Introduction
Colorectal adenocarcinoma is one of the most common
malignancies, of which the prevalence has been increasing recently.
With the second highest incidence among all cancers, it is also the
second most common cause of cancer-related mortality worldwide
(1,2). The carcinogenesis of colorectal
adenocarcinoma is a multi-step process that involves a number of
genomic alterations. Understanding the molecular mechanism of this
process may aid in cancer prevention, early diagnosis and effective
treatment.
Using a subtractive cDNA library and cDNA microarray
analysis, a total of 86 expressed sequence taqs were previously
identified from human colorectal adenocarcinoma tissues (3). In the present study, we mainly focused
on the FBXL20 gene (GenBank accession no., NM032875.2),
which was identified using the BLASTn program of NCBI from one
differentially expressed sequence taq (GenBank accession no.,
ES274070). This gene contains 10,381 base pairs, with a 1308-bp
open reading frame. It is located on the human chromosome 17q21.2,
predicted to encode a 436-amino acid protein containing an F-box
motif, which is the key feature of F-box proteins (FBPs) (4,5). FBPs
are defined by the presence of a 40-amino acid domain that can
interact with other proteins. It was originally identified in
cyclin F, therefore named F-box. Studies on different species have
shown that FBPs play a crucial role in the ubiquitin-mediated
degradation of cellular regulatory proteins (6). However, the function of FBXL20 remains
unknown.
In order to determine whether FBXL20 is involved in
the carcinogenesis of colorectal adenocarcinoma, the mRNA
expression of FBXL20 in human colorectal adenocarcinoma and
normal colorectal tissues was determined by real-time PCR (RT-PCR).
Two new cell lines, SW480−FBXL20 and
SW620−FBXL20, in which the FBXL20 gene was
permanently silenced, were generated to investigate the role of
FBXL20 in this pathogenic process.
Materials and methods
Collection of tissue samples
This study was approved by the Medical Research
Ethics Committee of Sichuan University, Chengdu, China. Written
informed consent was obtained from the patients. Colorectal
adenocarcinoma tissues and adjacent normal colorectal tissues were
obtained from 30 patients at the West China Hospital of Sichuan
University. Normal colorectal tissues were collected from the area
at least 5 cm away from the edge of the tumor tissues. Pathological
examination further confirmed the absence of cancer cells in the
collected normal tissues, which were snap-frozen and stored in
liquid nitrogen.
Quantitative RT-PCR (qRT-PCR)
According to the manufacturer’s protocol, total RNA
from tumor and matched normal tissues was isolated using the TRIzol
RNA isolation reagent (Invitrogen Life Technologies, USA). Total
RNA was reverse-transcribed using the M-MuLV Reverse Transcriptase
kit (Fermentas, Burlington, Canada). RT-PCR was performed using
SYBR Premix ExTaq (Takara, Otsu, Japan), according to the
manufacturer’s instructions and related international standards
(7). GAPDH was used as the
endogenous control. PCR primers specific for FBXL20 and
GAPDH are shown in Table
I.
 | Table IQuantitative real-time PCR. |
Table I
Quantitative real-time PCR.
| Name | Sequences | Product
location | Product length
(bp) |
|---|
| FBXL20 | Forward:
5′-gtcttcgtggatgtcttggag-3′ | 544–564 | 152 |
| Reverse:
3′-gtgcctgagtttggaacagaac-5′ | 695-674 | |
| GAPDH | Forward:
5′-ggaaggtgaaggtcggagt-3′ | 107–125 | 117 |
| Reverse:
3′-ctggggaaagtaactggagt-5′ | 223-205 | |
|
β-catenin | Forward:
5′-gctggtgacagggaagacatc-3′ | 1628–1648 | 116 |
| Reverse:
3′-ggtagtccatagtgaaggcgaac-5′ | 1743-1721 | |
|
E-cadherin | Forward:
5′-ttgctactggaacagggacact-3 | 1845–1866 | 154 |
| Reverse:
3′-ggagatgtattgggaggaaggtc-5′ | 1998-1976 | |
| c-Myc | Forward:
5′-tcaagaggcgaacacacaac-3′ | 1631–1550 | 110 |
| Reverse:
3′-ggccttttcattgttttcca-5′ | 1740-1721 | |
| cyclin
D1 | Forward:
5′-gtggcctctaagatgaaggaga-3′ | 534–555 | 169 |
| Reverse:
3′-ggaagtgttcaatgaaatcgtg-5′ | 702-681 | |
| PP2A | Forward: 5′
-agttggccaaatgtgtctcc -3′ | 1129–1148 | 145 |
| Reverse:
3′-gagttgcggtacaaggaagg-5′ | 1273-1254 | |
| SET | Forward:
5′-gctcaactccaaccacgac-3′ | 389–407 | 120 |
| Reverse:
3′-attacttgttcggtcactcct-5′ | 508-488 | |
| p53 | Forward:
5′-tagtgtggtggtgccctatgag-3′ | 524–545 | 129 |
| Reverser:
3′-agtgtgatgatggtgaggatgg-5′ | 652-631 | |
Cell culture
The human colon adenocarcinoma cell lines, SW480 and
SW620 (ATCC, USA), were cultured in DMEM (10% FBS, 100 U/ml
penicillin and 100 U/ml streptomycin) at 37°C with 5%
CO2.
Construction of the pGPU6/GFP/Neo-FBXL20
siRNA expression vector
The synthesized 59-mer oligonucleotide containing
the sequence specifically silencing the FBXL20 gene was
inserted into the pGPU6/GFP/Neo siRNA expression vector. The sense
sequence of this oligonucleotide was as follows:
5′-CACCGCCAAATGCTTAGCCAATCTTTC AAGAGAAGATTGGCTAAGCATTTGGTTTTTTG-3′;
and the antisense sequence of the oligonucleotide was as follows:
5′-GATCCAAAAAACCAAATGCTTAGCCAATCTTCTC TTGAAAGATTGGCTAAGCATTTGGC-3′.
The two annealed complementary oligonucleotides were subsequently
inserted into the BamHI/BbsI site of the
PGPU6/GFP/Neo siRNA expression vector. The engineered
PGPU6/GFP/Neo-FBXL20 siRNA expression vector was verified by
restriction digestion and then sequenced by the Beijing Genomic
Institute (Beijing, China). The pGPU6/GFP/Neo negative control
siRNA vector was provided by GenePharma Company (Shanghai,
China).
Establishing the SW480−FBXL20
and SW620−FBXL20 cell lines
According to the Lipofectamine 2000 (Invitrogen Life
Technologies) manufacturer’s instructions, the SW480 and SW620
cells were transfected with 1 μg of the pGPU6/GFP/Neo-FBXL20
siRNA expression vector. After 48 h, the transfected SW480 and
SW620 cells were treated with 800 μg/ml G418 (Sigma, St.
Louis, MO, USA). After 14 days, monoclonal cells were cultured in
the presence of 400 μg/ml G418. The new cell lines generated
from the corresponding monoclonal cells were named
SW480−FBXL20 and SW620−FBXL20. The
SW480−negatice control (SW480−NC) and
SW620−negatice control (SW620−NC) cell lines
were generated by the transfection of the pGPU6/GFP/Neo negative
control siRNA vector into the SW480 and SW620 cell lines.
SW480−FBXL20 and SW620−FBXL20 cells were
regarded as the experimental group, SW480−NC and
SW620−NC cells were regarded as the negative group, and
SW480 and SW620 cells were regarded as the blank group in the
following experiments.
MTT assay
MTT assay was performed to examine cell
proliferation. The experiment was repeated three times. Cells in
the logarithmic phase were cultured in 96-well culture plates.
Three groups were set up as follows: the blank, the negative and
the experimental group. Each group contained six replicates. The
MTT reagent (20 μl) (Sigma) was added at four time-points
(one, two, three and four days). After the cells were cultured for
an additional 4 h, the medium was removed, and 150 μl of
DMSO were added into each well. The plate was read at the
absorbance wavelength of 490 nm by a microplate reader (Bio-Rad,
Hercules, CA, USA).
Flow cytometry
Cell apoptosis was measured by flow cytometry
(FACSAria II cell sorter; BD Biosciences, San Jose, CA, USA). The
experiment was repeated three times. Cells in the logarithmic phase
were collected and cultured in six-well culture plates. Three
groups (each in triplicate) were set up as follows: the blank, the
negative and the experimental group. Briefly, cells were collected
48 h post-plating, after being washed twice with PBS. The cells
were suspended in 400 μl of binding buffer (BestBio,
Shanghai, China), after which 5 μl of Annexin V-FITC were
added followed by gentle agitation. The mixture was incubated for
10 min at room temperature. After incubation, 10 μl of PI
were added into the mixture, followed by 10 min of incubation on
ice. Cell apoptosis was then measured by flow cytometry. For the
cell cycle assay, the cells were collected 60 h post-plating and
washed twice using ice-cold PBS, after which the cells were fixed
in 75% ice-cold ethanol at −20°C for 48 h. Finally, the cell cycle
assay was performed using the Cell Cycle Detection kit
(BestBio).
RT-PCR for the detection of the mRNA
expression of β-catenin, SET, E-cadherin, c-Myc, cyclin D1, p53 and
PP2A
Real-time PCR was performed as stated. Quantitative
real-time PCR primers of these genes are shown in Table I.
Western blot analysis for the detection
of the protein expression of β-catenin, SET, c-Myc and
caspase-3
The cells were lysed in ice-cold RIPA buffer
(Beyotime, China). The total protein concentration was determined
using an Enhanced BCA Protein assay kit (Beyotime). The equivalent
amount of protein was then separated by 12% SDS-PAGE and was
transferred onto PVDF membranes (Bio-Rad). After blocking in
Tris-buffered saline (TBS) containing 5% non-fat milk, the
membranes were incubated with primary antibodies (at 4°C for 12 h,
followed by incubation with horseradish peroxidase (HRP) conjugated
anti-rabbit antibody (all from Santa Cruz Biotechnologies, Inc.,
Santa Cruz, CA, USA) at room temperature for 1 h. Signals were
detected on a gel imaging system using the ECL western blotting
substrate (Thermo Scientific, USA). The expression of β-actin was
used as the loading control.
Statistical analysis
The results presented in the current study are the
means ± standard error of the mean (SEM). Statistical analysis was
performed using the Student’s t-test, the Fisher’s exact
probability test and ANOVA with SPSS17.0 software. P<0.05 was
considered to indicate a statistically significant difference.
Results
mRNA expression level of FBXL20 in human
colorectal adenocarcinoma tissues
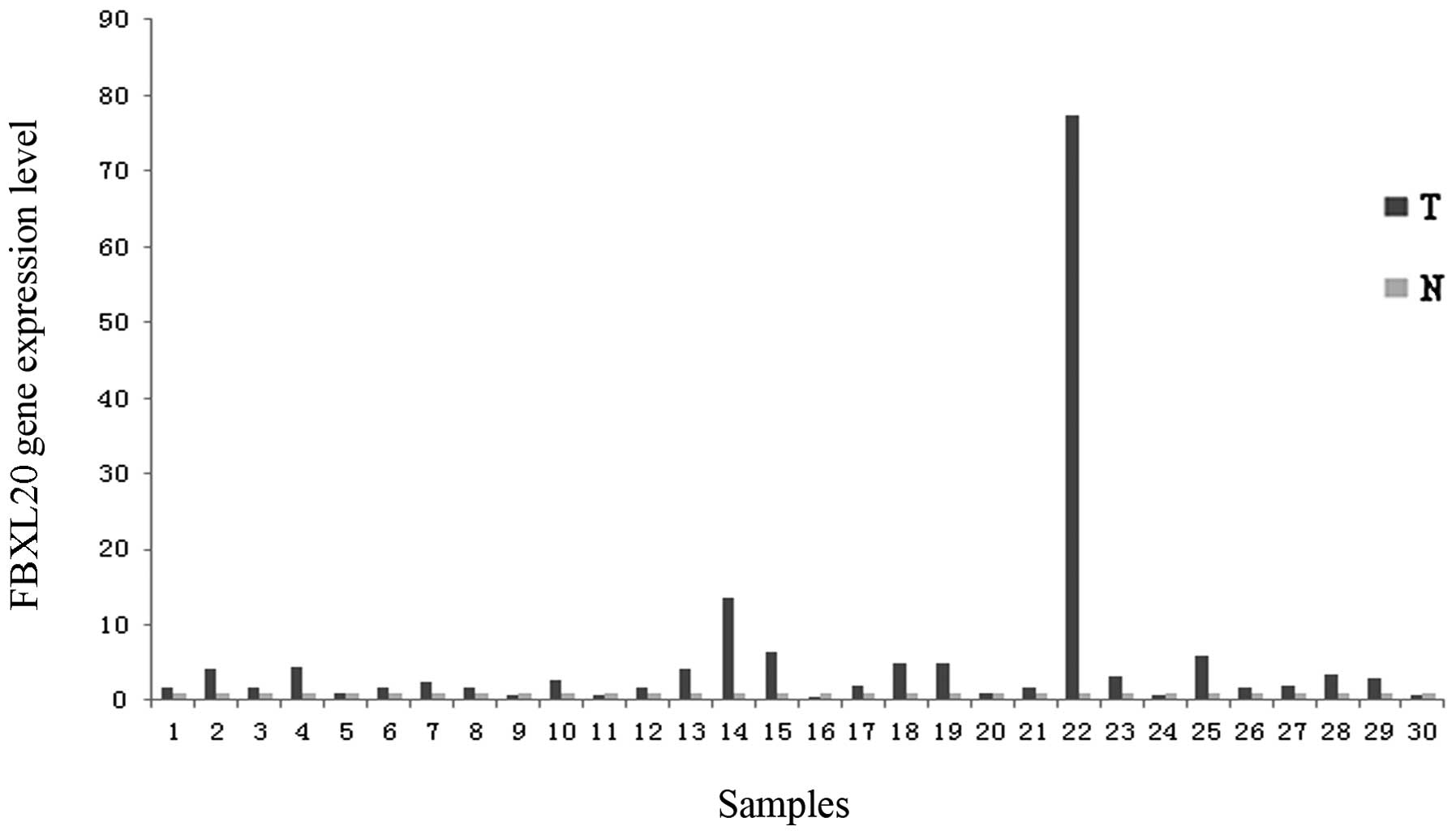
The mRNA expression level of FBXL20 was
upregulated in 76.7% of the tumor samples (23 out of 30 samples),
which was statistically significant (P=0.000, by the one-sample
t-test). Compared to the adjacent normal colorectal tissues, 14
samples exhibited a fold-change of >2, ranging from 2.445 to
77.35 (Fig. 1). Of note, the
FBXL20 expression in older patients (>50 years old) was
significantly higher than that in younger patients (<50 years
old) (P=0.047). No significant correlation was found between
FBXL20 mRNA expression and gender (P=0.815), Dukes’ stage
(P=0.284) or differentiation degree (P=0.643) by the ANOVA test
(Table II).
 | Table IIFBXL20 expression and patient
characteristics. |
Table II
FBXL20 expression and patient
characteristics.
| Gender | Age | Dukes’ Stage | Differentiation
degree |
|---|
|
|
|
|
|
|---|
| Expression | Male | Female | ≥50 | <50 | A | B | C | D | High | Middle | Low |
|---|
| Upregulation | 13 | 10 | 13 | 10 | 2 | 8 | 10 | 3 | 3 | 19 | 1 |
| Downregulation | 5 | 2 | 4 | 3 | 0 | 6 | 1 | 0 | 0 | 6 | 1 |
mRNA expression levels of FBXL20 in
SW480−FBXL20 and SW620−FBXL20 cell lines
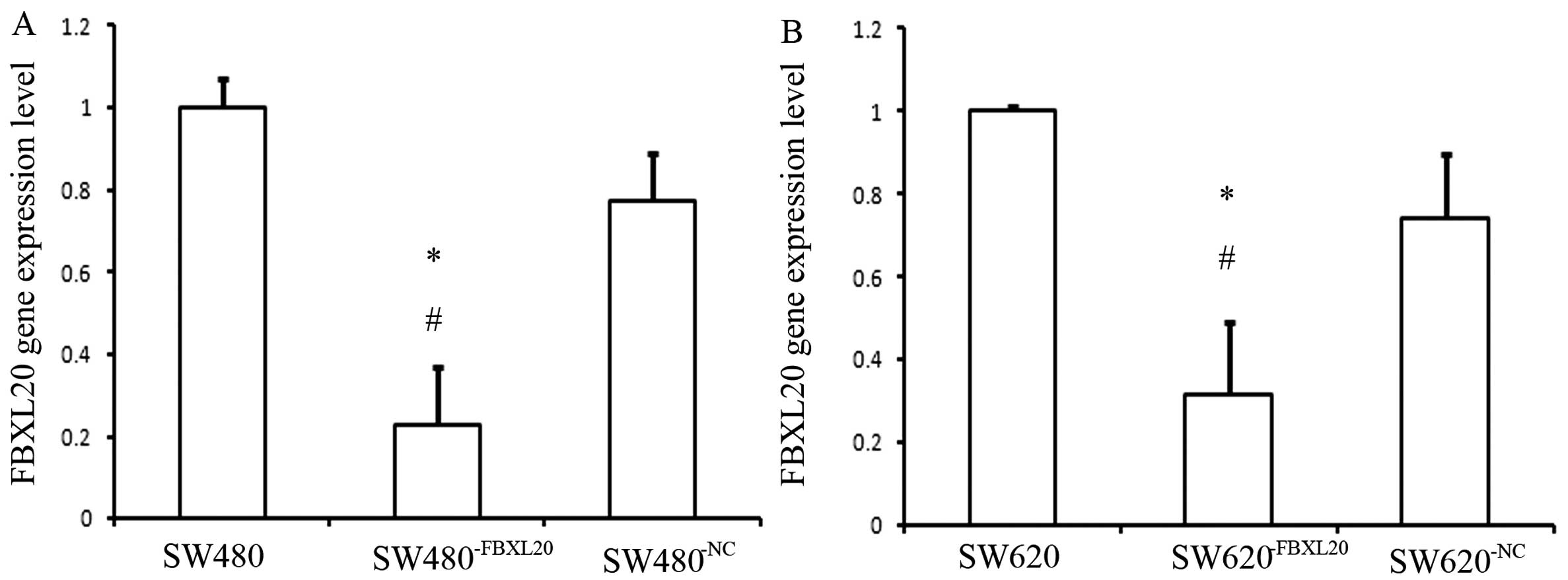
The mRNA expression level of FBXL20 in the
SW480−FBXL20 cell line was significantly lower than that
in the SW480 cell line. The reduction rate was 77.19%, which was
statistically significant as shown by the ANOVA test (P=0.002). The
FBXL20 expression level in the SW620−FBXL20 cell
line was significantly lower than that in the SW620 cell line. The
reduction rate was 68.12%, which was statistically significant as
shown by the ANOVA test (P=0.004) (Fig.
2).
Cell proliferation
The cell proliferation of the
SW480−FBXL20 and SW620−FBXL20 cells was
significantly inhibited, compared to that of the blank and the
negative group, measured by MTT assay. The inhibition rate was
33.3% in the SW480−FBXL20 cell line and 22.7% in the
SW620−FBXL20 cell line (Fig.
3). The inhibition was statistically significant as shown by
the ANOVA test.
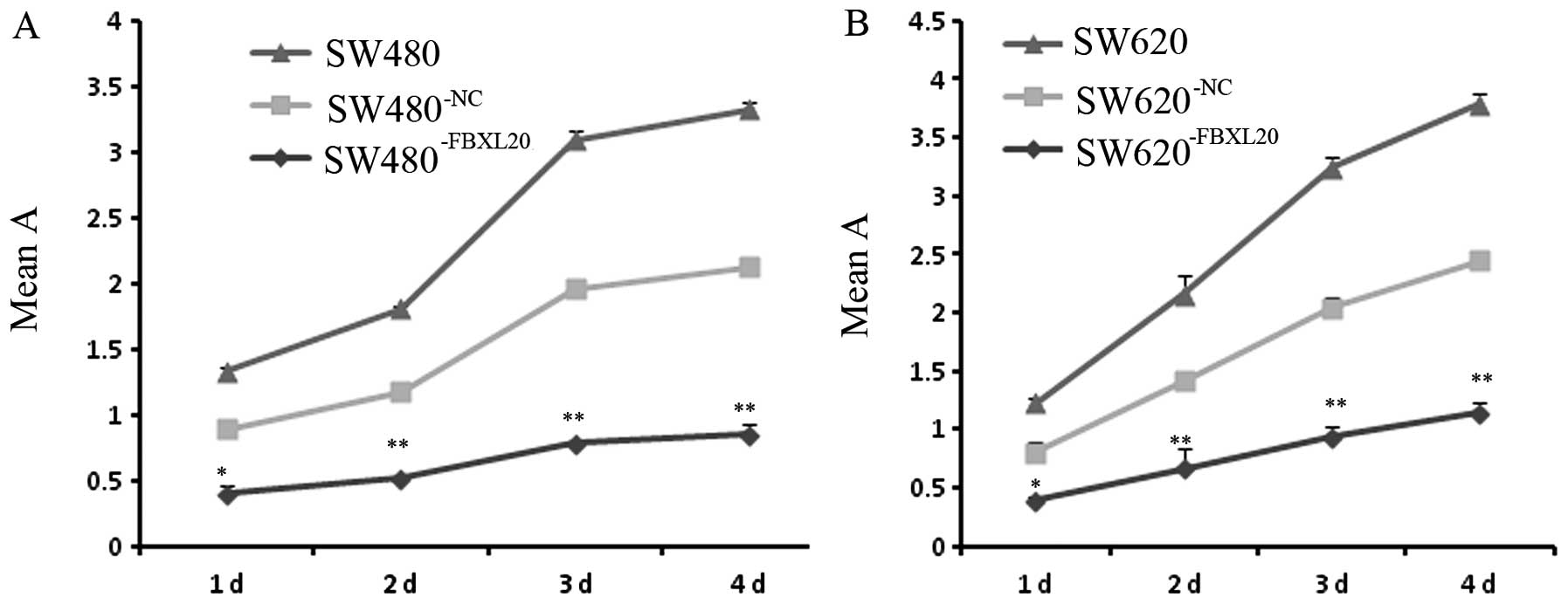 | Figure 3Growth inhibition of human colorectal
cell lines by silencing FBXL20 mRNA expression. (A) Cell
proliferation curves of the human colorectal cells,
SW480−FBXL20, SW480−NC and SW480. (B) Cell
proliferation curves of the human colorectal cells,
SW620−FBXL20, SW620−NC and SW620. Human
colorectal cell lines were incubated for the indicated time-periods
(one, two, three and four days). As a measurement of cell
viability, the relative absorbances (means ± SD, n=6), obtained
from MTT assay, are shown. Statistical differences, compared to the
blank group, are indicated as *P<0.05 and
**P<0.01. |
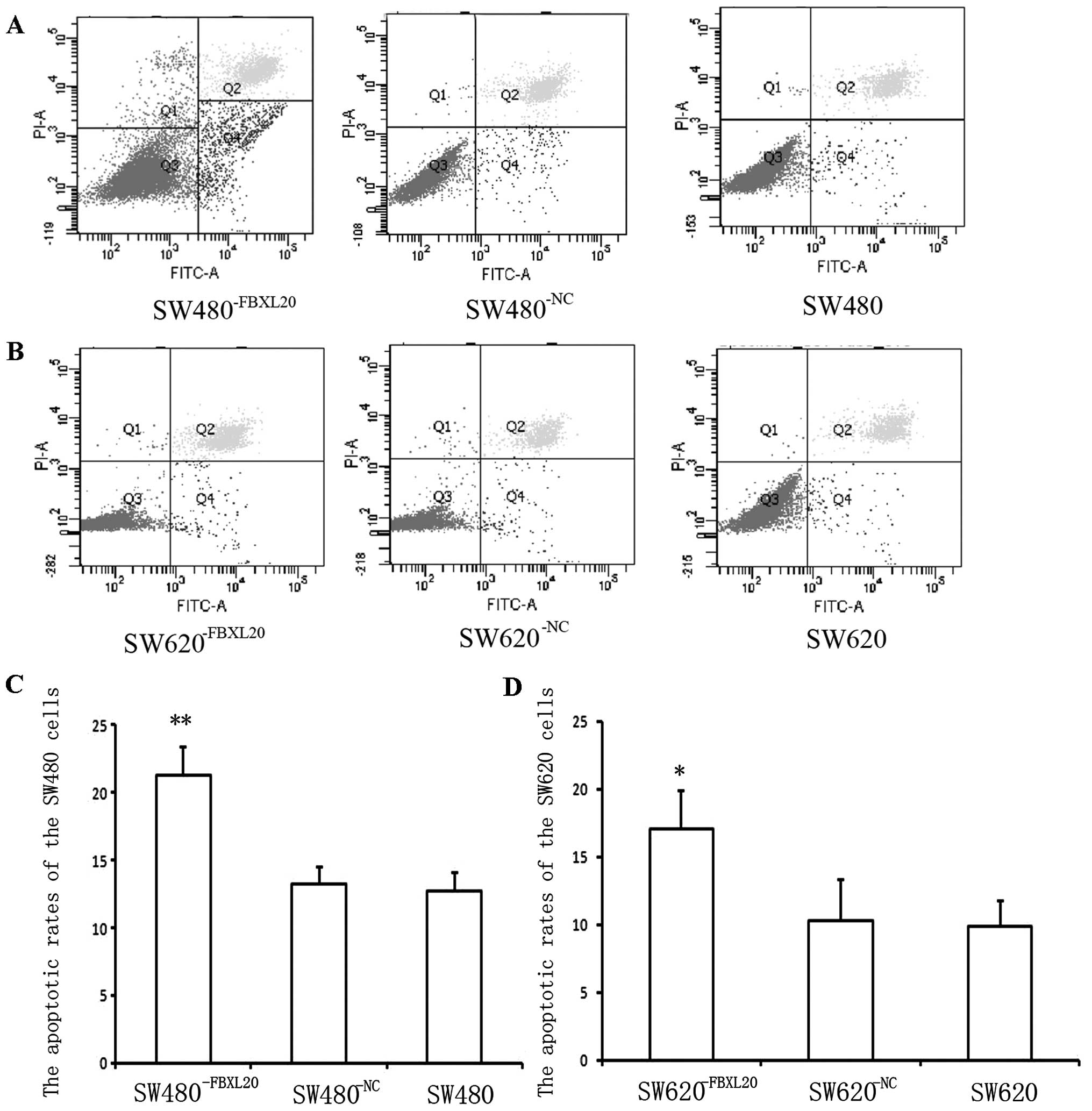
Cell apoptosis and cell cycle
After the cells were plated for 48 h, the apoptotic
rates of the SW480−FBXL20, SW480−NC and SW480
cells were 21.3, 13.3 and 12.7%, respectively, measured by flow
cytometry. The apoptotic rate of the SW480−FBXL20 cells
was significantly higher than that of the SW480 cells (P=0.003)
(Fig. 4A). The apoptotic rates of
the SW620−FBXL20, SW620−NC and SW620 cells
were 17.1, 10.32 and 9.9%, respectively. The difference between the
SW620−FBXL20 and SW620 cells was statistically
significant (P=0.036) (Fig.
4B).
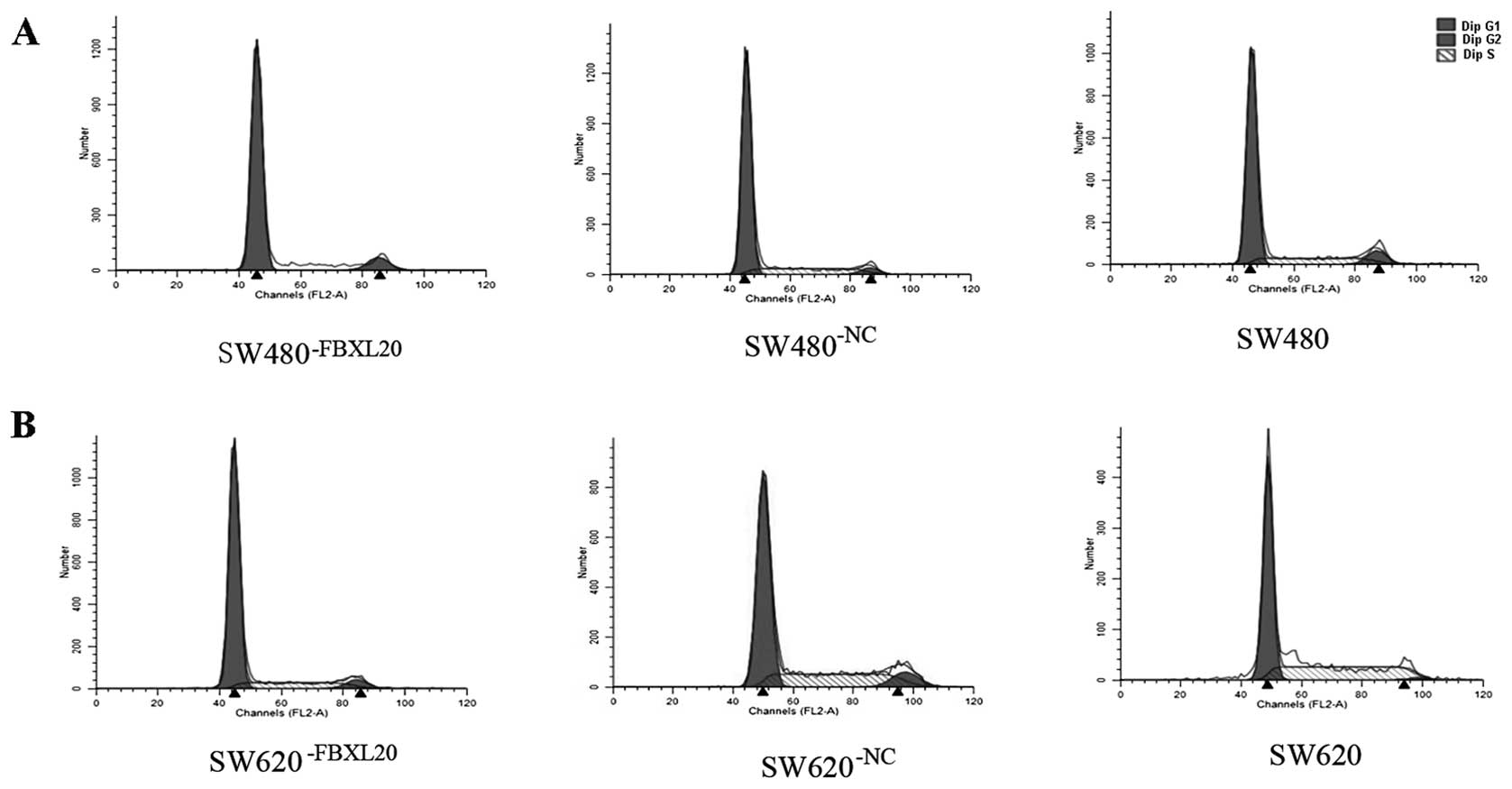
Cell cycle analysis showed that the percentages of
the SW480−FBXL20, SW480−NC and SW480 cells in
the G1/G0 phase were 90.13, 76.81 and 71.97%, respectively. The
difference between the SW480−FBXL20 and SW480 cells was
statistically significant (P=0.000) (Fig. 5A). The percentages of the
SW620−FBXL20, SW620−NC and SW620 cells in the
G1/G0 phase were 78.13, 60.22 and 58.33%, respectively. The
difference between the SW620−FBXL20 and SW620 cells was
statistically significant (P=0.001) (Fig. 5B).
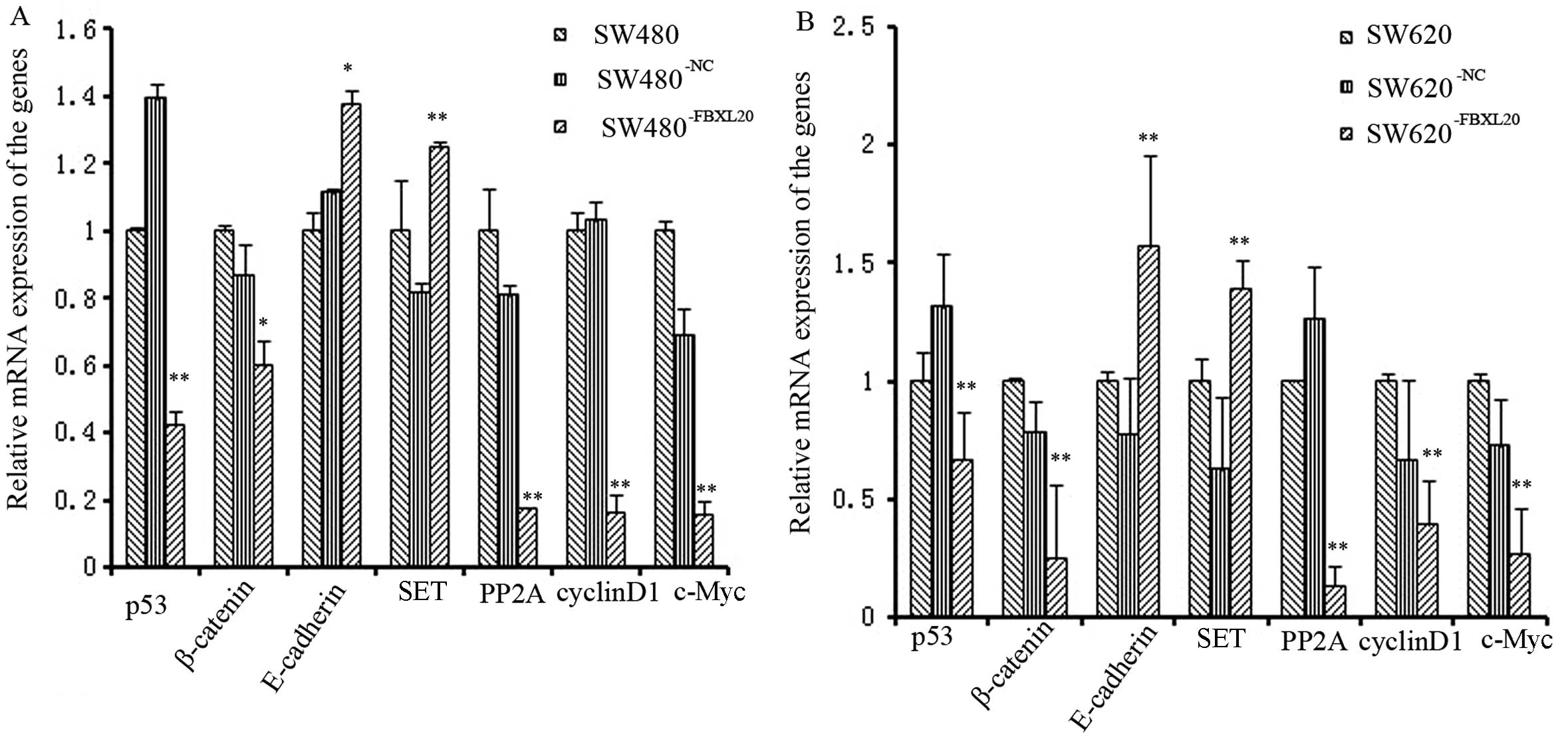
mRNA expression levels of p53, β-catenin,
PP2A, cyclin D1, c-Myc, E-cadherin and SET in
SW480−FBXL20 and SW620−FBXL20 cell lines
The mRNA expression levels of p53,
β-catenin, PP2A, cyclin D1 and c-Myc
were downregulated in the SW480−FBXL20 and
SW620−FBXL20 cell lines, compared to those in the
corresponding control cells. The respective reduction rates were
57.96, 40.13, 82.73, 84.25 and 84.54% for the
SW480−FBXL20 cells, which were all statistically
significant as shown by the ANOVA test (P=0.001, P=0.042, P=0.004,
P=0.000 and P=0.000). The respective reduction rates were 32.95,
25.06, 86.42, 60.95 and 74.57% for the SW620−FBXL20
cells, which were all statistically significant as well, as shown
by the ANOVA test (P=0.000, P=0.000, P=0.000, P=0.000 and P=0.000).
The expression of E-cadherin and SET was also
upregulated in the engineered cell lines. The increase rates were
37.5 and 24.8%, respectively, for the SW480−FBXL20 cells
(P=0.027 and P=0.009), whereas for the SW620−FBXL20
cells, the increase rates were 57.64 and 39.47%, respectively
(P=0.001 and P=0.000) (Fig. 6).
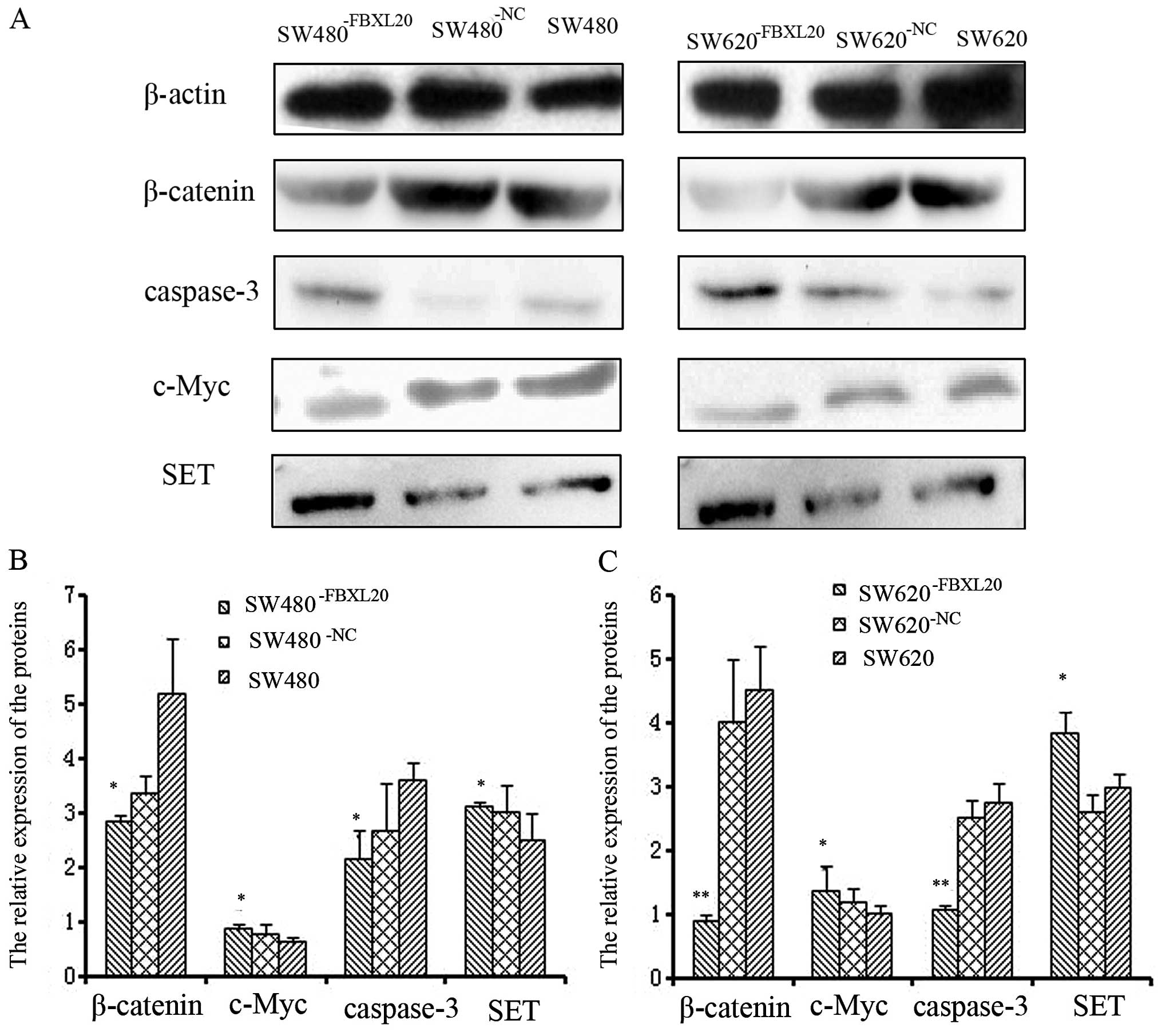
Protein expression levels of β-catenin,
c-Myc, SET and caspase-3 in SW480−FBXL20 and
SW620−FBXL20 cell lines
The protein expression of β-catenin and c-Myc was
downregulated in the SW480−FBXL20 and
SW620−FBXL20 cell lines, compared to that in the SW480
and SW620 cell lines. The inhibition rates were 45.14 and 40.08% in
the SW480−FBXL20 cells (P=0.029 and P=0.003), whereas in
the SW620−FBXL20 cells, the numbers were 80.25 and 62.06
%, accordingly (P=0.000 and P=0.000).
The protein expression of caspase-3 and SET was
upregulated in the SW480−FBXL20 and
SW620−FBXL20 cells lines, compared to that in the SW480
and SW620 cell lines. The increase rates were 37.55 and 24.83% in
the SW480−FBXL20 cells (P=0.019 and P=0.012), whereas in
the SW620−FBXL20 cells, the numbers were 35.78 and
28.39%, accordingly (P=0.025 and P=0.036) (Fig. 7).
Discussion
Among the 100 human FBPs identified so far, β-Trcp,
Skp2 and Fbw7 are well characterized components with matched
downstream substrates (8–10). A number of studies have reported
that high levels of FBPs correlate with poor prognosis in several
human malignancies, such as lung, renal, esophageal and colon
cancer (11,12). Our results showed that the mRNA
expression of FBXL20 was significantly upregulated in human
colorectal adenocarcinoma tissues, compared to that in the adjacent
normal colorectal tissues, and its expression level closely
correlated with the age of the patients. The upregulation rate in
older patients (>50 years old) was higher than that in younger
ones (<50 years old). It has been reported that sporadic
colorectal carcinoma frequently appears in older individuals
(>50 years old), suggesting the possible role of FBXL20 as a
pathological factor in sporadic colorectal carcinoma. Of note, we
did not observe any significant correlation between the FBXL20
level and other patient characteristics, such as gender, Dukes’
stage and tumor differentiation; however, in one female patient,
the FBXL20 level in the tumor tissues was 77.35-fold higher than
that in the adjacent normal colorectal tissues. Therefore, it is
possible that FBXL20 may be related to colorectal adenocarcinoma
praecox. Therefore, we can conclude that FBXL20 overexpression
correlates with colorectal carcinoma progression, and that it may
play a vital role in the pathogenesis of colorectal cancer.
In the present study, we showed that cell
proliferation was significantly inhibited, whereas cell apoptosis
was considerably increased in the SW480−FBXL20 and
SW620−FBXL20 cells, compared to that in the
corresponding control cells. In addition, a number of studies have
reported that the migration and proliferation capacities of
malignant cells were diminished when the FBP genes, such as RNF5,
gp78 and Skp2, were silenced (13,14,15).
Furthermore, von der Lehr et al (16) demonstrated that Skp2 activates the
c-Myc gene by inducing the degradation of related proteins through
the ubiquitin-proteasome pathway. It has also been reported that
Skp2 can also induce p27 protein degradation through the
ubiquitin-proteasome pathway (17).
Based on our observation of inhibited proliferation and increased
apoptosis in the SW480−FBXL20 and
SW620−FBXL20 cells, it is likely that FBXL20 has a
similar function to other FBP members in regulating cellular
activities.
The FBP family members function through different
signaling pathways. Shirane et al (10) found that β-TrCP can specifically
degrade IκB, leading to the activation of the NF-κB signaling
pathway. By contrast, a number of studies have reported that β-TrCP
can specifically degrade β-catenin, therefore inhibiting the signal
transduction of the Wnt pathway (18–20).
In general, the function of FBPs has not been fully elaborated. The
function of the FBXL20 gene has not yet been elucidated.
Numerous studies have confirmed that the development of colorectal
cancer closely correlates with the activation of the Wnt signaling
pathway (21). It has been
demonstrated that the level of c-Myc, a downstream molecule in the
Wnt signaling pathway, correlates with the proliferation capacity
of neoplastic cells (22). In the
present study, we also observed that c-Myc expression was
downregulated in the SW480−FBXL20 and
SW620−FBXL20 cell lines, compared to that in the control
cells. Of note, the proliferation capacity of these cells were also
reduced, as shown by MTT assay. These findings raised the
possibility that the reduced proliferation capacity of
SW480−FBXL20 and SW620−FBXL20 cells was
caused by the downregulation of c-Myc expression. It is known that
cyclin D1 is involved in cell cycle regulation, and it has been
reported that the cyclin D1-CDK complex promotes cell cycle
progression into the S phase (23).
Our flow cytometry results showed that the percentages of the
SW480−FBXL20 and SW620−FBXL20 cells in the
G1/G0 phase were higher than those of the control cells. Of note,
cyclin D1 expression was downregulated in the
SW480−FBXL20 and SW620−FBXL20 cells.
Therefore, we conclude that the G1/G0 arrest in the
SW480−FBXL20 and SW620−FBXL20 cell lines
correlates with the downregulation of cyclin D1 expression.
β-catenin is a key player in the signal transduction
of the Wnt pathway. Together with the Tcf/lef transcription
activator, β-catenin can activate the downstream targets, c-Myc and
cyclin D1, in the nucleus (24).
Our results showed that β-catenin expression was significantly
decreased in the SW480−FBXL20 and
SW620−FBXL20 cell lines, suggesting that the
downregulation of c-Myc and cyclin D1 in these cells may be due to
the decreased β-catenin expression. β-catenin also plays a role in
E-cadherin-mediated cell adhesion in epithelial cells (25). Tian et al (26) reported that the aberrant expression
of the E-cadherin/β-catenin complex is associated with a wide
variety of human malignancies and fibrotic disorders. In our study,
E-cadherin expression was significantly upregulated in the
SW480−FBXL20 and SW620−FBXL20 cell lines,
suggesting that the suppression of the Wnt signaling pathway in
these cells may have resulted from E-cadherin upregulation, which
then led to the downregulation of c-Myc and cyclin D1 expression
through the inhibition of the nuclear translocation of
β-catenin.
Our previous study showed that the SET gene
is overexpressed in human colorectal adenocarcinoma, and that the
inhibition of SET expression effectively suppresses cell
proliferation and promotes cell apoptosis (27). SET can inhibit the tumor suppressor,
PP2A (27,28). It has also been found that SET can
increase the activity of the splitting enzyme in CYP17 and prompt
the production of γ-interferon in natural killer cells by
inhibiting PP2A expression. PP2A plays a crucial role in the cell
cycle, DNA replication, signal transduction, cell differentiation
and malignant transformation (28).
It has been reported that β-catenin can be degraded by the
Axin-APC-CK1-GSK3β complex. Moreover, Kim et al (25) found that PP2A can dephosphorylate
Axin, which in turn leads to the destabilization and degradation of
Axin. In the present study, we observed that SET expression was
upregulated, whereas PP2A expression was downregulated. Therefore,
one possible explanation could be that the low level of β-catenin
was caused by the accumulation of the Axin-APC-CK1-GSK3β complex,
resulting from the decreased PP2A level.
FBXL20 expression was inhibited by 77.19% in the
SW480−FBXL20 and by 68.12% in the
SW620−FBXL20 cells, compared to that in the
corresponding control cells. We hypothesized that the increased
E-cadherin and SET expression may be caused by the silencing of the
FBXL20 gene, and that FBXL20 may play a role in the
degradation of E-cadherin and SET. Low FBXL20 expression leads to
the accumulation of E-cadherin, which in turn downregulates
β-catenin. Low FBXL20 expression may also increase the expression
of SET, which leads to the downregulation of PP2A. The
downregulation of PP2A inhibits the degradation of Axin, leading to
the downregulation of β-catenin and therefore a decreased β-catenin
level in the nucleus. The decreased nuclear β-catenin level in
cancer cells leads to the downregulation of c-Myc and cyclin D1,
which reduces the proliferation capacity of the
SW480−FBXL20 and SW620−FBXL20 cells.
Therefore, FBXL20 may be involved in multiple signaling
pathways.
p53 is a tumor suppressor, of which many mutations
have been found in more than 50% of malignancies (24). A number of studies have shown that
p53 not only inhibits cell growth but also induces apoptosis. By
contrast, we observed by flow cytometry that cell apoptosis was
significantly increased, whereas the p53 expression level was
significantly decreased in the SW480−FBXL20 and
SW620−FBXL20 cell lines, indicating that this increased
cell apoptosis was not induced by p53, directly. However, we did
find that the protein level of activated caspase-3 was increased in
the SW480−FBXL20 and SW620−FBXL20 cell lines,
suggesting that FBXL20 may activate caspase-3 directly or
indirectly, which in turn induces apoptosis in the
SW480−FBXL20 and SW620−FBXL20 cells. However,
the underlying mechanism warrants further investigation.
In conclusion, our study shows that the
FBXL20 gene is overexpressed in human colorectal
adenocarcinoma. Moreover, the inhibition of FBXL20 expression can
effectively suppress cell proliferation and promote apoptosis in
colorectal carcinoma cells, possibly by inducing the degradation of
SET and E-cadherin through caspase activation. In conclusion,
FBXL20 expression correlates with the pathogenesis of colorectal
cancer. However, further studies are required to validate this
correlation and to elucidate the underlying mechanism.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81072023) and by research funds from
the Oral Disease Key Laboratory of Sichuan University
(SKLODSCU20090021).
References
|
1
|
Bosetti C, Levi F, Rosato V, et al: Recent
trends in colorectal cancer mortality in Europe. Int J Cancer.
129:180–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng S and Cai SR: Colorectal cancer
epidemiology and prevention study in China. Chin Ger J Clin Oncol.
2:72–75. 2003. View Article : Google Scholar
|
|
3
|
Chen Y, Zhang YZ, Zhou ZG, et al:
Identification of differentially expressed genes in human
colorectal adenocarcinoma. World J Gastroenterol. 12:1025–1032.
2006.
|
|
4
|
Zhang C and Chen Y: Electronic cloning and
validating of the suppression subtractive hybridization EST
ES274070 of human colorectal adenocarcinoma. US Chin J Lymphol
Oncol. 6:83–88. 2007.
|
|
5
|
Zhu JJ, Jiang Q, Yuan LX, et al: Screening
and verification of the siRNA targeted to the FBXL20 gene. Chin J
Biochem Mol Biol. 27:237–243. 2011.
|
|
6
|
Skowyra D, Craig KL, Tyers M, et al: F-box
proteins are receptors that recruit phosphorylated substrates to
the SCF ubiquitin-ligase complex. Cell. 91:209–219. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai C, Sen PK, Hofman K, et al: SKP1
connects cell cycle regulators to the ubiquitin proteolysis
machinery through a novel motif, the F-box. Cell. 86:263–274. 1996.
View Article : Google Scholar
|
|
9
|
Craig KL and Tyers M: The F-box: a new
motif for ubiquitin dependent proteolysis in cell cycle regulation
and signal transduction. Prog Biophys Mol Biol. 72:299–328. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shirane M, Hatakeyama S, Hattori K, et al:
Common pathway for the ubiquitination of IkappaBalpha, IkappaBbeta,
and IkappaBepsilon mediated by the F-box protein FWD1. J Biol Chem.
274:28169–28174. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Frescas D and Pagano M: Deregulated
proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the
scales of cancer. Nat Rev Cancer. 8:438–449. 2008.
|
|
12
|
Kudo Y, Guardavaccaro D, Santamaria PG, et
al: Role of F-Box protein beta-Trcp1 in mammary gland development
and tumorigenesis. Mol Cell Biol. 24:8184–8194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bromberg KD, Kluger HM, Delaunay A, et al:
Increased expression of the E3 ubiquitin ligase RNF5 is associated
with decreased survival in breast cancer. Cancer Res. 67:8172–8179.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai YC, Mendoza A, Mariano TM, et al: The
ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAl1
for degradation. Nat Med. 13:1504–1509. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yokoi S, Yasui K, Saito-Ohara F, et al: A
novel target gene, SKP2, within the 5p13 amplicon that is
frequently detected in small cell lung cancers. Am J Pathol.
161:207–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
von der Lehr N, Johansson S and Larsson
LG: Implication of the ubiquitin/proteasome in Myc-regulated
transcription. Cell Cycle. 2:403–407. 2003.PubMed/NCBI
|
|
17
|
Masuda TA, Inoue H, Sonoda H, et al:
Clinical and biological significance of S-phase kinase-associated
protein 2 (Skp2) gene expression in gastric carcinoma: modulation
of malignant phenotype by Skp2 overexpression, possibly via p27
proteolysis. Cancer Res. 62:3819–3825. 2002.
|
|
18
|
Gnarra JR and Dressler GR: Expression of
Pax-2 in human renal cell carcinoma and growth inhibition by
antisense oligonucleotides. Cancer Res. 55:4092–4098.
1995.PubMed/NCBI
|
|
19
|
Kim M, Yan Y, Lee K, et al: Ectopic
expression of Von Hippel-Lindau tumor suppressor induces apoptosis
in 786-O renal cell carcinoma cells and regresses tumor growth of
786-O cells in nude mouse. Biochemical Biophys Res Commun.
320:945–950. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maxwell PH, Wiesener MS, Chanq GW, et al:
The tumour suppressor protein VHL targets hypoxia-inducible factors
for oxygen-dependent proteolysis. Nature. 399:271–275. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Segditsas S and Tomlinson I: Colorectal
cancer and genetic alterations in the Wnt pathway. Oncogene.
25:7531–7537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jung P, Menssen A, Mayr D and Hermeking H:
AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci
USA. 105:15046–15051. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chu IM, Hengst L and Slingerland JM: The
Cdk inhibitor p27 in human cancer: prognostic potential and
relevance to anticancer therapy. Nat Rev Cancer. 8:253–267. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Koldobskiy MA, Chakraborty A, Werner JK
Jr, et al: p53-mediated apoptosis requires inositol
hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 107:20947–20951.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim Y, Reifenberger G, Lu D, et al:
Influencing the Wnt signaling pathway in multiple myeloma.
Anticancer Res. 31:725–730. 2011.PubMed/NCBI
|
|
26
|
Tian X, Liu Z, Niu B, et al:
E-cadherin/β-catenin complex and the epithelial barrier. J Biomed
Biotechnol. 2011:5673052011.
|
|
27
|
Jiang Q, Zhang C, Zhu J, et al: The set
gene is a potential oncogene in human colorectal adenocarcinoma and
oral squamous cell carcinoma. Mol Med Rep. 4:993–999.
2011.PubMed/NCBI
|
|
28
|
Willert K, Shibamoto S and Nusse R:
Wnt-induced dephosphorylation of axin releases beta-catenin from
the axin complex. Genes Dev. 13:1768–1773. 1999. View Article : Google Scholar : PubMed/NCBI
|