Introduction
Nasopharyngeal carcinoma is a common head and neck
malignancy. Despite recent advances in cancer treatment, the
prognosis for patients with nasopharyngeal carcinoma, particularly
for those at the advanced stages of the disease, remains poor.
Therefore, it is essential to investigate the mechanisms involved
in the development and progression of nasopharyngeal carcinoma.
microRNAs (miRNAs) comprise a large group of
endogenous non-coding RNAs that can block mRNA translation
onegatively regulate mRNA stability and thereby play a central role
in the regulation of gene expression. miRNAs can function as
oncogenic miRNAs or tumor suppressor miRNAs, playing crucial roles
in the development and progression of cancer (1,2).
Recent studies have indicated that deregulated miRNA expression is
a common feature of human nasopharyngeal carcinoma (3–5).
Let-7a has been shown to be significantly downregulated in human
nasopharyngeal squamous cancer tissues, and functions as a
potential tumor suppressor in human nasopharyngeal cancer (6). However, the involvement of deregulated
miRNAs in nasopharyngeal carcinoma remains unknown and warrants
further investigation.
In the present study, we determined the function of
let-7a in nasopharyngeal carcinoma and investigated the mechanims
involved. Our results demonstrate that the enhancer of zeste
homolog 2 (EZH2) expression in nasopharyngeal carcinoma is directly
and negatively regulated by let-7a. Let-7a is downregulated in
nasopharyngeal carcinoma cells, resulting in increased EZH2
expression, thus leading to enhanced proliferation and the
inhibition of apoptosis in nasopharyngeal carcinoma cells.
Materials and methods
Clinical sample collection
The 10 nasopharyngeal carcinoma tissues used in this
study were obtained from the Taizhou People’s Hospital in China.
Specimens were snap-frozen in liquid nitrogen. Fresh normal
nasopharyngeal mucosal tissues were obtained from another 5
patients undergoing biopsies for other non-neoplastic diseases. The
collection and use of the patient samples were reviewed and
approved by the Institutional Ethics Committees and written
informed consent from all patients was appropriately obtained.
In silico analysis
mRNA expression profiling containing 31
nasopharyngeal carcinoma and 10 normal tissues samples (GSE12452)
was carried out (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12452).
Using the AmiGO tool (7) of the
Gene Ontology project (8), lists of
transcripts associated with the biological processes, proliferation
(GO:0008283) and apoptosis (GO:0006915), were obtained. Lists of
unique transcripts were prepared by removing duplicate entries.
Subsequently, the microarray dataset was queried for the genes in
each of these ontologies. Samples were sorted according to the EZH2
expression level for the nasopharyngeal carcinoma and 10 normal
tissues samples separately and the average expression levels scaled
on a gene by gene basis for genes significantly correlating with
EZH2 expression (absolute value Pearson’s correlation >0.6) were
plotted as a heatmap.
Cell culture and transfection
CNE-2 cells maintained in RPMI-1640 medium
supplemented with 10% fetal bovine serum. Hsa-let-7a, siEZH2 and
negative control oligonucleotides were purchased from GenePharma
Co., Ltd. (Shanghai, China). For the expression plasmid construct,
wild-type EZH2 cDNA sequence without 3’UTR was selected and cloned
into the pGenesil-1 vector. Cells were transfected using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at 50–60%
confluency. Forty-eight hours after transfection, the cells were
harvested for further experiments.
Quantitive RT-PCR
Real-time quantification of hsa-let-7a was performed
by stem-loop RT-PCR. All the primers of miRNAs for TaqMan miRNA
assays were purchased from GenePharma Co., Ltd. and were as
follows: human EZH2 forward, 5′-CCTG AAGATGTCGGCATCGAAAGAG-3′ and
reverse, 5′-TGCAA AAATTCACTGGTACAAAACACT-3′; and human GAPDH
forward, 5′-GTCGGAGTCAACGGATT-3′ and reverse, 5′-AAG
CTTCCCGTTCTCAG-3′. Real-time PCR was performed according to the
manufacturer’s instructions. All experiments were performed using
biological triplicates and experimental duplicates. The relative
expression was calculated via the 2-ΔΔCt method.
MTT assay
Cells were plated at 104 cells/well in
96-well plates with 6 replicate wells. After transfection as
previously described, 20 μl of MTT (5 g/l, Sigma, St. Louis, MO,
USA) were added into each well on 4 consecutive days after
treatment and the cells were incubated for an additional 4 h. The
supernatant was subsequently discarded. DMSO (200 μl) was added to
each well to dissolve the precipitate. Optical density (OD) was
measured at a wavelength of 550 nm. The data are presented as the
means ± SD, which are derived from triplicate samples of at least 3
independent experiments.
Cell cycle analysis
Cells were washed with PBS and fixed with 70%
ethanol for at least 1 h. After extensive washing, the cells were
suspended in Hank’s balanced salt solution (HBSS) containing 50
μg/ml PI and 50 μg/ml RNase A and incubated for 1 h at room
temperature and analyzed by FACScan (Becton-Dickinson, Franklin
Lakes, NJ, USA). Cell cycle analysis was analyzed by ModFit
software. Experiments were performed in triplicate. The results
were presented as a percentage of cells in a particular phase.
Cell apoptosis assay
Cells were plated into 6-well plates at
1×105 cells/well. Forty-eight hours after transfection,
the cells were harvested by trypsinization and washed with PBS.
Annexin V and PI double-staining (BD Biosciences, San Jose, CA,
USA) were used to detect and quantify cellular apoptosis by flow
cytometry. Annexin V− and PI− cells were used
as the controls. Annexin V+ and PI− cells
were designated as apoptotic; Annexin V+ and
PI+ cells were considered necrotic. All the tests were
performed in triplicate.
Western blot analysis
Equal amounts of protein per lane were separated by
8% SDS-polyacrylamide gel and transferred onto a PVDF membrane. The
membrane was blocked in 5% skim milk for 1 h and then incubated
with a specific antibody for 2 h. The antibodies used in this study
were as follows: antibodies to EZH2 (Santa Cruz Biotechnology,
Santa Cruz, CA, USA). The antibody against GAPDH (Santa Cruz
Biotechnology) was used as the control. The specific protein was
detected by using a SuperSignal protein detection kit (Pierce,
Rockford, IL, USA). The band density of specific proteins was
quantified after normalization with the density of GAPDH.
Luciferase reporter assay
The human EZH2 3’UTR were amplified and cloned into
the XbaI site of the pGL3-control vector (Promega, Madison,
WI, USA), downstream of the luciferase gene, to generate the
plasmids pGL3-WT-EZH2-3’UTR. pGL3-MUT-EZH2-3’UTR plasmids were
generated from pGL3-WT-EZH2-3’UTR by deleting the binding site for
let-7a ‘CUACCUC’. For the luciferase reporter assay, cells were
cultured in 96-well plates, transfected with the plasmids and
let-7a using Lipofectamine 2000. Forty-eight hours after
transfection, luciferase activity was measured using the Luciferase
assay system (Promega).
Statistical analysis
Statistics was determined by ANOVA, or the t-test
using SPSS11.0. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Let-7a expression in nasopharyngeal
carcinoma
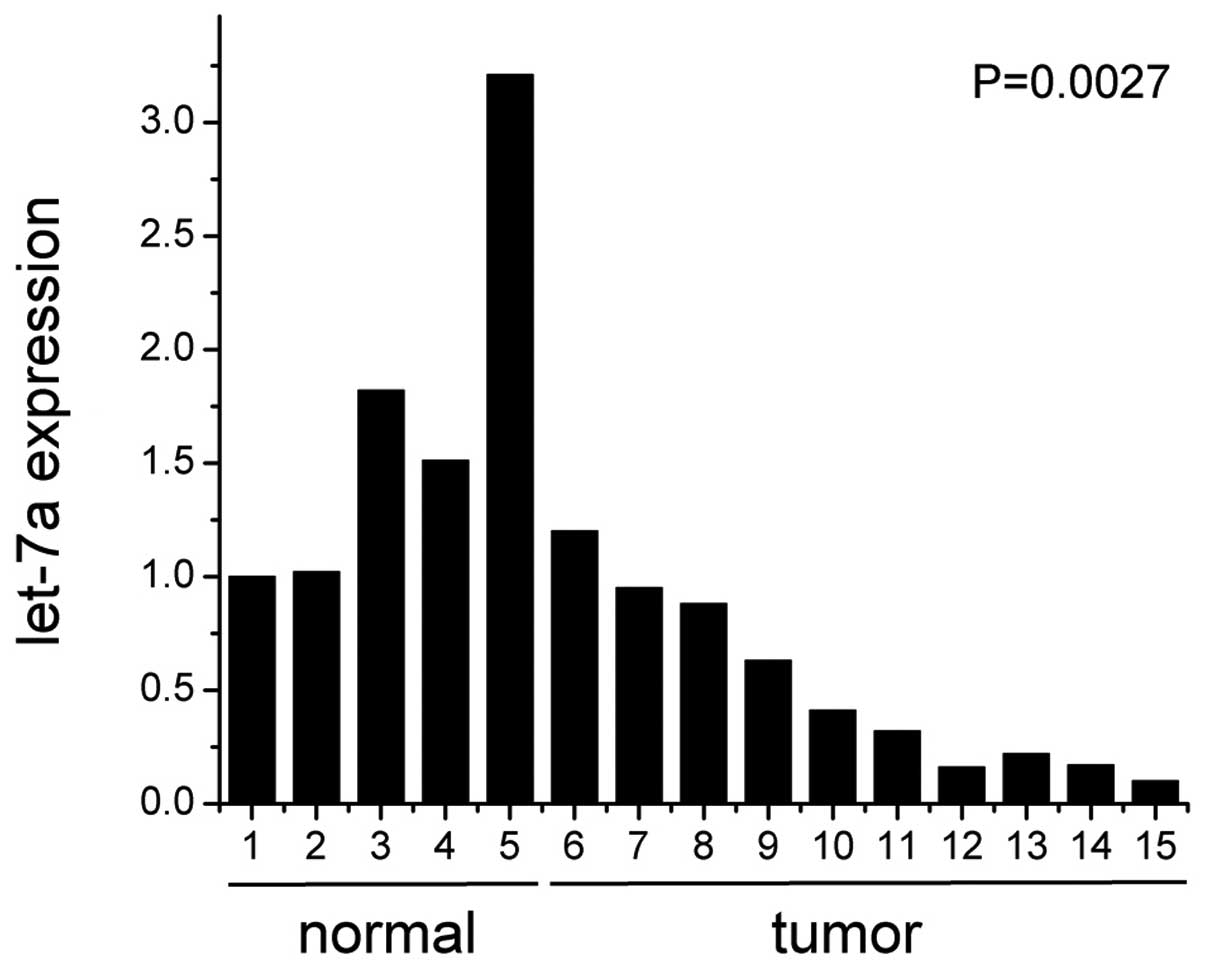
To explore let-7a expression in nasopharyngeal
carcinoma, we examined 10 human nasopharyngeal carcinoma specimens
and 5 normal nasopharyngeal mucosal tissues using real-time PCR. As
shown in Fig. 1, the levels of
let-7a decreased markedly in the nasopharyngeal carcinoma in
comparison to the normal tissue samples (P<0.01).
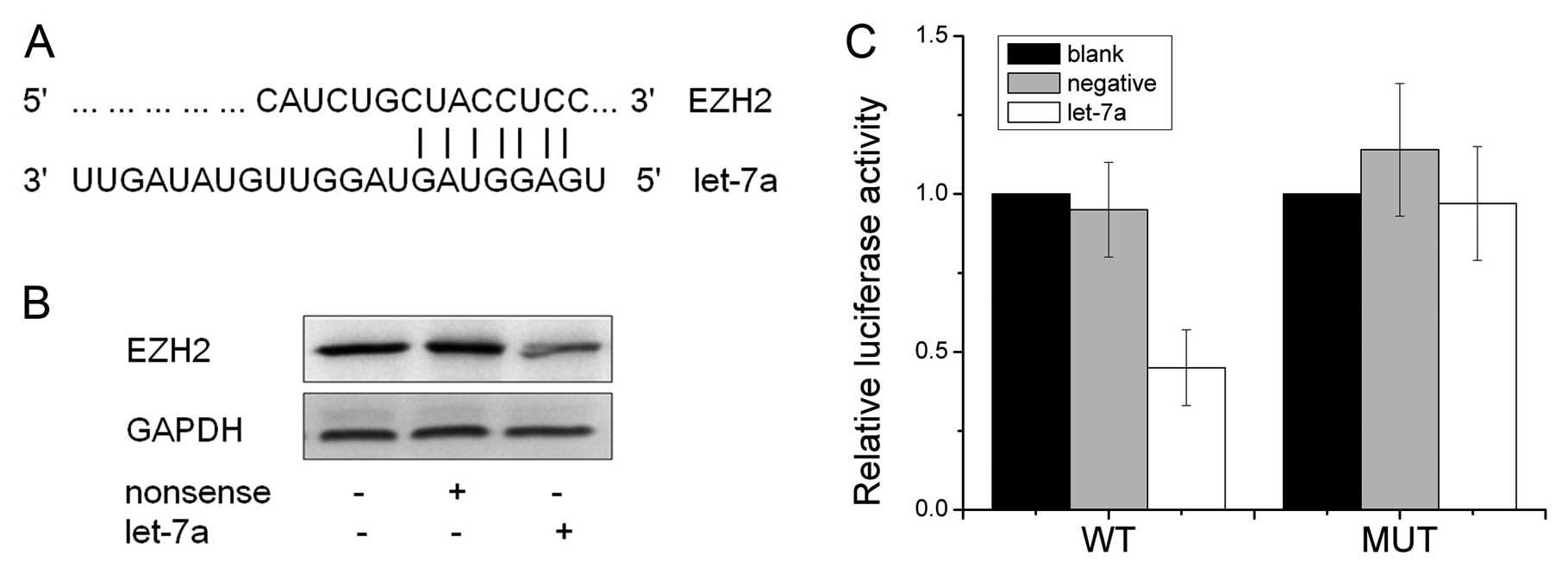
EZH2 is a direct target of let-7a
By performing bioinformatics analysis of let-7a
potential target genes, we found that the tumor suppressor EZH2
contained the highly conserved putative let-7a binding sites. To
determine whether EZH2 is directly regulated by let-7a (Fig. 2A), western blot analysis and
luciferase reporter assay were employed. Western blot analysis
demonstrated a significant reduction in EZH2 expression following
the overexpression of let-7a in the CNE-2 cells (Fig. 2B). Furthermore, we created the
pGL3-WT-EZH2-3’UTR and pGL3-MUT-EZH2-3’UTR plasmids. Reporter assay
revealed that the overexpression of let-7a triggered a marked
decrease in the luciferase activity of the pGL3-EZH2-EZH2-3’UTR
plasmid in the CNE-2 cells; however, no change in the luciferase
activity of the pGL3-MUT-EZH2-3’UTR plasmid was observed (Fig. 2C).
Inverse correlation of expression of
let-7a and EZH2 in nasopharyngeal carcinoma tissues
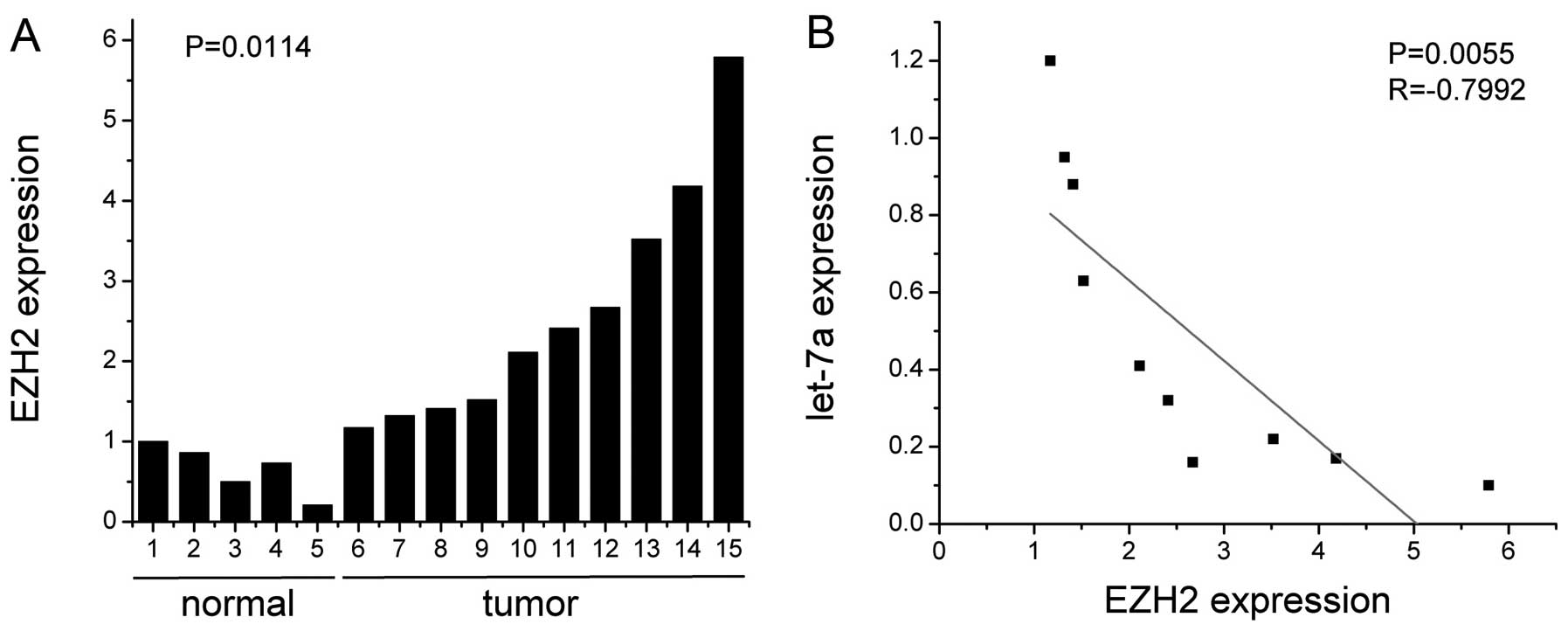
We further explored the correlation between let-7a
and EZH2 expression in nasopharyngeal carcinoma. We examined 10
human nasopharyngeal carcinoma specimens using real-time PCR. The
levels of EZH2 increased markedly in the nasopharyngeal carcinoma
in comparison to the normal tissues (P<0.02) (Fig. 3A). Additionally, Pearson’s
correlation showed that a significant negative correlation existed
between let-7a and EZH2 expression in the nasopharyngeal carcinoma
tissues (R=-0.7992, P<0.01) (Fig.
3B). These data indicate that EZH2 is a direct target of let-7a
in nasopharyngeal carcinoma.
Let-7a-mediated EZH2 expression affects
cell proliferation in nasopharyngeal carcinoma
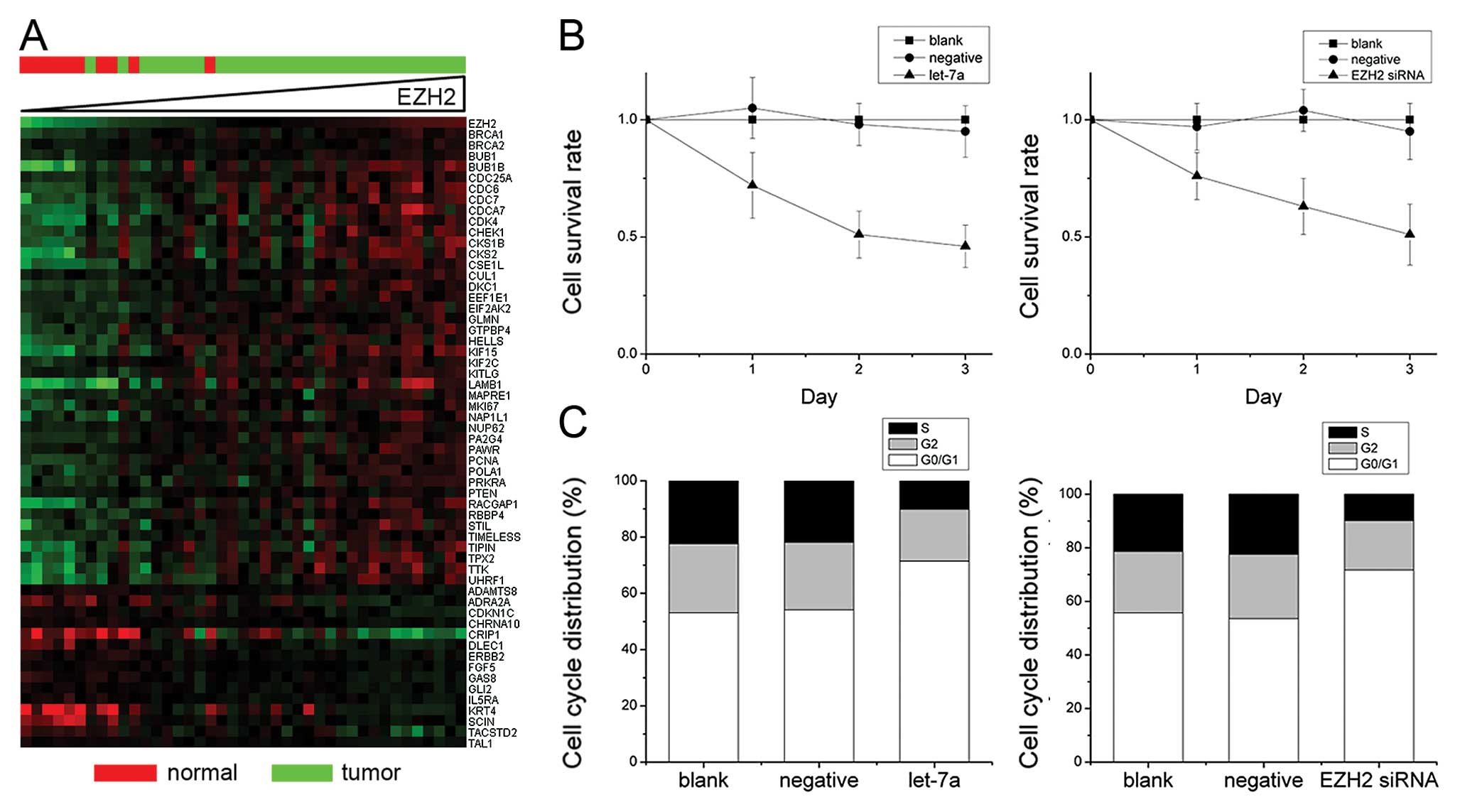
To determine the EZH2-mediated effects of let-7a on
nasopharyngeal carcinoma cell proliferation, we first analyzed
which genes associated with cell proliferation correlated with EZH2
expression in nasopharyngeal carcinoma. First, EZH2 was found to be
overexpressed in the majority of the nasopharyngeal carcinoma
samples as compared to the normal tissues. However, in a few
samples the EZH2 mRNA expression was found to be in the same range
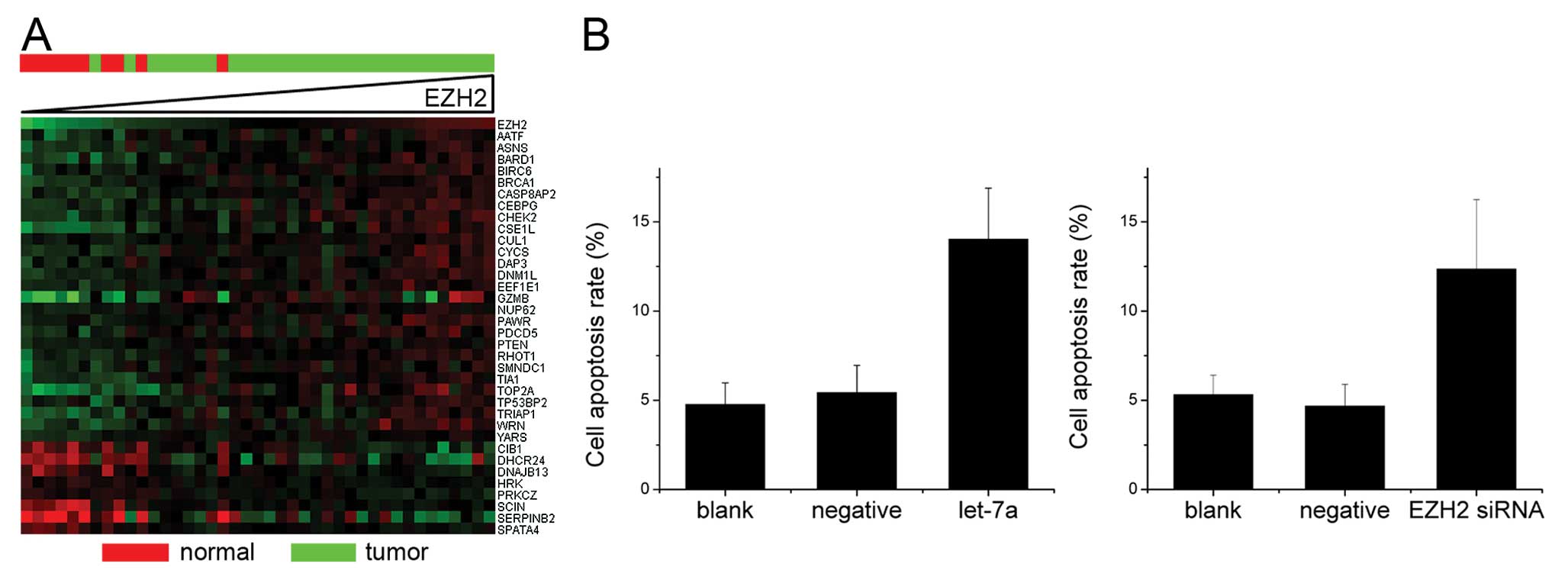
as in the normal tissues (Fig. 4A).
Out of the 1,134 genes that were linked to cell proliferation as
determined by AmiGO (7), 57 genes
showed a clear correlation (>60%) with EZH2 expression in
nasopharyngeal carcinoma (Fig. 4A).
Of note, the nasopharyngeal carcinoma samples with normal EZH2
expression levels also showed a decreased expression of the genes
associated with cell proliferation. Cellular proliferation was then
examined in nasopharyngeal carcinoma cell cultures in order to
determine whether EZH2 influences the proliferation of
nasopharyngeal carcinoma cells. Let-7a induction and EZH2 knockdown
by siRNA significantly reduced cellular proliferation in CNE-2
cells (Fig. 4B). Furthermore, the
let-7a-treated cells demonstrated a significant increase in the
number of cells in the G0/G1 phase in comparison to the untreated
CNE-2 cells (Fig. 4C).
Let-7a-mediated EZH2 expression affects
cell apoptosis in nasopharyngeal carcinoma
To determine the EZH2-mediated effects of let-7a on
nasopharyngeal carcinoma cell apoptosis, we analyzed which genes
associated with apoptosis correlated with EZH2 expression in the
nasopharyngeal carcinoma and normal tissues. A significant
correlation between the expression of 35 out of 424 genes
associated with cell apoptosis and EZH2 expression was observed
(Fig. 5A). In order to determine
whether the let-7a upregulation or EZH2 inhibition also affected
nasopharyngeal carcinoma cell apoptosis, Annexin V and PI
double-staining was performed. The upregulation of let-7a resulted
in a significant increase of apoptosis in the CNE-2 cells (Fig. 5B).
Core role of EZH2 in let-7a-mediated cell
proliferation and apoptosis
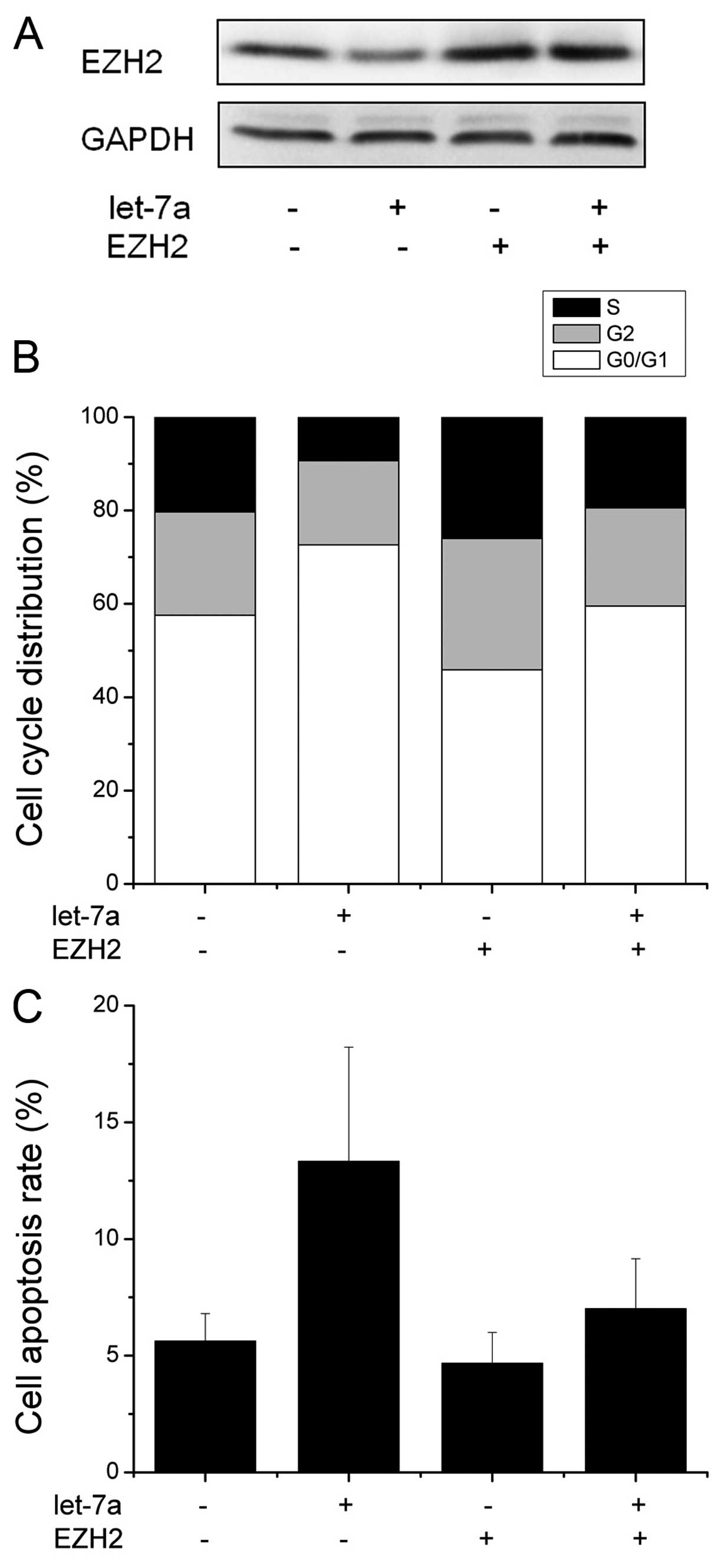
Having demonstrated EZH2 as a direct target of
let-7a, we then examined the importance of EZH2 in let-7a-mediated
cell proliferation and apoptosis. We first transfected EZH2 without
3’UTR into the CNE-2 cells. Western blot analysis showed that the
transfection with EZH2 without 3’UTR overrided EZH2 expression
targeted by let-7a (Fig. 6A). As
shown in Fig. 6B, when we
transfected the cells with EZH2 without 3’UTR and let-7a, the
expression of EZH2 significantly abrogated the effect of let-7a on
cell cycle distribution and apoptosis (Fig. 6B and C). These findings suggest that
EZH2 is a major target of miR-106b involved in nasopharyngeal
carcinoma cell proliferation and apoptosis.
Discussion
Recent evidence has indicated that let-7a plays a
role in the development and progression of human tumors, such as
breast cancer, renal cell carcinoma, gastric and hepatocellular
cancer (9–12). In this study, the overexpression of
let-7a led to the downregulation of EZH2 expression via targeting
the 3’UTR of EZH2, resulting in the inhibition of cell
proliferation, cell cycle arrest at the G0/G1 phase and the
induction of cell apoptosis in the nasopharyngeal carcinoma
cells.
Polycomb repressive complex 2 (PRC2) is recognized
as an important epigenetic group of regulators which function as
transcriptional repressors that silence specific sets of genes
through chromatin modification (13,14).
PRC2 includes EZH2, the suppressor of zeste 12 (SUZ12) and
embryonic ectoderm development (EED). EZH2 is the catalytically
active component of PRC2 and is capable of trimethylating lysine 27
of histone H3 (H3K27) (15–17). Recently, an increasing number of
reports have documented the overexpression of EZH2 in a variety of
human cancers, including prostate cancer, lymphoblastic leukemia,
neuroblastoma and lung cancer (18–21).
Compared with normal nasopharyngeal tissue, the expression levels
of EZH2 are significantly upregulated in nasopharyngeal carcinoma
specimens (22). The increased
expression of EZH2 in nasopharyngeal carcinoma is closely
associated with an aggressive and poor prognostic phenotype
(23). These data suggest the
involvement of EZH2 in disease progression and tumor
aggressiveness, and also suggest that EZH2 may be used as a
prognostic indicator in patients with nasopharyngeal carcinoma. In
this study, we show that EZH2 is overexpressed in nasopharyngeal
carcinoma tissues, and that the suppression of EZH2 expression
inhibits nasopharyngeal carcinoma cell proliferation and induces
apoptosis, consistent with previous data.
EZH2 may be directly regulated by several miRNAs,
such as miR-124, miR-138 and miR-214 (24–26).
In nasopharyngeal carcinoma, miR-26a has been shown to inhibit cell
growth and cell cycle progression by targeting EZH2. The
introduction of EZH2 cDNA abrogates the suppressive effect of
miR-26a (27). In another study of
nasopharyngeal carcinoma, EZH2 has been shown to be a direct target
for miR-98, miR-26a and miR-101 (28). In this study, we show that let-7a
regulates EZH2 expression via targeting the 3’UTR of EZH2, and that
the expression of EZH2 significantly abrogates let-7a-mediated cell
proliferation and apoptosis in nasopharyngeal carcinoma cells.
In conclusion, our results show that let-7a is a
tumor suppressor miRNA in nasopharyngeal carcinoma and that EZH2 is
a novel and critical target of let-7a. Our data suggest that let-7a
and EZH2 may be useful as potential therapeutic targets for
nasopharyngeal carcinoma and warrant further investigation.
Acknowledgements
This study was supported by the China Natural
Science Foundation (81000963), the 333 Talent Program of Jiangsu
Province (BRA2011046), the Kunshan Social Development Foundation
(grant nos. KS1006 and KS1009) and the Suzhou Social Development
Foundation (SYS201063).
References
|
1
|
Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu
Z and You Y: hsa-mir-181a and hsa-mir-181b function as tumor
suppressors in human glioma cells. Brain Res. 1236:185–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Wang X, Zhang J, Sun G, Luo H,
Kang C, Pu P, Jiang T, Liu N and You Y: MicroRNAs involved in the
EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 106:217–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei
RR, Zhang MY, Sun Y, Huang BJ, Chen M, et al: Prognostic value of a
microRNA signature in nasopharyngeal carcinoma: a microRNA
expression analysis. Lancet Oncol. 13:633–641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li T, Chen JX, Fu XP, Yang S, Zhang Z,
Chen K-H and Li Y: microRNA expression profiling of nasopharyngeal
carcinoma. Oncol Rep. 25:1353–1363. 2011.PubMed/NCBI
|
|
5
|
Chen SJ, Chen GH, Chen YH, Liu CY, Chang
KP, Chang YS and Chen HC: Characterization of Epstein-Barr virus
miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One.
5:pii. e127452010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wong TS, Man OY, Tsang CM, Tsao SW, Tsang
RK, Chan JY, Ho WK, Wei WI and To VS: MicroRNA let-7 suppresses
nasopharyngeal carcinoma cells proliferation through downregulating
c-Myc expression. J Cancer Res Clin Oncol. 137:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carbon S, Ireland A, Mungall CJ, Shu S,
Marshall B and Lewis S: AmiGO: online access to ontology and
annotation data. Bioinformatics. 25:288–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW,
Nam SJ and Chun KH: MicroRNA let-7a suppresses breast cancer cell
migration and invasion through downregulation of C-C chemokine
receptor type 7. Breast Cancer Res. 14:R142012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Yin B, Zhang C, Zhou L and Fan J:
Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma
cell lines by targeting c-myc. Biochem Biophys Res Commun.
417:371–375. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Q, Jie Z, Cao H, Greenlee AR, Yang C,
Zou F and Jiang Y: Low-level expression of let-7a in gastric cancer
and its involvement in tumorigenesis by targeting RAB40C.
Carcinogenesis. 32:713–722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus x protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Richly H, Aloia L and Di Croce L: Roles of
the Polycomb group proteins in stem cells and cancer. Cell Death
Dis. 2:e2042011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Margueron R and Reinberg D: The Polycomb
complex PRC2 and its mark in life. Nature. 469:343–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang CJ and Hung MC: The role of EZH2 in
tumour progression. Br J Cancer. 106:243–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao Y: Enhancer of zeste homolog 2: a
potential target for tumor therapy. Int J Biochem Cell Biol.
43:474–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Simon JA and Lange CA: Roles of the EZH2
histone methyltransferase in cancer epigenetics. Mutat Res.
647:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin YJ and Kim JH: The role of EZH2 in
the regulation of the activity of matrix metalloproteinases in
prostate cancer cells. PLoS One. 7:e303932012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ntziachristos P, Tsirigos A, Van
Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D,
da Ros V, Tang Z, Siegle J, et al: Genetic inactivation of the
polycomb repressive complex 2 in T cell acute lymphoblastic
leukemia. Nat Med. 18:298–301. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei
JS, Marquez VE, Bates SE, Jin Q, Khan J, et al: EZH2 Mediates
epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU,
RUNX3, and NGFR. Cancer Res. 72:315–324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huqun, Ishikawa R, Zhang J, Miyazawa H,
Goto Y, Shimizu Y, Hagiwara K and Koyama N: Enhancer of zeste
homolog 2 is a novel prognostic biomarker in nonsmall cell lung
cancer. Cancer. 118:1599–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hwang CF, Huang HY, Chen CH, Chien CY, Hsu
YC, Li CF and Fang FM: Enhancer of zeste homolog 2 overexpression
in nasopharyngeal carcinoma: an independent poor prognosticator
that enhances cell growth. Int J Radiat Oncol Biol Phys.
82:597–604. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ,
Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, et al: EZH2 supports
nasopharyngeal carcinoma cell aggressiveness by forming a
co-repressor complex with HDAC1/HDAC2 and Snail to inhibit
E-cadherin. Oncogene. 31:583–594. 2012.PubMed/NCBI
|
|
24
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Wang C, Chen Z, Jin Y, Wang Y,
Kolokythas A, Dai Y and Zhou X: MicroRNA-138 suppresses
epithelial-mesenchymal transition in squamous cell carcinoma cell
lines. Biochem J. 440:23–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Juan AH, Kumar RM, Marx JG, Young RA and
Sartorelli V: Mir-214-dependent regulation of the polycomb protein
Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell.
36:61–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu J, He ML, Wang L, Chen Y, Liu X, Dong
Q, Chen YC, Peng Y, Yao KT, Kung HF and Li XP: MiR-26a inhibits
cell growth and tumorigenesis of nasopharyngeal carcinoma through
repression of EZH2. Cancer Res. 71:225–233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alajez NM, Shi W, Hui AB, Bruce J,
Lenarduzzi M, Ito E, Yue S, O’Sullivan B and Liu FF: Enhancer of
Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal
carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell
Death Dis. 1:e852010. View Article : Google Scholar : PubMed/NCBI
|