Introduction
Malignant lymphomas (MLs) are the lymph node and/or
extranodal lymphoid malignancies. The lymphoma, in histopathology,
is broadly divided into Hodgkin's lymphoma (HD) and non-Hodgkin's
lymphoma (NHL). Within both subsets are numerous variations with
distinct biological, molecular, and cytogenetic characteristics
(1). NHL is the most common
malignant tumor with rapid incidence growth, a high mortality rate
in children and adolescents and a serious harm to human health
(2). One type of NHL is T-cell
lymphoma (T-NHL). In recent years, with the rapid development of
molecular biology and the completion of the Human Genome Project
indicating that all tumors are the result of abnormal gene
expressions, the causes of lymphoma are now better understood
(3). Studies have shown that
abnormal gene expression caused by oncogenes (4) and tumor suppressor gene mutations
(5), chromosomal translocations
(6) and viral infections are also
significant causes of T-NHL (7).
Astrocyte elevated gene-1 (AEG-1) was originally
characterized as a human immunodeficiency virus (HIV)-1-inducible
gene in primary human fetal astrocytes (8). The AEG-1 gene is located on 8q22.1,
there are 3611 pairs of bases, including 11 introns and 12 exons,
encoding 582 amino acids. Its expression product MTDH distributed
in the perinuclear cytoplasm is a 64-kDa transmembrane protein
(9). In the study of molecular
signaling pathways, AEG-1 is a transmembrane protein in the
endoplasmic reticulum, nuclear membrane and nucleolus (10). In previous in-depth studies of
AEG-1, it was found to promote the occurrence of neurodegeneration,
as well as tumor progression and metastasis. AEG-1 expression is
markedly induced by oncogenic Ha-ras, activating the
phosphatidylinositol 3-kinase signaling pathway that augments
binding of c-Myc to key E-box elements in the AEG-1 promoter,
thereby regulating AEG-1 transcription (11). AEG-1 expression is elevated in
diverse neoplastic states, it cooperates with Ha-ras and c-myc to
promote transformation, and its overexpression augments invasion of
transformed cells. These results strongly suggest that AEG-1 may
play a functionally critical role in Ha-ras-mediated tumorigenesis;
therefore, the role of AEG-1 in tumor growth, angiogenesis, and
metastasis is studied extensively (12–14).
In the present study, we used immunohistochemistry
to detect AEG-1 expression in T-NHL, and RNA interference (RNAi)
technology to silence the expression of AEG-1 in the human T-NHL
cell lines Hut 78 (cutaneous T-cell lymphoma) and Jurkat (adult
T-cell leukemia/lymphoma). In vitro experiments described
herein demonstrate that AEG-1 abnormal expression in T-NHL and the
capability of tumor growth and metastasis is reduced, whereas
apoptosis is induced in AEG-1-siRNA transfected Jurkat and Hut-78
cells. In addition, we demonstrate for the first time the molecular
mechanisms of the antitumor effects of AEG-1 knockdown, which could
lay a foundation for genetic therapy for T-NHL.
Materials and methods
Cell lines
The T-NHL cell lines Jurkat and Hut-78 were used in
these experiments. Jurkat and Hut-78 were maintained in the Key
Clinical Laboratory of Henan Province. Cells were cultured in
RPMI-1640 medium supplemented with 10% heat-inactivated FBS (fetal
bovine serum), 50 U/ml penicillin and 50 U/ml streptomycin
(Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a humidified
atmosphere containing 5% CO2.
Clinical samples
From 2008 to 2011, 129 patients who were diagnosed
with T-NHL and received standard chemotherapy after lymph node
biopsy at The First Affiliated Hospital of Zhengzhou University
were included in the study after obtaining their oral and written
informed consent. The biopsy specimen of each patient was prepared
by the Department of Clinical Pathology for paraffin-embedded tumor
tissue sections. Information on histopathological diagnosis was
extracted from medical records and reviewed by a specialist in
gynecologic tumor pathology. The control group consisted of 17
samples of lymph node that were obtained from standard operations.
This study was reviewed and approved by the ethics committee of the
medical faculty at the First Affiliated Hospital of Zhengzhou
University.
Antibodies and reagents
Antibodies against AEG-1, survivin, Bcl-2 and Bax
were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
β-actin antibody and secondary goat anti-rabbit and goat anti-mouse
alkaline phosphatase antibodies were also purchased from Santa Cruz
Biotechnology. All antibodies were used for western blot analysis
at a dilution of 1:2,000 and 1:5,000, respectively. RPMI-1640, FBS,
Lipofectamine™ 2000 and TRIzol reagent were purchased from
Invitrogen Co. (Carlsbad, CA, USA). MTT and DMSO were obtained from
Sigma.
Immunohistochemistry
Standard immunoperoxidase procedures were used to
visualize AEG-1 expression. Briefly, paraffin sections were
deparaffinized in xylene, followed by a graded series of alcohols
(100, 95 and 75%) and re-hydrated in water followed by
Tris-buffered saline. Following antigen retrieval, slides were
incubated with 3% H2O2 to inhibit endogenous
peroxidase. Slides were then blocked with 5% normal serum and
incubated with anti-AEG-1 antibody. After washing, the tissue
sections were treated with biotinylated anti-rabbit secondary
antibody (Zymed Laboratories Inc., South San Francisco, CA, USA),
followed by further incubation with streptavidin-horseradish
peroxidase complex (Zymed). Tissue sections were then immersed in
3,3′-diaminobenzidine and counterstained with 10% Mayer's
hematoxylin, dehydrated and mounted.
AEG-1 expression in paraffin-embedded sections was
viewed and scored separately by two independent pathologists, who
were blinded to the histopathological features and patient data,
and AEG-1 expression was determined by combining the proportion of
positively stained tumor cells and the intensity of staining
(15).
MTT cell viability assay
To quantify cell proliferation, Jurkat and Hut-78
cells and stably transfected cells (Jurkat-RNAi and Hut-78-RNAi)
were seeded in a 96-well plate at a concentration of
2.5×104/ml (100 μl/well). Six parallel wells were
assigned to each group. Then, 20 μl/well of 5 mg/ml MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was
added at 1, 2, 3, 4, 5 and 6 days after seeding and were then
incubated for another 4 h. The supernatant was removed and the
product converted from MTT was dissolved by adding 150 μl/well
dimethylsulfoxide (DMSO). The plate was gently shaken for 15 min at
room temperature and an enzyme-linked immunosorbent assay reader
was used to measure the absorbance of each well at 570 nm. The
growth curve was drawn according to the cell growth rate.
RNA extraction, reverse transcription and
real-time PCR
Total-RNA from cultured cells was extracted using
the TRIzol reagent according to the manufacturer's instructions.
The cDNA synthesis was carried out according to the protocol of the
Takara Reverse Transcription System for real-time PCR [Takara
Biotechnology (Dalian) Co., Ltd., China] with a starting amount of
2 μg RNA and reverse transcription performed with random primers.
Real-time PCR primers were designed according to www.ncbi.nlm.nih.gov. Primer sequences are: AEG-1
forward, 5′-CTCGGGCTGCTGCTGCTGTT-3′, and reverse, 5′-CAGC
AAGGCCAGGTCGTCGG-3′; Bax forward, 5′-GGCCGG GTTGTCGCCCTTTT-3′, and
reverse, 5′-CCGCTCCCGGA GGAAGTCCA-3′; Bcl-2 forward, 5′-GGACGGGGTGA
ACTGGGGGA-3′, and reverse, 5′-GGTGTGCAGGTGCCGG TTCA-3′; survivin
forward, 5′-CGCTTTGCCACGGTGGT GGA-3′, and reverse,
5′-CAGGGGCGACATCTCCC GGT-3′; β-actin forward,
5′-CGACAACGGCTCCGGCATGT-3′, and reverse,
5′-TGGGCCTCGTCGCCCACATA-3′. Real-time PCR analysis was carried out
on a LightCycler real-time PCR instrument using SYBR Green I kit
(Tiangen Biotech Co., Ltd., Beijing, China) according to the
manufacturer's instructions. Each reaction was performed in
triplicate. Data were analyzed with the Sequence detector software
(v1.9, Applied Biosystems) and analyzed using the 2−ΔΔCT
method as previously described (16).
Western blot analysis
Total proteins were extracted by lysing cells in
buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP-40, 50 mM
NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl
fluoride, 25 mg/ml leupeptin and 25 mg/ml aprotinin. The lysates
were cleared by centrifugation and the supernatants were collected.
Proteins were extracted using the protein extraction kit according
to the manufacturer's instructions. Protein concentration was
determined using protein assay reagent (Bio-Rad, Hercules, CA,
USA). Equal amounts of protein were separated on SDS-PAGE,
transferred to PVDF membranes, incubated with antibodies against
AEG-1, survivin, Bcl-2 and Bax, β-actin, followed by incubation
with the secondary antibodies. The membrane was then washed three
times with TBST and visualized with diaminobenzidine.
Quantification of the proteins was detected with the ECL system
(Pierce Biotechnology Inc., Rockford, IL, USA). Each value
represents the mean of triple experiments, and is presented as the
relative density of protein bands normalized to β-actin.
Plasmid vector constructs,
transfection
The plasmid constructs carrying siRNA
(pSUPER-EGFP-AEG-1) against AEG-1 were designed and constructed as
previously described (17). The
following siRNA sequences were cloned into the pSUPER-EGFP plasmid:
forward, 5′-GCTGATCCCAACTCTGATTTTCAAGAGAAATCAGAGTTGGGATCAGC-3′, and
reverse, 5′-CGACTAGGGTTGAGACTAAAAGTTCTCTTTAGTCTCAACCCTAGTCG-3′. The
recombinant plasmids were transformed into competence bacillus
coli and cultured; they were then extracted, purified,
identified, measured and stored at −20°C. The T-NHL cell line
Jurkat and Hut-78 cells were transfected with pSUPER-EGFP-AEG-1
using Lipo-fectamine 2000 (Invitrogen) according to the
manufacturer's instructions. Forty-eight hours after transfection,
Jurkat and Hut-78 cells were diluted and selected in the medium
containing 500 μg/ml G418 for 4 weeks. Then, the positive clones
were picked up and expanded to establish cell lines after
maintaining to select for 4 weeks. The stable transfection cell
clones Jurkat-RNAi and Hut-78-RNAi were verified for RT-PCR and
western blot analysis.
Cell cycle analysis
Flow cytometric analysis of PI-stained cells was
performed to demonstrate the cell cycle of the different cell
groups. Each different group of cells was cultured in a 10 cm dish,
respectively. After culturing for 48 h, the cells were harvested,
washed and fixed in 70% ethanol for 30 min. After fixation, cells
were resuspended in 1 ml propidium iodide (PI) staining buffer
(0.1% Triton X-100, 100 mg ml-1 RNase A, 500 mg ml-1 PI in PBS) at
37°C for 30 min and the proportion of cells in a particular phase
of the cell cycle was determined by flow cytometry.
Annexin V-FITC flow cytometric
analysis
Annexin V-FITC apoptosis detection kit (BD
Biosciences, San Jose, CA, USA) was used to detect early apoptosis.
In brief, after culturing for 48 h, each group of cells was
harvested, washed twice with pre-chilled PBS and resuspended in
binding buffer (HEPES-NaOH 10 mM pH 7.4, 144 mM NaCl and 25 mM
CaCl2) at a concentration of 1×106 cells/ml.
One hundred microliters of this solution (1×105 cells)
was mixed with 5 μl of Annexin V-FITC and 5 μl of PI (BD
Biosciences) according to the manufacturer's instructions. The
mixed solution was gently vortexed and incubated in the dark at
room temperature (25°C) for 15 min. Four hundred microliters of 1X
dilution buffer were added to each tube and cell apoptosis analysis
was performed by flow cytometry (BD FACSCalibur) within 1 h. At
least 10,000 events were recorded and represented as dot plots.
Cells that stained positive for Annexin V-FITC and negative for PI
were counted as apoptotic cells.
Analysis of MMP-2 and MMP-9 activities by
gelatin zymography
The activities of MMP-2 and MMP-9 tumor cells were
assayed by gelatin zymography. Briefly, each group of cells was
incubated with serum-free medium for 24 h. The conditioned medium
was then harvested and concentrated by ultra-filtration
centrifugation. The sample (20 μg) was mixed with loading buffer
and subjected to gelatin zymogram gel (10% SDS-polyacrylamide gel,
0.1% gelatin). Electrophoresis was performed at 100 V for 3 h at
4°C. Following electrophoresis, gels were washed with washing
buffer (2.5% Triton X-100 in ddH2O) at room temperature
to remove SDS, followed by incubation at 37°C in reaction buffer
(40 mM Tris-HCl, pH 8.0, 10 mM CaCl2, 0.02%
NaN3). After 16 h, the gels were stained with Coomassie
blue R-250 (0.125% Coomassie blue R-250, 50% methanol, 10% acetic
acid) for 3 h and destained with destaining solution (20% methanol,
10% acetic acid, 70% ddH2O) until the clear bands were
visualized. MMP-2 and MMP-9 (gelatinase) activity was visible as
clear bands against the dark blue background, indicating
proteolysis of the substrate protein, gelatin.
Statistical analysis
Each experiment was performed at least twice;
results are expressed as mean ± SEM using Excel software
(Microsoft, Redmond, WA, USA) for calculation. The SPSS13.0
software package (SPSS, Inc., Chicago, IL, USA) was used for all
statistical analyses. Statistical analysis was performed by ANOVA.
Student's t-test was also performed and the results were considered
statistically significant at P<0.05.
Results
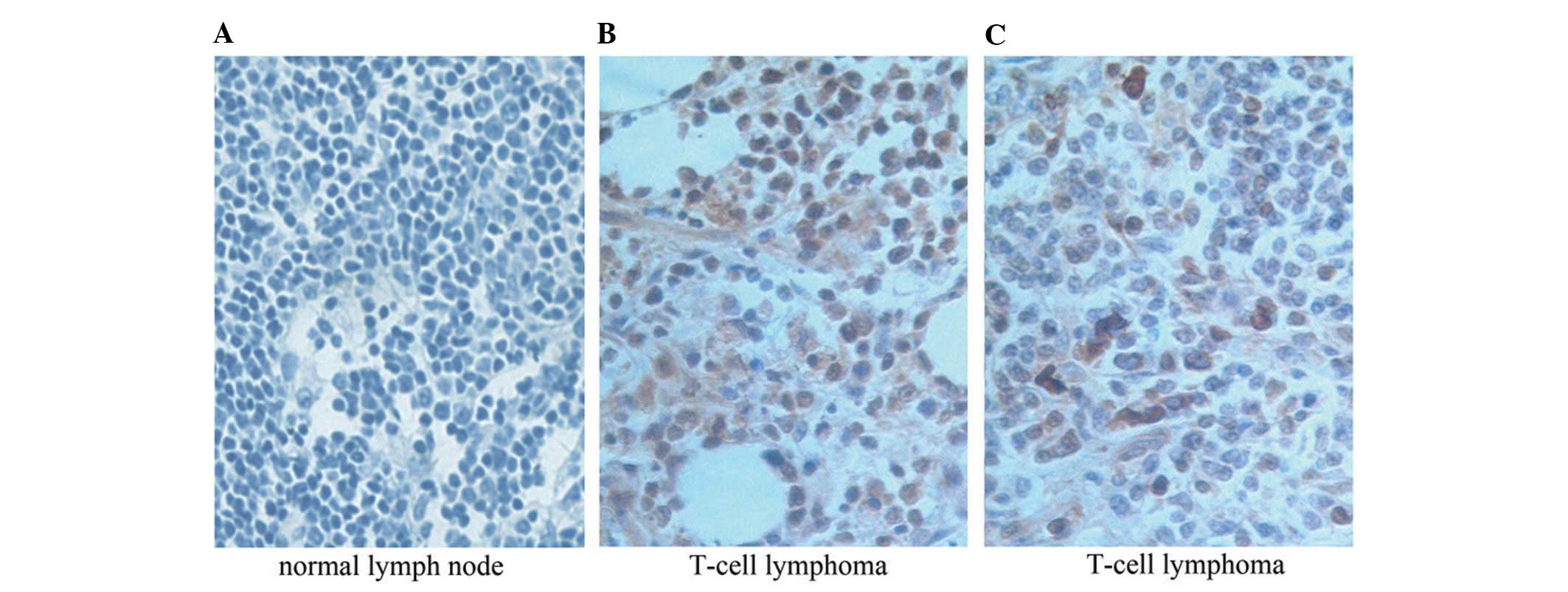
Upregulation of AEG-1 in T-NHL
To examine whether the AEG-1 protein is upregulated
in T-NHL tissues, 129 paraffin-embedded, archived T-NHL tissues
were examined by immunohistochemical staining with an antibody
against human AEG-1. As shown in Fig.
1, the AEG-1 protein was found to be upregulated in T-NHL
compared with normal lymph node tissues. High levels of AEG-1
expression were present in areas containing tumor cells of the
T-NHL. By contrast, AEG-1 was barely detectable in the normal lymph
node tissues (P<0.01, Fig.
1).
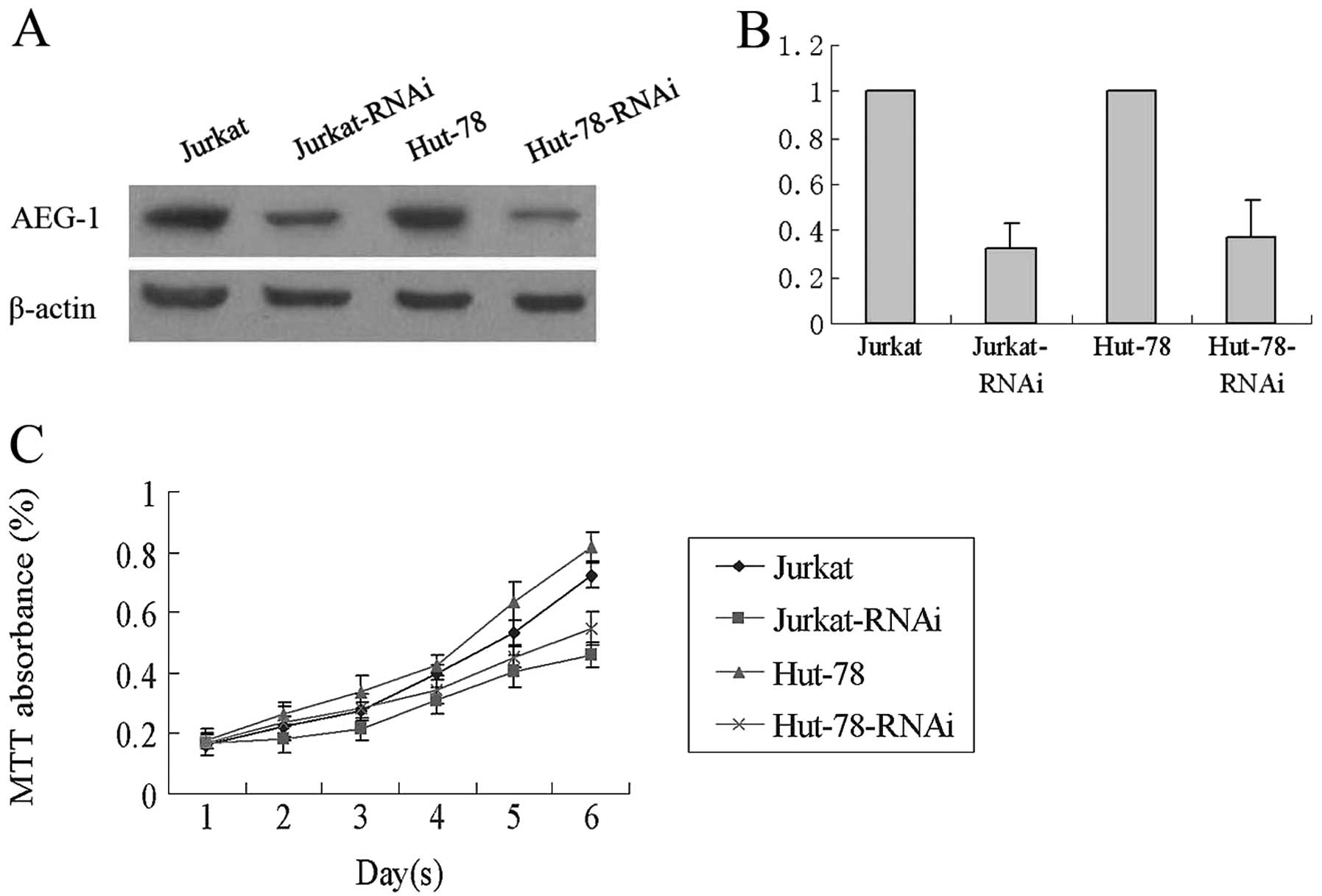
AEG-1-siRNA inhibits Jurkat and Hut-78
cell proliferation
Previous studies have shown that the expression of
AEG-1 is associated with T-NHL growth. To further investigate the
biological role of AEG-1 expression in T-NHL progression, the T-NHL
cell lines Jurkat and Hut-78 were established to stably
low-expressed AEG-1. Western blotting and RT-PCR assay results
indicated that AEG-1-infected Jurkat-RNAi and Hut-78-RNAi cells
have low AEG-1 mRNA and protein expression (P<0.01, Fig. 2A and B). The effect of AEG-1-siRNA
on T-NHL cell proliferation was determined by MTT assay. As shown
in Fig. 2, cell proliferation was
remarkably inhibited in the Jurkat-RNAi and Hut-78-RNAi groups,
when compared with the Jurkat and Hut-78 groups (P<0.05,
Fig. 2C).
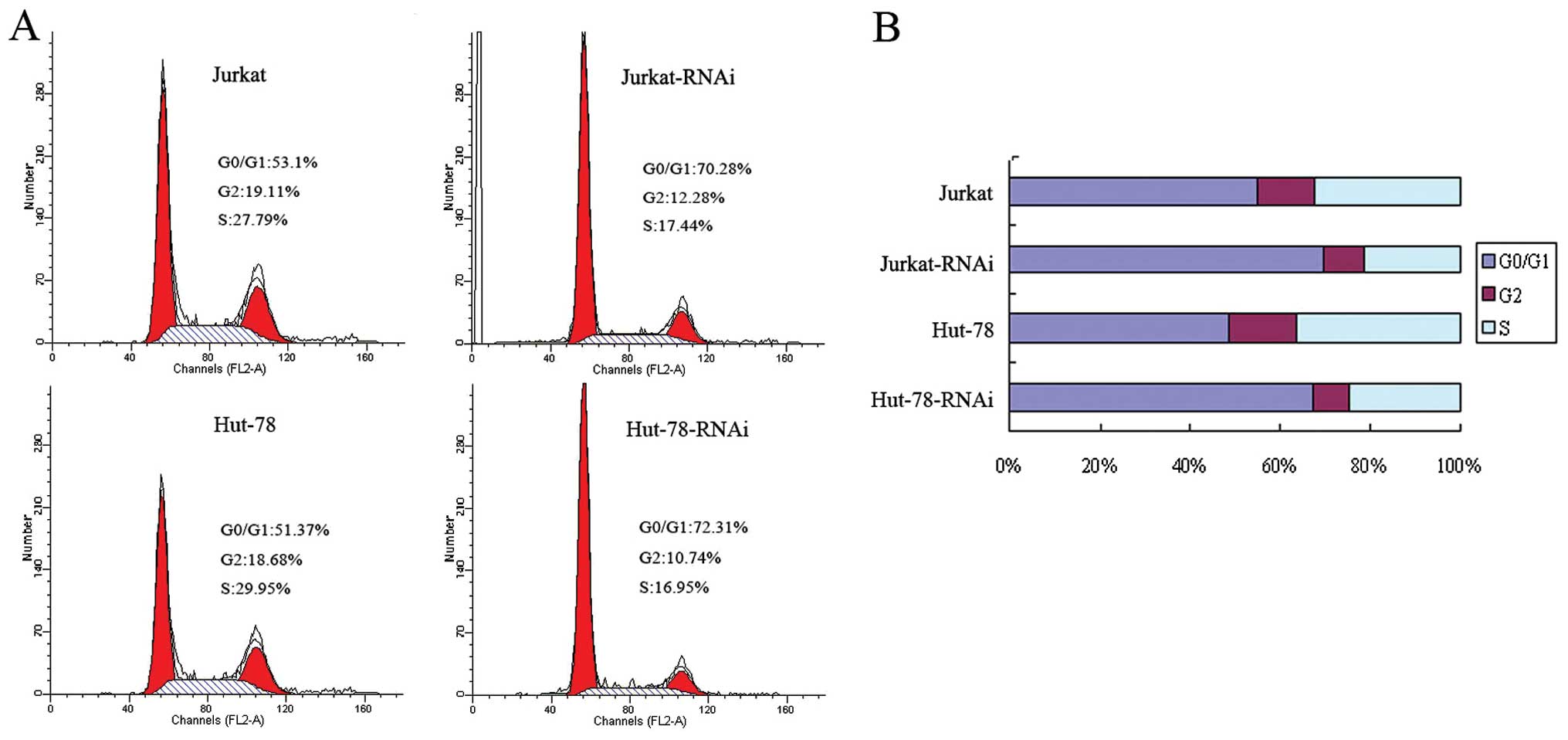
AEG-1-siRNA arrests Jurkat and Hut-78
cells at the G0/G1 phase
To analyze the mechanisms by which AEG-1-siRNA
inhibits cell proliferation, the cell cycle of Jurkat and Hut-78
cells as well as transfected Jurkat-RNAi and Hut-78-RNAi cells were
analyzed by flow cytometry. As shown in Fig. 3, the percentage of Jurkat and Hut-78
cells at the G0/G1 phase was increased from 54.95 and 48.52 to
69.52 and 67.19%; the S-phase cells were decreased from 32.63 and
36.4 to 21.36 and 24.44% (P<0.05, Fig. 3). These data show that silencing of
AEG-1 can arrest cells at the G0/G1 phase.
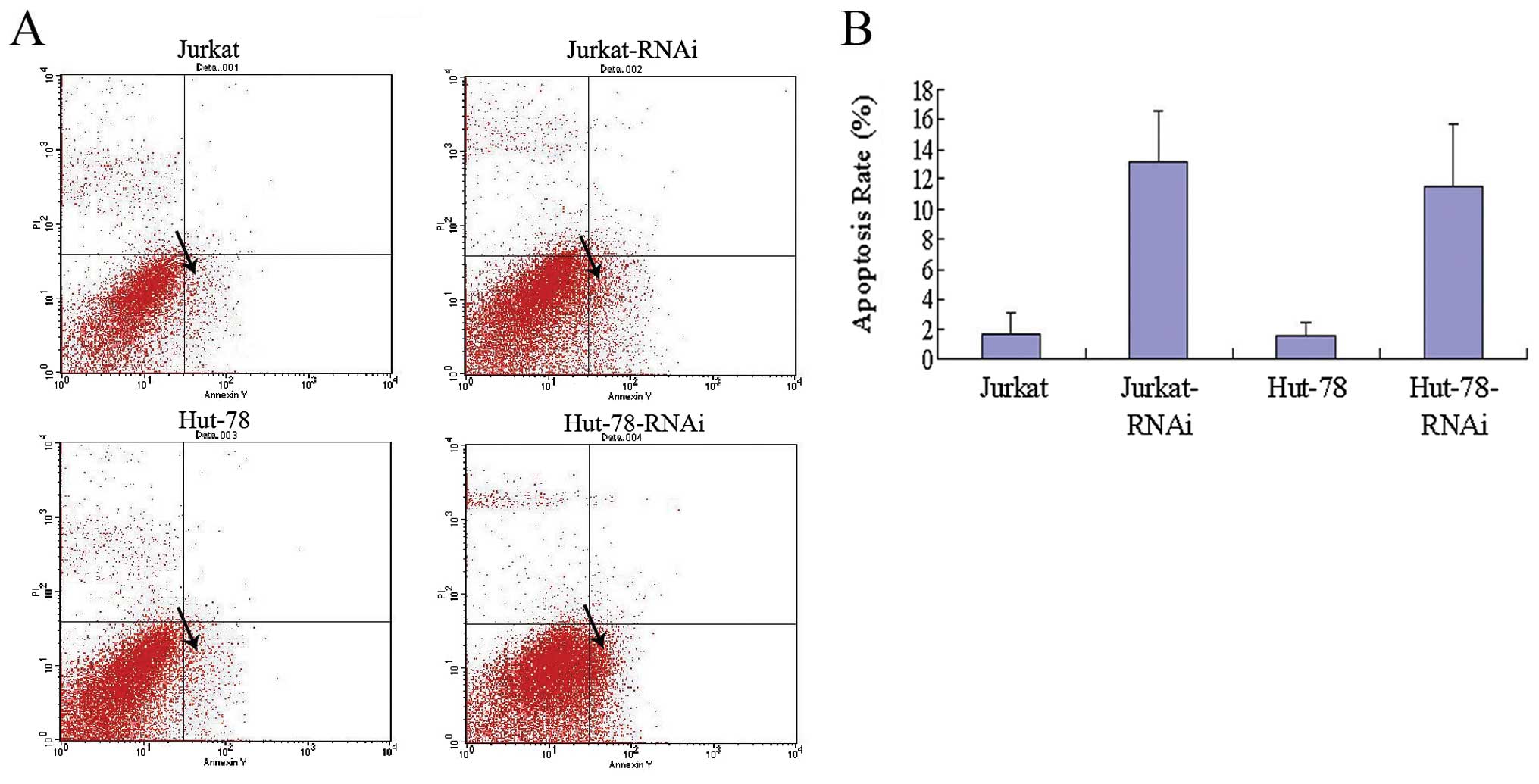
AEG-1-siRNA promotes Jurkat and Hut-78
cell apoptosis
To further evaluate whether knockdown of AEG-1
induces Jurkat and Hut-78 cell apoptosis, at 48 h after culture,
the cells were harvested and analyzed by flow cytometry. As shown
in Fig. 4, the apoptosis rates of
the Jurkat-RNAi and Hut-78-RNAi groups were 13.2 and 11.51%,
respectively, and were >1.64% in the Jurkat and 1.51% in the
Hut-78 group (P<0.01, Fig.
4).
Molecular mechanisms of the antitumor
effects by AEG-1-siRNA
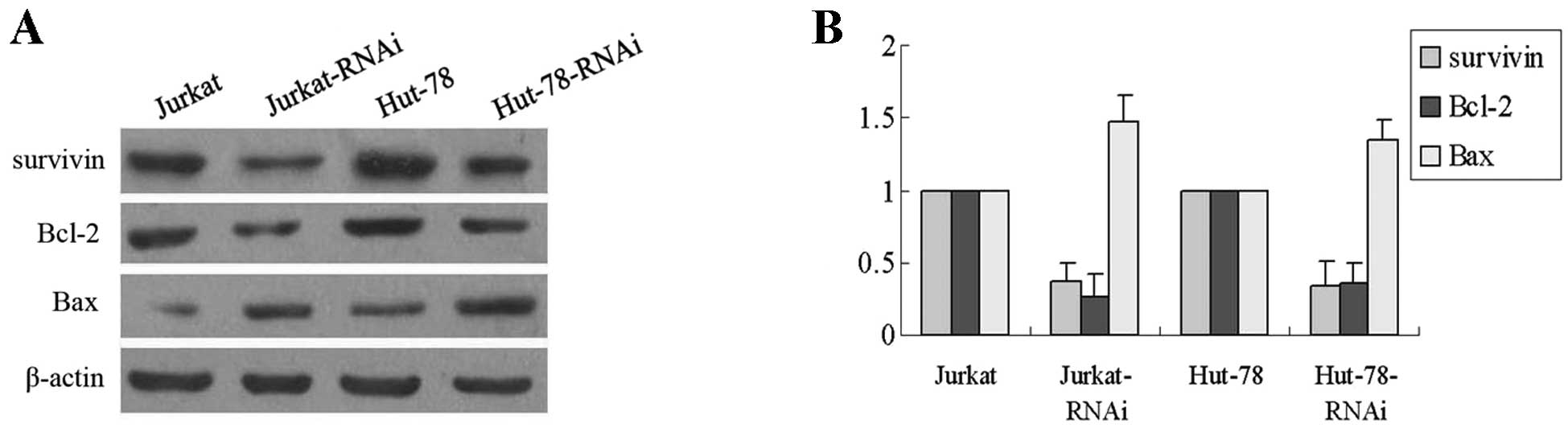
Since deregulation of the AEG-1 expression appears
tightly linked to the apoptosis of Jurkat and Hut-78 cells, we
further investigated whether survivin, Bcl-2 and Bax could be
regulated by AEG-1. Western blot analysis revealed that ectopic
expression of AEG-1 affects the expression of survivin, Bcl-2 and
Bax, whereas the expression of Bax is upregulated and survivin and
Bcl-2 is downregulated in Jurkat-RNAi and Hut-78-RNAi cells
compared with control cells (P<0.01, Fig. 5A). Furthermore, real-time PCR
results indicated that the modulation of survivin, Bcl-2 and Bax by
AEG-1 were regulated at the transcriptional level (P<0.01,
Fig. 5B). These data demonstrate
that knockdown of AEG-1 may affect survivin, Bcl-2 and Bax
expression, which all play an important role in tumor
progression.
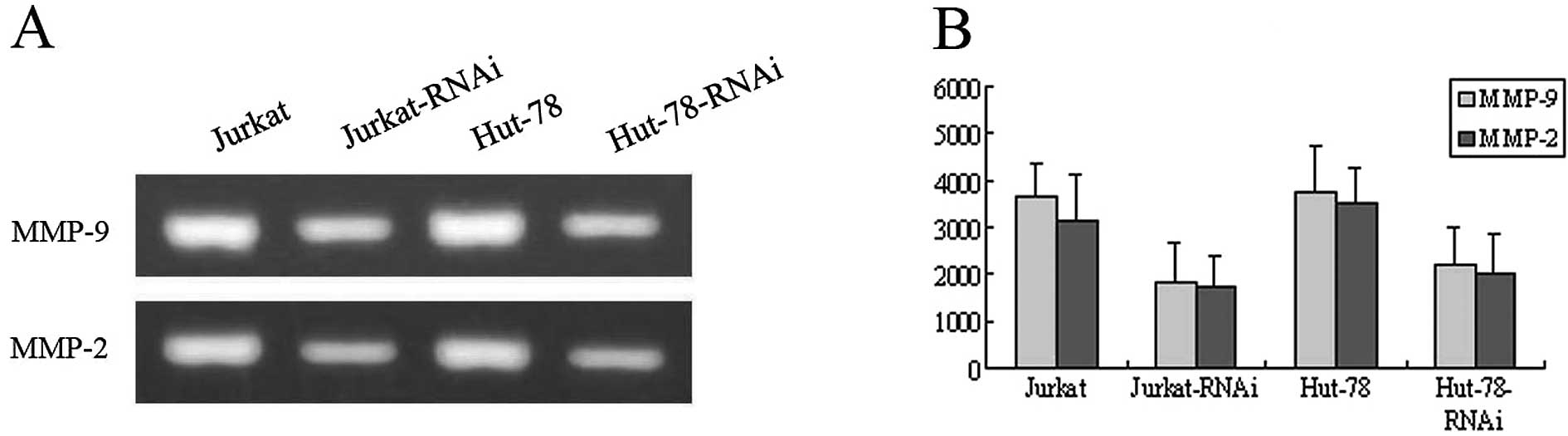
AEG-1-siRNA arrests MMP-2 and MMP-9
activities in Jurkat and Hut-78 cells
Since MMP-2 and MMP-9 play a critical role in tumor
cell invasion, we examined the effects of AEG-1 silencing on the
enzyme activities of MMP-2 and MMP-9 using gelatin zymography. Each
group of cells was incubated in serum-free medium. After culturing
for 24 h, conditioned media in each group were collected and the
expression levels of interest were detected using zymography. The
gelatinolytic activities of both MMP-2 and MMP-9 were found to be
markedly reduced in the Jurkat-RNAi and Hut-78-RNAi groups compared
with the control groups (P<0.01, Fig. 6). There was a significant difference
between the groups (P<0.05).
Discussion
T-NHL is a group of malignant tumors with high
heterogeneity from the lymphatic system accounting for 10–15% of
all NHL, with incidence rates increasing year by year (18). Despite advances in the medical
treatments, and due to its intrinsic properties, T-NHL has a poorer
prognosis compared to its B-cell counterparts (19,20).
T-NHL progression includes tumor cell proliferation, migration and
invasion. Therefore, effectively inhibiting the proliferative and
metastatic biological behavior of T-NHL cells is a key problem to
improving the outcome (21–24).
Recent studies have indicated that AEG-1 plays an
important role in the biological processes of many types of cancer,
and the incidences of tumor development, invasion, metastasis and
resistance are closely related (25–28).
In human gallbladder carcinoma (GBC), there is increased AEG-1
expression in GBC samples and highly invasive GBC-SD cell lines at
both the protein and mRNA levels, and this is strongly correlated
with differentiation degree, Nevin stage, Ki-67 expression and
survival time (29). AEG-1 is
overexpressed in a great portion of epithelial ovarian cancer (EOC)
patients with peritoneal dissemination and/or lymph node metastasis
and this may be clinically useful for predicting metastasis in EOC
(30). AEG-1 overexpression was
also positively correlated with the International Federation of
Gynecology and Obstetrics stage, depth of myometrial invasion,
lymph node metastasis, lymph vascular space invasion, and
recurrence. Patients with high AEG-1 expression had a significantly
poor overall and disease-free survival (31). In this study, we used
immunohistochemical analysis to show that AEG-1 is highly expressed
in human T-NHL and it is confined in the endochylema of the tumor
cells, suggesting that this protein is significantly involved in
T-NHL growth.
Although AEG-1 does not affect MDR1 gene
transcription, it facilitates association of MDR1 mRNA to
polysomes, resulting in increased translation, and AEG-1 also
inhibits ubiquitination and subsequent proteasome-mediated
degradation of the MDR1 protein. This suggests that inhibition of
AEG-1 might provide a means of more effectively using chemotherapy
to treat human hepatocellular carcinoma (HCC). Inhibition of AEG-1
expression by short hairpin RNA can increase doxorubicin toxicity
of chemotherapy drugs to aggressive human HCC cells compared with
either agent alone (32).
Inhibition of AEG-1 expression also clearly inhibited SGC-7901 cell
growth and enhanced cell apoptosis by reducing phosphorylation of
AKT and glycogen synthase kinase (GSK)-3β (Ser 9) and decreasing
the level of β-catenin, lymphoid enhancer binding factor 1 (LEF1),
and cyclin D1 (33). These reports
demonstrate that silencing the expression of AEG-1 in cancer
affects the different types of tumor cell proliferation, apoptosis,
cell cycle, chemoresistance, invasion and metastasis, but the
effect of AEG-1-siRNA on T-NHL has yet to be explored. In this
study, we showed that chemically synthesized siRNAs specifically
targeting AEG-1 successfully knocked down the expression of AEG-1
in both protein and mRNA levels by 64–68% in human T-NHL Jurkat and
Hut-78 cells. The assays herein described detected the effects on
the biological behavior of A549 cells in vitro. Using MTT,
we were able to first show that the proliferation of the
AEG-1-siRNA transfected T-NHL cells is significantly inhibited
in vitro. Using flow cytometric analysis, we revealed that
knockdown of AEG-1 expression induced apoptosis in Jurkat and
Hut-78 cells. Based on these findings, we support that AEG-1 plays
a key role in the proliferation of Jurkat and Hut-78 cells.
The mechanism of AEG-1 on tumor formation and
development is associated with a wide range of downstream signaling
pathways, which makes AEG-1 a potentially significant molecule and
a complex network of signaling pathways in malignant progression of
human tumors (34–36). However the molecular mechanism of
apoptosis induced by the inhibition of AEG-1 expression remains
unclear. In the current study, we established that downregulation
of AEG-1 expression in Jurkat and Hut-78 cells suppressed the
protein expression of survivin and Bcl-2, whereas it upregulated
the protein expression of Bax, which are well known for mediating
cell apoptosis (37–39). Therefore, this result explains, at
least in part, why AEG-1-siRNA inhibited cell proliferation and
induced apoptosis in Jurkat and Hut-78 cells.
At the molecular level, this study showed that
upregulated AEG-1 in glioma cells interacted with MMP-9 promoter
and transactivated MMP-9 expression, whereas knockdown of AEG-1
expression reduced the level of MMP-9 (40). Mechanistic studies conducted in
vitro and in vivo revealed that AEG-1-mediated
carcinogenesis and invasiveness might be through upregulation of
MMP-2 (41). Our study also found
the activities of MMP-2/-9 were also downregulated in AEG-1-siRNA
transfected Jurkat and Hut-78 cells, and this may suggest that
MMP-2/-9 are the downstream products of the AEG-1-induced cell
signaling. MMPs are a family of enzymes that degrade proteins in
tissue extracellular matrices, which are clearly involved in cancer
progression, including tumor cell degradation of basement membranes
and stroma and blood vessel penetration (42,43).
Consequently, the reduction of MMP-2/-9 activities by AEG-1-siRNA
results in attenuating the metastatic potency of Jurkat and Hut-78
cells.
In conclusion, our findings clearly demonstrate that
AEG-1 plays a crucial role in T-NHL growth and metastasis. Our
study is the first to show that silencing of AEG-1 expression in
Jurkat and Hut-78 cells by AEG-1-siRNA could inhibit cell growth
and metastasis in vitro, and the antitumor effects may be
associated with inhibition of survivin, the Bcl-2/Bax and MMP-2/-9
signal pathways in Jurkat and Hut-78 cells. Therefore, RNA
interference targeting AEG-1 may be a beneficial potential tool for
the gene therapy of T-NHL as well as other types of cancer at high
levels of AEG-1 expression.
Abbreviations:
|
AEG-1
|
astrocyte elevated gene-1
|
|
T-NHL
|
T-cell non-Hodgkin's lymphoma
|
References
|
1
|
Ramsdale E, van Besien K and Smith SM:
Personalized treatment of lymphoma: promise and reality. Semin
Oncol. 38:225–235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hadzi-Pecova L, Petrusevska G and
Stojanovic A: Non-Hodgkin's lymphomas: immunologic prognostic
studies. Prilozi. 28:39–55. 2007.
|
|
3
|
Alizadeh AA, Ross DT, Perou CM and van de
Rijn M: Towards a novel classification of human malignancies based
on gene expression patterns. J Pathol. 195:41–52. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, de Reyniès A, de Leval L, Ghazi
B, Martin-Garcia N, Travert M, Bosq J, Brière J, Petit B, Thomas E,
et al: Gene expression profiling identifies emerging oncogenic
pathways operating in extranodal NK/T-cell lymphoma, nasal type.
Blood. 115:1226–1237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kleppe M, Tousseyn T, Geissinger E,
Kalender Atak Z, Aerts S, Rosenwald A, Wlodarska I and Cools J:
Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin's
lymphoma and T-cell non-Hodgkin's lymphoma. Haematologica.
96:1723–1727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernberg P, Chang ET, Duvefelt K, Hjalgrim
H, Eloranta S, Sørensen KM, Porwit A, Humphreys K, Melbye M and
Ekström Smedby K: Genetic variation in chromosomal translocation
breakpoint and immune function genes and risk of non-Hodgkin
lymphoma. Cancer Causes Control. 21:759–769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramos JC and Lossos IS: Newly emerging
therapies targeting viral-related lymphomas. Curr Oncol Rep.
13:416–426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Emdad L, Sarkar D, Su ZZ, Randolph A,
Boukerche H, Valerie K and Fisher PB: Activation of the nuclear
factor kappaB pathway by astrocyte elevated gene-1: implications
for tumor progression and metastasis. Cancer Res. 66:1509–1516.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anttila V, Stefansson H, Kallela M, et al:
Genome-wide association study of migraine implicates a common
susceptibility variant on 8q22.1. Nat Genet. 42:869–873. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky
DJ and Fisher PB: Cloning and characterization of HIV-1-inducible
astrocyte elevated gene-1, AEG-1. Gene. 353:8–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee SG, Su ZZ, Emdad L, Sarkar D and
Fisher PB: Astrocyte elevated gene-1 (AEG-1) is a target gene of
oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc.
Proc Natl Acad Sci USA. 103:17390–17395. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Emdad L, Sarkar D, Su ZZ, Lee SG, Kang DC,
Bruce JN, Volsky DJ and Fisher PB: Astrocyte elevated gene-1:
recent insights into a novel gene involved in tumor progression,
metastasis and neurodegeneration. Pharmacol Ther. 114:155–170.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu G, Wei Y and Kang Y: The multifaceted
role of MTDH/AEG-1 in cancer progression. Clin Cancer Res.
15:5615–5620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoo BK, Emdad L, Lee SG, Su ZZ,
Santhekadur P, Chen D, Gredler R, Fisher PB and Sarkar D: Astrocyte
elevated gene-1 (AEG-1): A multifunctional regulator of normal and
abnormal physiology. Pharmacol Ther. 130:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu C, Chen K, Zheng H, Guo X, Jia W, Li M,
Zeng M, Li J and Song L: Overexpression of astrocyte elevated
gene-1 (AEG-1) is associated with esophageal squamous cell
carcinoma (ESCC) progression and pathogenesis. Carcinogenesis.
30:894–901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan B, Wang YX, Yao T and Zhu YC: p38
mitogen-activated protein kinase mediates hypoxia-induced vascular
endothelial growth factor release in human endothelial cells. Acta
Physiol Sin. 57:13–20. 2005.
|
|
17
|
Shen YM, Yang XC, Song ML, Qin CH, Yang C
and Sun YH: Growth inhibition induced by short hairpin RNA to
silence survivin gene in human pancreatic cancer cells.
Hepatobiliary Pancreat Dis Int. 9:69–77. 2010.PubMed/NCBI
|
|
18
|
Vose JM: Peripheral T-cell non-Hodgkin's
lymphoma. Hematol Oncol Clin North Am. 22:997–1005. 2008.
View Article : Google Scholar
|
|
19
|
Fornari A, Piva R, Chiarle R, Novero D and
Inghirami G: Anaplastic large cell lymphoma: one or more entities
among T-cell lymphoma? Hematol Oncol. 27:161–170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Windsor R, Stiller C and Webb D:
Peripheral T-cell lymphoma in childhood: population-based
experience in the United Kingdom over 20 years. Pediatr Blood
Cancer. 50:784–787. 2008.PubMed/NCBI
|
|
21
|
Kossakowska AE, Edwards DR, Prusinkiewicz
C, Zhang MC, Guo D, Urbanski SJ, Grogan T, Marquez LA and
Janowska-Wieczorek A: Interleukin-6 regulation of matrix
metalloproteinase (MMP-2 and MMP-9) and tissue inhibitor of
metalloproteinase (TIMP-1) expression in malignant non-Hodgkin's
lymphomas. Blood. 94:2080–2089. 1999.PubMed/NCBI
|
|
22
|
Chow KU, Boehrer S, Geduldig K, Krapohl A,
Hoelzer D, Mitrou PS and Weidmann E: In vitro induction of
apoptosis of neoplastic cells in low-grade non-Hodgkin's lymphomas
using combinations of established cytotoxic drugs with
bendamustine. Haematologica. 86:485–493. 2001.
|
|
23
|
Sebti Y, Le Maux A, Gros F, De Guibert S,
Pangault C, Rouas-Freiss N, Bernard M and Amiot L: Expression of
functional soluble human leucocyte antigen-G molecules in
lymphoproliferative disorders. Br J Haematol. 138:202–212. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang J, Wang S, Zhao G and Sun B: Effect
of chemokine receptors CCR7 on disseminated behavior of human T
cell lymphoma: clinical and experimental study. J Exp Clin Cancer
Res. 30:51–59. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sarkar D, Emdad L, Lee SG, Yoo BK, Su ZZ
and Fisher PB: Astrocyte elevated gene-1: far more than just a gene
regulated in astrocytes. Cancer Res. 69:8529–8535. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Ke Z, Shi H, Yang S and Wang L:
Overexpression of AEG-1 in renal cell carcinoma and its correlation
with tumor nuclear grade and progression. Neoplasma. 57:522–529.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song H, Li C, Li R and Geng J: Prognostic
significance of AEG-1 expression in colorectal carcinoma. Int J
Colorectal Dis. 25:1201–1209. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li C, Li R, Song H, Wang D, Feng T, Yu X,
Zhao Y, Liu J, Yu X, Wang Y and Geng J: Significance of AEG-1
expression in correlation with VEGF, microvessel density and
clinicopathological characteristics in triple-negative breast
cancer. J Surg Oncol. 103:184–192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun W, Fan YZ, Xi H, Lu XS, Ye C and Zhang
JT: Astrocyte elevated gene-1 overexpression in human primary
gallbladder carcinomas: An unfavorable and independent prognostic
factor. Oncol Rep. 26:1133–1142. 2011.PubMed/NCBI
|
|
30
|
Li C, Liu J, Lu R, Yu G, Wang X, Zhao Y,
Song H, Lin P, Sun X, Yu X, Zhang Y, Ning X and Geng J: AEG-1
overexpression: a novel indicator for peritoneal dissemination and
lymph node metastasis in epithelial ovarian cancers. Int J Gynecol
Cancer. 21:602–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song H, Li C, Lu R, Zhang Y and Geng J:
Expression of astrocyte elevated gene-1: a novel marker of the
pathogenesis, progression, and poor prognosis for endometrial
cancer. Int J Gynecol Cancer. 20:1188–1196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoo BK, Chen D, Su ZZ, Gredler R, Yoo J,
Shah K, Fisher PB and Sarkar D: Molecular mechanism of
chemoresistance by astrocyte elevated gene-1. Cancer Res.
70:3249–3258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu JB, Wu H, He YL, Zhang CH, Zhang LJ,
Cai SR and Zhan WH: Astrocyte-elevated gene-1 overexpression is
associated with poor prognosis in gastric cancer. Med Oncol.
28:455–462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song L, Li W, Zhang H, Liao W, Dai T, Yu
C, Ding X, Zhang L and Li J: Over-expression of AEG-1 significantly
associates with tumour aggressiveness and poor prognosis in human
non-small cell lung cancer. J Pathol. 219:317–326. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Khuda II, Koide N, Noman AS, Dagvadorj J,
Tumurkhuu G, Naiki Y, Komatsu T, Yoshida T and Yokochi T: Astrocyte
elevated gene-1 (AEG-1) is induced by lipopolysaccharide as
toll-like receptor 4 (TLR4) ligand and regulates TLR4 signalling.
Immunology. 128:700–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche
H, Sarkar D and Fisher PB: Astrocyte elevated gene-1 (AEG-1)
functions as an oncogene and regulates angiogenesis. Proc Natl Acad
Sci USA. 106:21300–21305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kehinde EO, Maghrebi MA and Anim JT: The
importance of determining the aggressiveness of prostate cancer
using serum and tissue molecular markers. Can J Urol. 15:3967–3974.
2008.PubMed/NCBI
|
|
38
|
Nardiello T, Jungbluth AA, Mei A, et al:
MAGE-A inhibits apoptosis in proliferating myeloma cells through
repression of Bax and maintenance of survivin. Clin Cancer Res.
17:4309–4319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jamieson NB, Carter CR, McKay CJ and Oien
KA: Tissue biomarkers for prognosis in pancreatic ductal
adenocarcinoma: a systematic review and meta-analysis. Clin Cancer
Res. 17:3316–3331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu L, Wu J, Ying Z, Chen B, Han A, Liang
Y, Song L, Yuan J, Li J and Li M: Astrocyte elevated gene-1
upregulates matrix metalloproteinase-9 and induces human glioma
invasion. Cancer Res. 70:3750–3759. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang F, Ke ZF, Sun SJ, Chen WF, Yang SC,
Li SH, Mao XP and Wang LT: Oncogenic roles of astrocyte elevated
gene-1 (AEG-1) in osteosarcoma progression and prognosis. Cancer
Biol Ther. 12:539–548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nelson AR, Fingleton B, Rothenberg ML and
Matrisian LM: Matrix metalloproteinases: biologic activity and
clinical implications. J Clin Oncol. 18:1135–1149. 2000.PubMed/NCBI
|
|
43
|
Peschos D, Damala C, Stefanou D, Tsanou E,
Assimakopoulos D, Vougiouklakis T, Charalabopoulos K and Agnantis
NJ: Expression of matrix metalloproteinase-9 (gelatinase B) in
benign, premalignant and malignant laryngeal lesions. Histol
Histopathol. 21:603–608. 2006.PubMed/NCBI
|