–209.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Frasa MA, Maximiano FC, Smolarczyk K, et
al: Armus is a Rac1 effector that inactivates Rab7 and regulates
E-cadherin degradation. Curr Biol. 20:198–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Steffan JJ, Williams BC, Welbourne T and
Cardelli JA: HGF-induced invasion by prostate tumor cells requires
anterograde lysosome trafficking and activity of
Na+-H+ exchangers. J Cell Sci. 123:1151–1159.
2010. View Article : Google Scholar : PubMed/NCBI
|