Introduction
Osteosarcoma is a tumor characterized by the
production of osteoid by malignant cells. This tumor commonly
afflicts patients of 20 to 30 years of age, and the number has
increased markedly in recent years. Tumor is a disease
characterized by abnormal cell cycle phases. The first gap phase,
G1, is unique among cell cycle phases because it is the point at
which cells are responsive to extracellular cues. Cyclin D1 plays a
key regulatory role during the G1 phase and its gene is amplified
and overexpressed in many cancers, such as non-small cell lung
cancer (1), stomach cancer, and
mantle cell lymphoma (2). However,
the mechanism that would explain CCND1 deregulation in these cancer
cells is yet to be discovered. MicroRNAs are a new class of small
non-coding RNAs found in both animals and plants. They bind to the
complementary sequences in the 3′UTR of the protein coding genes
and induce mRNA degradation or translational repression (3). Growing evidence has indicated that
microRNAs control basic cell functions, including development,
differentiation, apoptosis, and proliferation (4,5).
miR-15a and miR-16-1 are highly conserved RNAs that
form a cluster at the chromosomal region 13q14.3 (6), which is frequently deleted in cancer.
A number of studies have reported that miR-15a and miR-16-1 are
missed or deleted in chronic lymphocytic leukemia (CLL), non-small
cell lung carcinoma, liver cancer, breast cancer, ovarian cancer,
prostatic cancer, stomach cancer, pituitary adenoma, multiple
myeloma, and osteosarcoma (7-13).
Numerous studies have reported that miR-15a and miR-16-1 induce
apoptosis and inhibit cell proliferation by targeting multiple
genes, but their specific mechanisms remain unclear.
In the current study, experimental evidence which
shows that miR-15a and miR-16-1 induce apoptosis and cell cycle
arrest in osteosarcoma was provided. In addition, a
post-transcriptional regulatory mechanism of CCND1 expression
mediated by miR-15a and miR-16-1 through direct interaction with
the CCND1 mRNA at the 3′-UTR was discovered. CCND1 protein level
expression is suppressed by miR-15a and miR-16-1, thus providing a
potential strategy to prevent osteosarcoma proliferation by
targeting the CCND1 oncogene.
Materials and methods
Cell lines
The human osteosarcoma cell lines SOSP-9607 and MG63
were grown in the RPMI-1640 medium and MEM medium with 10%
heat-inactivated fetal bovine serum, 2 g/l sodium bicarbonate, and
4 g/l HEPES (Sigma) at 37°C in an environment containing 5%
CO2.
microRNA transfection
Cells were grown in the appointed medium 12–16 h
before transfection. The cells were transfected with 100 nm/l of
miR-15a, miR-16-1, miR-15a inhibitor, miR-16-1 inhibitor, or
negative control precursor using lipofectamine 2000 (Invitrogen)
according to the protocol of the manufacturer. The microRNA mimics,
inhibitors, and negative control precursor were from GenePharma.
The efficiency of the transfection rate was measured using 5′-FAM
marked negative control 5 h after transfection. The cells were
harvested and processed for flow cytometry, cell cycle analysis,
TUNEL, RT-PCR, immunocytochemistry, and western blot after 48
h.
Apoptosis and cell cycle assays
Cultured cells were grown in 6-, 24-, and 96-well
plates, respectively. They were divided into experimental groups,
inhibitor groups, and control groups. The experimental groups
consisted of the miR-15a and miR-16-1 groups. The inhibitor groups
contained the miR-15a/miR-16-1 inhibitor groups, whereas the
control groups contained negative and blank controls. Apoptosis and
cell cycle were measured by flow cytometry. MTT was performed in
24, 48, 72, and 96 h, respectively. The absorbance at 492 nm was
measured after incubation with 20 μl MTT for 4 h. The curve of cell
proliferation was then drawn and the proliferation efficiency was
examined. The in situ cell death detection kit (Roche
Diagnostics, Indianapolis, IN, USA) was used for the TUNEL assay,
and the procedures were performed as described by the
manufacturer.
Target screening
In the current study, four publicly available search
engines for target prediction were used: MiRanda, http://www.microrna.org (ref. 8505); PicTar,
http://pictar.mdc-berlin.de (ref. 746);
TargetScan, http://genes.mit.edu/targetscan (ref. 968); and DIANA
LAB, http://diana.cslab.ece.ntua.gr/microT (ref. 684). The
putative targets common to the different algorithms were obtained
by sequentially inputting MiRanda hits into PicTar, followed by
TargetScan, and finally into Diana lab. In accordance with other
articles and repeated RT-PCR verifications, the CCND1 gene was
finally chosen as the target.
RT-PCR and immunocytochemistry
RT-PCR and immunocytochemistry procedures were
performed as described. Total RNA was extracted using TRIzol
reagent (Invitrogen) according to the protocol of the manufacturer,
and then transcribed into cDNA using BioRT Two Step RT-PCR kit
(Bioer). The following primer set was used for RT-PCR: CCND1-F,
5′-CTGTGCATCTACACCGACAACT-3′ and CCND1-R,
5′-GCATTTTGGAGAGGAAGTGTTC-3′. GAPDH was chosen as the normalization
procedure. The primers were GAPDH-F, 5′-AGGTCCACCACTGACACGTT-3′ and
GAPDH-R, 5′-GCCTCAAGATCATCAGCAAT-3′. SP-0024 Histostain™-Plus kits
(BIOS) were used for immunocytochemistry. Mouse monoclonal (DCS-6)
antibody to cyclin D1 antibody (Abcam) were utilized for
immunocytochemistry (1:1000) and western blotting (1:200).
Western blot analysis
Protein extracts were prepared through a modified
RIPA buffer with 0.5% sodium dodecyl sulfate (SDS) in the presence
of proteinase inhibitor cocktail (Complete Mini, Roche
Diagnostics). The procedure was as follows. Equal amounts of
protein were resolved by 12% SDS-PAGE, then immunoblotted to the
nitrocellulose membranes. The blocked membranes were soaked in 5%
evaporated skimmed milk for 2 h, probed with primary antibody
against cyclin D1 (Abcam) overnight at 4°C, washed extensively with
20% Tween-20 in TBS, and incubated with goat anti-mouse monoclonal
antibody conjugated with goat horseradish peroxidase (1:3000
dilution) for 2 h. Mouse monoclonal anti-β-actin antibody (1:500
CWBIO) was used for normalization. The signals were visualized with
enhanced chemiluminescence (Bio-Rad).
Luciferase report of CCND1
Through an analysis of the complementary sequences
between the microRNAs and CCND1 mRNA, both miR-15a and miR-16-1
have been found to contain seven nucleotides that complement bases
1961 to 1967 of the CCND1 mRNA. Moreover, miR-15a has 12
nucleotides complementing bases 2033–2039, which is not a
continuous complement. Apart from this finding, miR-16-1 has 11
nucleotides imperfectly complementing both bases 1961-1967 and
2033–2039, which facilitates the formation of the stem-loop
structure (Fig. 4A). Thus, two
3′UTR segments of 195 bp (1820–2014) and 265 bp (1967 to2237) of
the CCND1 gene were amplified by PCR from the total cDNA of
SOSP-9607 cells and inserted into the pMIR-REPORT™ Luciferase
(pMIR-R-L) control vector (Ambion) using HindIII and
SpeI as restriction sites. The primer set used to generate
specific fragments are shown in Fig.
4D. Two inserts with deletions of 7 bp (3′Ma) and 8 bp (3′Mb),
respectively, were also generated from the perfect complementarity
site using the QuikChange XL Site-Directed Mutagenesis kit
(Stratagene). Target segments and mutant inserts were confirmed by
sequencing. Renilla luciferase vector was used for normalization.
The cells were co-transfected in 24-well plates using lipofectamine
2000 according to the protocol of the manufacturer with 0.2 μg
pMIR-R-L vector and 0.04 μg control vector. Moreover, 100 nm/l
microRNA mimics or inhibitors were used for each well. pMIR-R-L and
Renilla luciferase activities were measured consecutively using the
dual-luciferase reporter assay system (Promega) 24 h after
transfection.
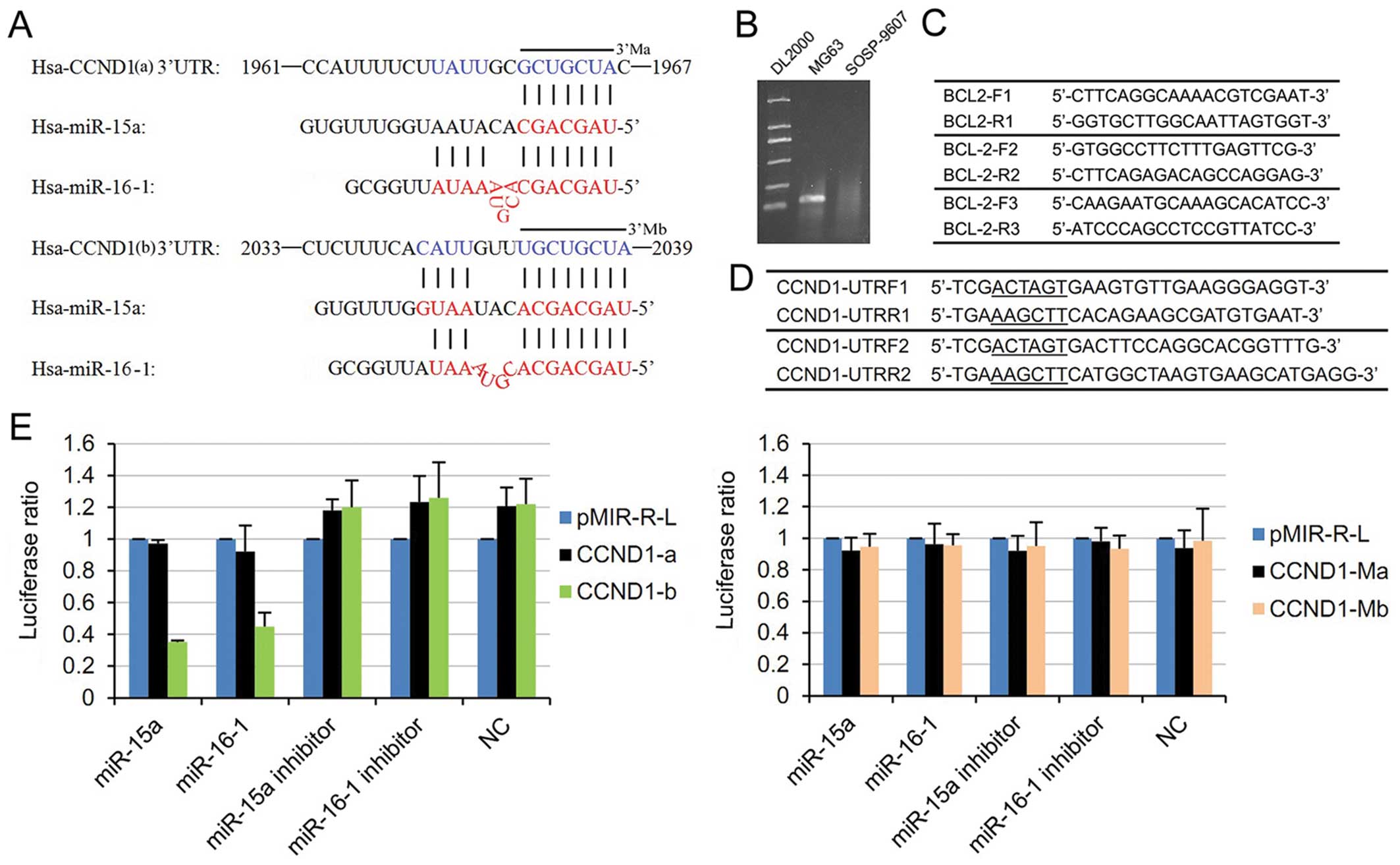 | Figure 4miR-15a and miR-16-1 may regulate the
expression of CCND1 by targeting putative binding site CCND1-b. (A)
Sites of complementarity sequences between microRNAs and CCND1
mRNA. (B) Using the first primer set of (C), MG63 has significant
production length of 148 bp (middle), but SOSP-9607 has no product
(right). The other two primer sets also showed the same results.
(C) Three BCL-2 primer set segments used for RT-PCR. The production
lengths are 148, 213, and 712 bp. (D) Two couples of primers
designed for RT-PCR. The products were used for luciferase report.
(E) Luciferase assays indicated that miR-15a and miR-16
downregulate the expression of CCND1 by targeting putative target
site CCND1-b. Relative repression of the firefly Luciferase
expression was standardized for transfection control, Renilla
luciferase. PMIR-REPORT™ luciferase (pMIR-R-L, Promega) was used as
empty vector. microRNA mimics, inhibitors, and negative control
were used for transfections. Putative target segments CCND1-a,
CCND1-b (Left), and mutant inserts CCND1-Ma and CCND1-Mb (Right)
were used to construct luciferase reporter vector. All experiments
were performed twice in triplicate (n=6). |
Statistical analysis
All values were expressed as means ± standard
deviation (SD). Statistical significance was determined using
one-way ANOVA for multiple comparisons and Student’s t-test was
used to compare two groups. P<0.05 was considered
significant.
Results
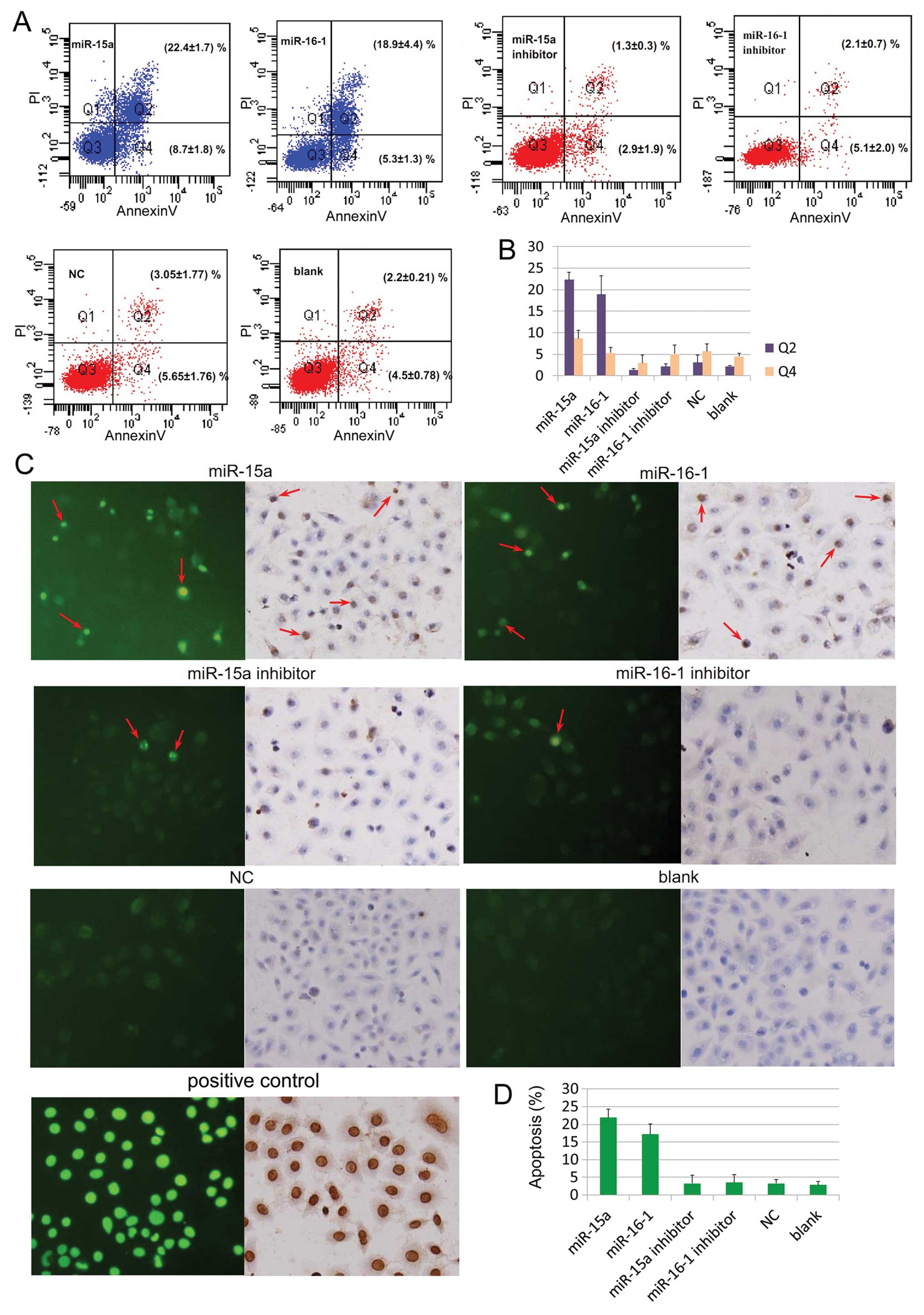
Apoptosis of human osteosarcoma cell line
SOSP-9607 following transfection with miR-15a and miR-16-1
Two different assays were used to identify the
biological effects of miR-15a and miR-16-1. Flow cytometry which
was used to examine phosphatidylserine translocation showed that
the cells transfected with miR-15a and miR-16-1 mimics obtain
higher apoptosis ratio compared with the inhibitor and control
groups (Fig. 1A and B, Q2
P<0.05). There was no observed difference between the inhibitor
and control groups (Fig. 1A and B,
P>0.05). TUNEL assay which was used to confirm DNA breakage and
chromatin condensation showed that more apoptotic cells are
significantly found in miR-15a and miR-16-1 groups (Fig. 1C left and D, P<0.05), and no
difference is found among the levels of apoptosis in the inhibitor,
negative, and blank control groups (Fig. 1C left and D, P>0.05).
Furthermore, converter-POD and haematoxylin staining demonstrated
that there are more apoptotic cells in the experiment group
compared with the other groups (Fig.
1C, right).
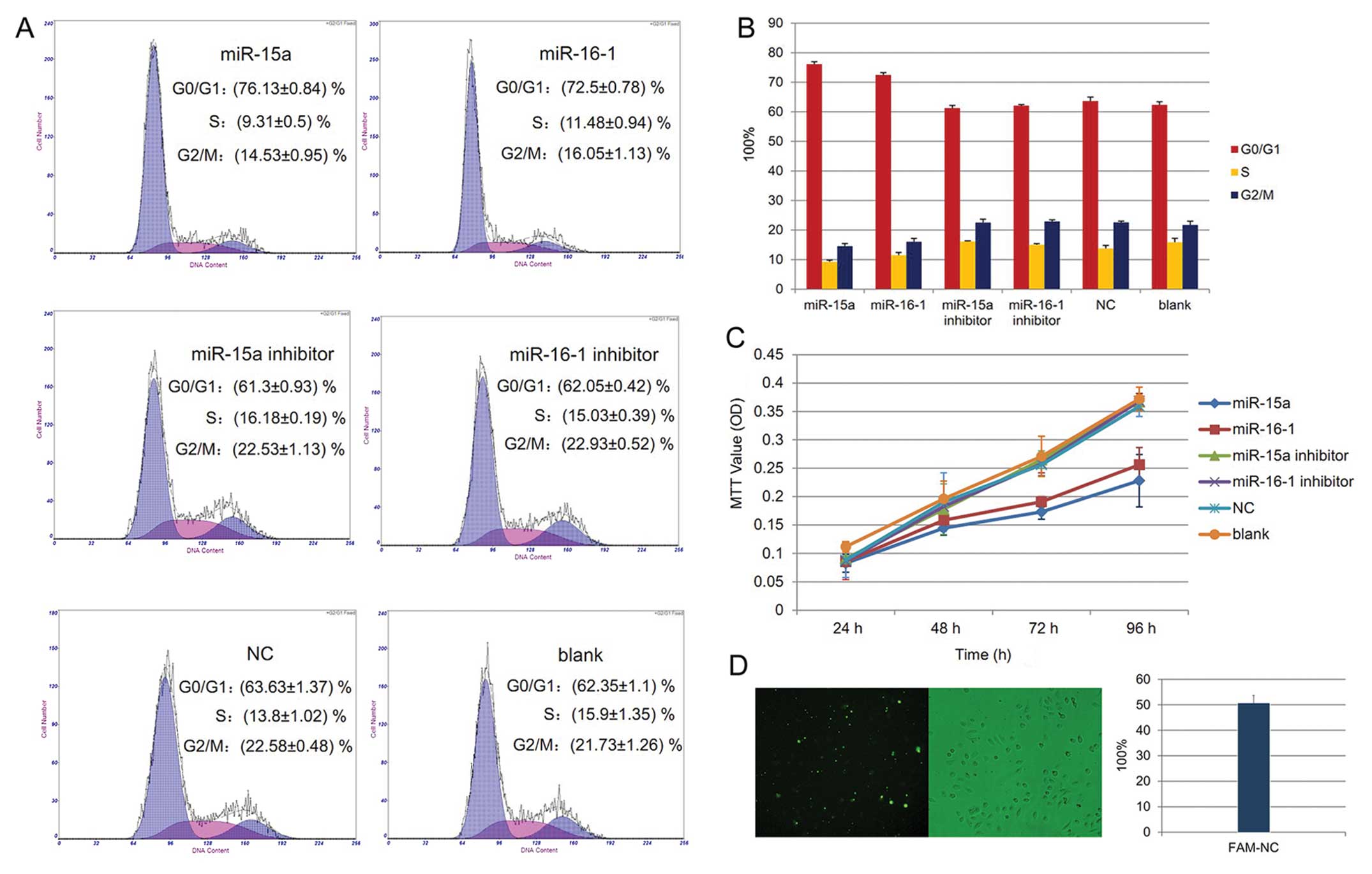
Alteration of cell cycle and cell
proliferation by miR-15a and miR-16-1 in human osteosarcoma cell
line SOSP-9607
Cell cycle analysis showed that more cells are in
the G0/G1 phase in the microRNAs treated cells than in the controls
(Fig. 2A and B, P<0.05). The MTT
assay indicated that the proliferation curves of cells transfected
with miR-15a and miR-16-1 are slower compared with that of the
other groups. However, no differences are found compared with the
inhibitor, negative control and blank groups, respectively
(Fig. 2C, P>0.05).
miR-15a and miR-16-1 downregulate the
protein levels of CCND1
After 48 h of transfection, the total RNA of each
group was extracted and then used for RT-PCR. The results showed
that miR-15a and miR-16-1 inhibit the mRNA levels of CCND1, but do
not degrade them (Fig. 3A,
P>0.05). Western blot was used to further confirm the
post-transcriptional repression of CCND1. The results proved that
miR-15a and miR-16-1 could downregulate the protein levels of CCND1
(Fig. 3B, P<0.05).
Immunocytochemistry also showed that cells transfected with miR-15a
and miR-16-1 have fewer CCND1 proteins (brown color) than the other
groups (Fig. 3C).
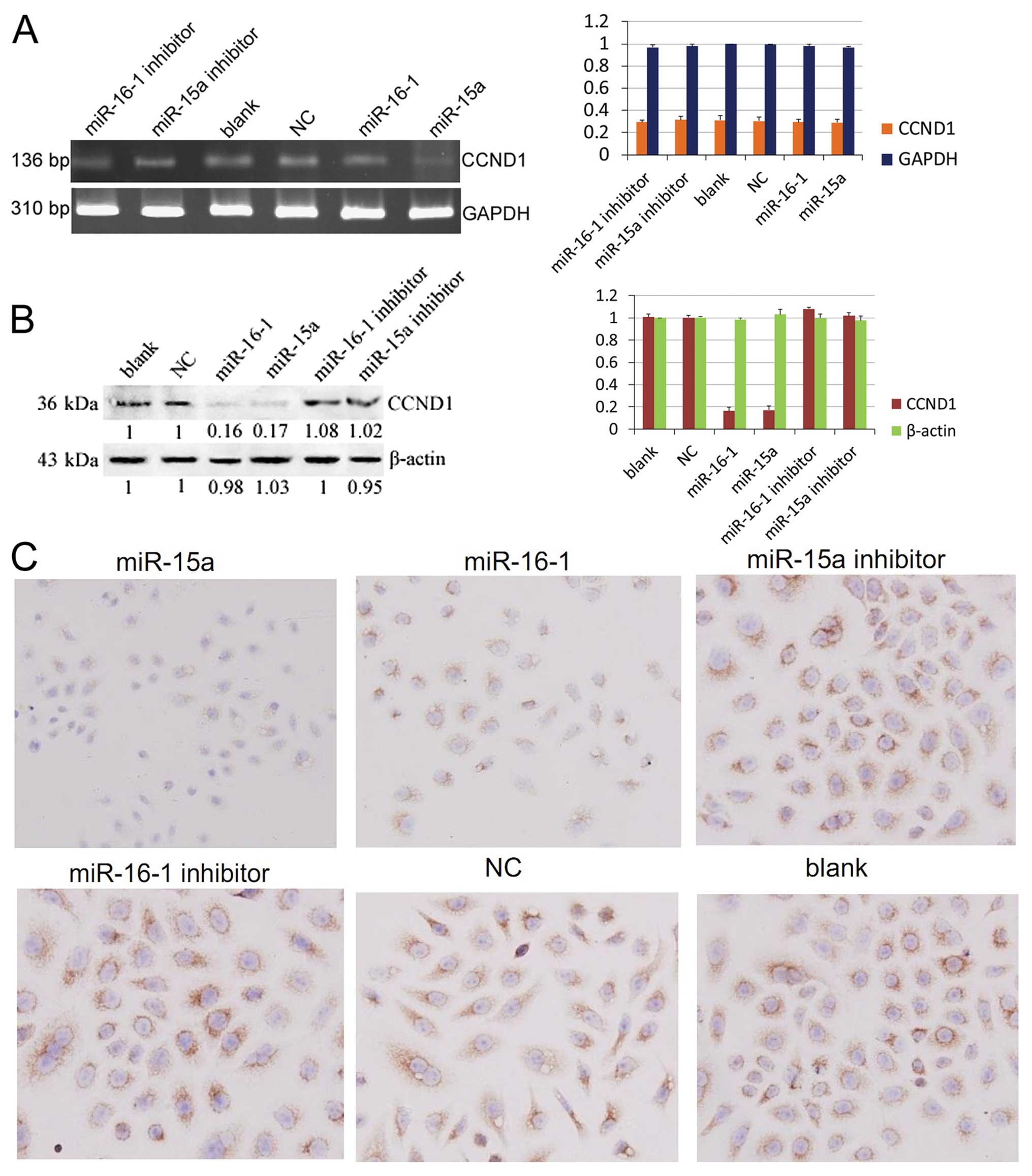 | Figure 3CCND1 is a target gene of miR-15a and
miR-16-1. (A) Results of RT-PCR showed no difference on mRNA levels
of CCND1 in each group (P>0.05). The production length of CCND1
and GAPDH are 136 and 310 bp, respectively. DL2000 was used as
marker. (B) Western blot analysis of miR-15a/miR-16-1,
miR-15a/miR-16-1 inhibitor, negative control, and blank control.
The normalization was done using mouse monoclonal anti β-actin
antibody. (C) Cyclin D1 immunocytochemistry staining of miR-15a,
miR-16, miR-15a inhibitor, miR-16-1 inhibitor, negative control,
and blank control. The brown color in the cytoplasm and nucleus are
the locations of cyclin D1. The blue color in the nucleus is the
staining of haematoxylin (magnification, ×200). |
miR-15a and miR-16-1 binding to the 3′UTR
segment of CCND1
The results of the luciferase assays revealed that
overexpression of miR-15a and miR-16-1 could reduce the luciferase
activity from the segment containing CCND1-b (Fig. 4E left, P<0.05), whereas CCND1-a
has no effect on luciferase activity (Fig. 4E left, P>0.05). A control
experiment with two types of mutated target mRNA sequences lacking
seven (3′Ma) or eight (3′Mb) complementary bases of CCND1 cDNA was
also performed. As expected, both mutants completely abolish the
interaction between the two miRNAs and the 3′UTR of CCND1 (Fig. 4E right, P>0.05). The experimental
data indicated that miR-15a and miR-16-1 regulate the expression of
CCND1 by targeting the putative binding site CCND1-b.
Discussion
Various studies have shown the downregulation of
miR-15a and miR-16-1 in osteosarcoma (14,15).
Moreover, these two miRNAs have been reported to induce apoptosis
by targeting BCL-2 (16). Cimmino
et al (17) showed that
these microRNAs directly target the BCL-2 3′-untranslated region
and significantly correlate with BCL-2 protein levels in CLL both
in vitro and in vivo. Furthermore, BCL-2 protein
levels are downregulated in response to miR-15a or miR-16-1
expression, leading to significant reduction in CLL cell
proliferation. Lerner et al (18) demonstrated that putative tumor
suppressor DLEU2 acts as a target gene of miR-15a and miR-16-1.
DLEU2 overexpression blocks cellular proliferation and inhibits the
colony-forming ability of tumor cell lines in a
miR-15a/miR-16-1-dependent manner. However, in the current study,
RT-PCR did not show the BCL-2 mRNA expression in the SOSP-9607 cell
line (Fig. 4B). This cell line was
primarily cultured from the left tibia of a 17-year-old male
patient (19–21). In the current study, miR-15a and
miR-16-1 induced the apoptosis of SOSP-9607 cells, indicating that
miR-15a and miR-16-1 induce apoptosis by targeting many genes.
CCND1 was found through target screening and RT-PCR verification.
This gene has been widely discussed due to its cell cycle
functions. One of the aims of the current research is to
investigate whether cyclin D1 correlates to the apoptosis induced
by miR-15a and miR-16-1.
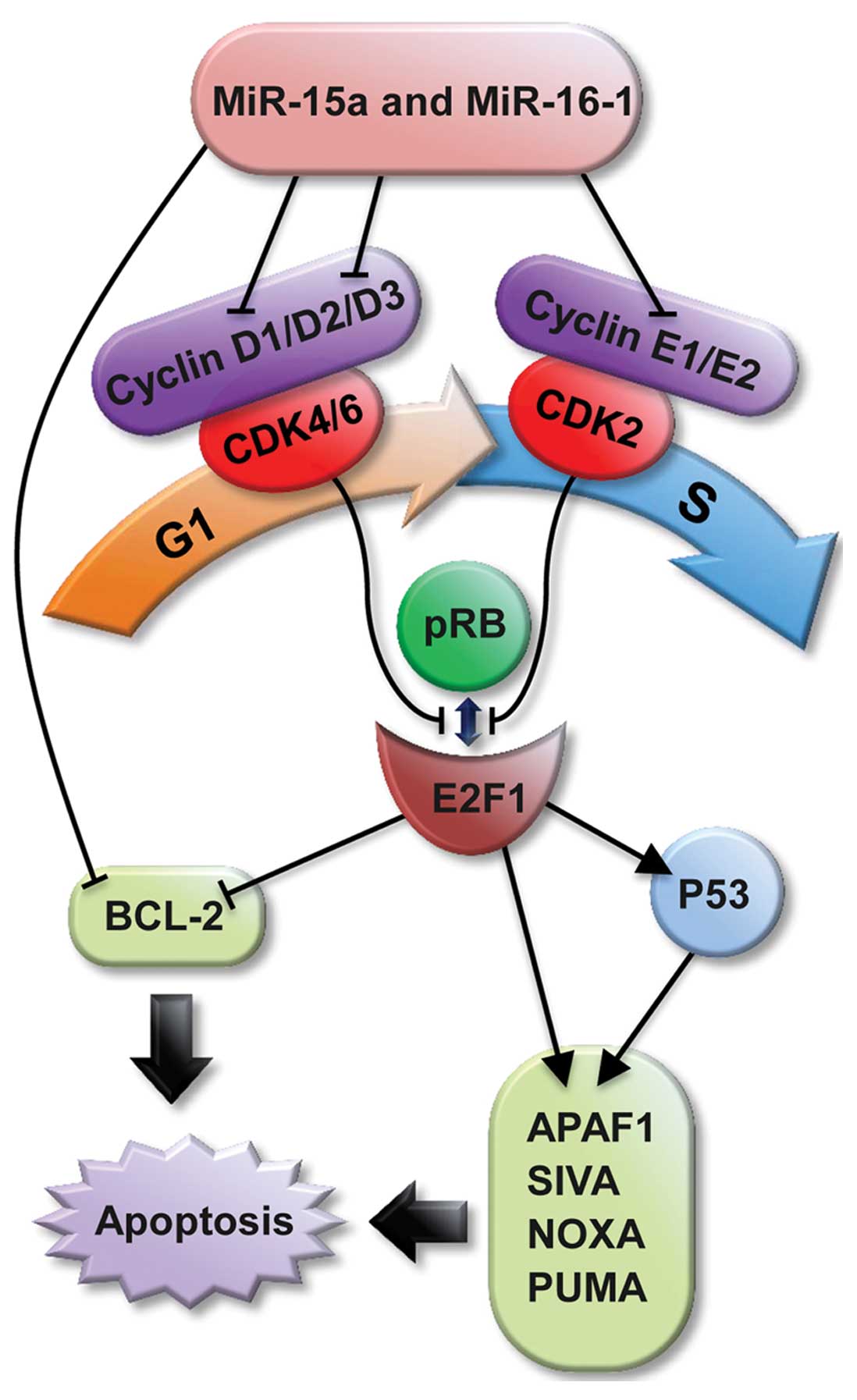
Cyclin D1 is an important regulator of cell cycle
progression (22). It was cloned
and identified from parathyroid adenoma by Motokura et al
(23) in 1991. CCND1 is located in
a cluster at 11q13 with high percentage of G/C components. The
56–141 nucleotides sequence of cyclin D1 is a conserved sequence
called ‘cyclin box’, which is the idio-combination areas to
cyclin-dependent kinases (CDK). The G1/S phase transition is
regulated primarily by D-type cyclins (D1, D2, or D3) belonging to
the CDK4/CDK6 complex and the E-type cyclins (E1 or E2) of the CDK2
complex. These complexes cooperate in phosphorylating and
preventing pRB binding to E2F, thus activating E2F-mediated
transcription and driving cells from G1 into S phase (24).
Furthermore, various studies have reported that the
E2F family can mediate apoptosis. For example, the ectopic
expression of E2F1 leads to apoptosis in tissue-culture cells.
Transgenic mice and E2F1 knockout mice develop tumors, partially
due to suppressed apoptosis (25).
Moreover, E2F1 and P53 cooperate to induce apoptosis (26,27).
Intensive research has identified several molecular mechanisms
underlying this cooperation. First, E2F1 induces the stabilization
and activation of P53. Second, E2F1 and P53 separately
transactivate a plethora of crucial pro-apoptotic genes, raising
the possibility that one or more of their respective targets
cooperate to induce apoptosis. Third, several pro-apoptotic genes,
including APAFI, SIVA, and the BH3 protein-encoding genes NOXA and
PUMA, seem to be transcriptionally regulated by both E2F1 and P53
(28–33). Taken together with the activation of
P53 by E2F1, this pattern of regulation constitutes a modified
feed-forward loop that modulates E2F1 activity and apoptosis
progression (34).
Activated CCND1 may have the ability to induce
apoptosis through the cyclin D/E-CDK2/4/6-pRB-E2F1 pathway
(Fig. 5). In the current research,
flow cytometry and TUNEL assay showed higher apoptosis ratios in
experimental groups, demonstrating that miR-15a and miR-16-1 could
induce apoptosis in osteosarcoma cells. From the cell cycle
analysis, the experiment groups have longer G1 phase compared with
the control groups, illustrating that miR-15a and miR-16-1 can
induce cell cycle arrest. Moreover, the curve reflected that these
two microRNAs could reduce cell proliferation. Thus, miR-15a and
miR-16-1 induce apoptosis and cell cycle arrest in
osteosarcoma.
The current study showed for the first time that
miR-15a and miR-16-1 do not degrade the mRNA levels of CCND1, but
regulate the expression of CCND1 by targeting putative binding site
CCND1-b. Putting these findings together, it can be concluded that
in the SOSP-9607 cell line, miR-15a and miR-16-1 induce apoptosis
and cell cycle arrest by targeting CCND1. Nevertheless, miR-15a and
miR-16-1 may have several target genes. During the experiment,
other predicted target gene interference points should be avoided,
a requirement which is both important and difficult. The data
presented in the current research are of considerable therapeutic
significance because miR-15 and miR-16-1 are natural anti-sense
CCND1 interactors that could be used for therapy in tumor
overexpressing CCND1.
In conclusion, the current experimental results
reveal that miR-15a and miR-16-1 arrest cell cycle and induce
apoptosis and negatively regulate CCND1 expression. These findings
may have important significance on the development of therapeutic
approaches for osteosarcoma.
Acknowledgements
This study was supported by the Department of
Orthopedic Surgery Center and Orthopedic Oncology Institute of
People’s Liberation Army and by the National Natural Science
Foundation of China under Grant No. 81072194. We sincerely thank
Jia-yong Fan for his assistance in polishing the standard of
English in this manuscript.
References
|
1
|
Kosacka M, Piesiak P, Porebska I, et al:
The cyclin A, B1, D1, and E expression in advanced non-small cell
lung cancer - stages IIIB-IV (preliminary report). Pol Merkur
Lekarski. 30:253–258. 2011.(in Polish).
|
|
2
|
Chen RW, Bemis LT, Amato CM, et al:
Truncation in CCND1 mRNA alters miR-16-1 regulation in mantel cell
lymphoma. Blood. 112:822–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miska EA: How microRNAs control cell
division, diffentiation and death. Curr Opin Genet Dev. 15:563–568.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carè A, Catalucci D, Felicetti F, et al:
MicroRNA-133 controls cardiac hypertrophy. Nat Med. 13:613–618.
2007.
|
|
6
|
Bandi N, Zbinden S, Gugger M, et al:
miR-15a and miR-16-1 are implicated in cell cycle regulation in a
Rb-dependent manner and are frequently deleted or down-regulated in
non-small cell lung cancer. Cancer Res. 69:5553–5559. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsang WP and Kwok TT: Epigallocatechin
gallate up-regulation of miR-16 and induction of apoptosis in human
cancer cells. J Nutr Biochem. 21:140–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo CJ, Pan Q, Jiang B, Chen GY and Li DG:
Effects of upregulated expression of microRNA-16 on biological
properties of culture-activated hepatic stellate cells. Apoptosis.
14:1331–1340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu F, Zhang X, Lei Y, et al: Loss of
repression of HuR translation by miR-16 may be responsible for the
elevation of HuR in human breast carcinoma. J Cell Biochem.
111:727–734. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhattacharya R, Nicoloso M, Arvizo R, et
al: MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer.
Cancer Res. 69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonci D, Coppola V, Musumeci M, et al: The
miR-15a-miR-16-1 cluster controls prostate cancer by targeting
multiple oncogenic activities. Nat Med. 14:1271–1277. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia L, Zhang DX, Du R, et al: miR-15b and
miR-16-1 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bottoni A, Piccin D, Tagliati F, et al:
miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell
Physiol. 204:280–285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nedelcu T, Kubista B, Koller A, et al:
Livin and Bcl-2 expression in high-grade osteosarcoma. J Cancer Res
Clin Oncol. 134:237–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao J, Yang TT, Qiu XC, et al: Cloning and
identification of microRNA from human osteosarcoma cell line
SOSP-9607. Ai Zheng. 26:561–565. 2007.(In Chinese).
|
|
16
|
Tsang TY, Tang WY, Chan JY, et al:
P-glycoprotein enhances radiation-induced apoptotic cell death
through the regulation of miR-16 and Bcl-2 expressions in
hepatocellar carcinoma cells. Apoptosis. 16:524–535. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cimmino A, Calin GA, Fabbri M, et al:
miR-15 and miR-16 induce apoptosis by targeting BCL-2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lerner M, Harada M, Lovén J, et al: DLEU2,
frequently deleted in malignancy, functions as a critical host gene
of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp
Cell Res. 315:2941–2952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan LQ, Ma S, Qiu XC, et al: A novel
recombinant immune-tBid with a furin site effectively suppresses
the growth of HER2-positive osteosarcoma cells in vitro.
Oncol Rep. 25:325–331. 2011.PubMed/NCBI
|
|
20
|
Wang LF, Zhou Y, Xu YM, et al: A caspase-6
and anti-HER2 antibody chimeric tumor-targeted proapoptotic
molecule decreased metastasis of human osteosarcoma. Cancer Invest.
27:774–780. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Yang TT, Wang W, et al:
Establishment and characterization of human osteosarcoma cell lines
with different pulmonary metastatic potentials. Cytotechnology.
61:37–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sherr CJ: D-type cyclins. Trends Biochem
Sci. 20:187–190. 1995. View Article : Google Scholar
|
|
23
|
Motokura T, Bloom T, Kim HG, et al: A
novel cyclin encoded by a bell-linked candidate oncogene. Nature.
350:512–515. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Q, Fu H, Sun F, et al: miR-16-1 family
induces cell cycle arrest by regulating multiple cell cycle genes.
Nucleic Acids Res. 36:5391–5404. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iaquinta PJ and Lees JA: Life and death
decisions by the E2F transcription factors. Curr Opin Cell Biol.
19:649–657. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X and Levine AJ: p53 and E2F-1
cooperate to mediate apoptosis. Proc Natl Acad Sci USA.
91:3602–3606. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han EK, Ng SC, Arber N, Begemann M and
Weinstein IB: Roles of cyclin D1 and related genes in growth
inhibition, senescence and apoptosis. Apoptosis. 4:213–219. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moroni MC, Hickman ES, Lazzerini Denchi E,
et al: Apaf-1 is a transcriptional target for E2F and p53. Nat Cell
Biol. 3:552–558. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han J, Flemington C, Houghton AB, et al:
Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by
diverse cell death and survival signals. Proc Natl Acad Sci USA.
98:11318–11323. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakano K and Vousden KH: PUMA, a novel
proapoptotic gene, is induced by p53. Mol Cell. 7:683–694. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oda E, Ohki R, Murasawa H, et al: Noxa, a
BH3-only member of the Bcl-2 family and candidate mediator of
p53-induced apoptosis. Science. 288:1053–1058. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fortin A, MacLaurin JG, Arbour N, et al:
The proapoptotic gene SIVA is a direct transcriptional target for
the tumor suppressors p53 and E2F1. J Biol Chem. 279:28706–28714.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hershko T and Ginsberg D: Up-regulation of
Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J
Biol Chem. 279:8627–8634. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Polager S and Ginsberg D: P53 and E2f:
partners in life and death. Nat Rev Cancer. 9:738–748. 2009.
View Article : Google Scholar : PubMed/NCBI
|