Introduction
Leukemia is the leading cause of mortality worldwide
in patients with malignant tumors under the age of 35 years.
Patients with acute myeloid leukemia (AML) who have relapsed or are
refractory to conventional chemotherapy have a poorer prognosis and
response to chemotherapy than those with de novo AML, which
remains a formidable therapeutic challenge even with the
introduction of several new therapeutic strategies (1–3). M5 is
largely incurable with high relapse rates, infiltration and a
median remission duration of only six months, approximately
(4). Moreover, M5 has been reported
to have a worse prognosis than other subtypes of AML (5). Thus, a vaccine or a new drug against
M5 is required as a strategic tool for the control of this disease,
but none are currently available for practical use.
The MLAA-34 gene (GenBank no. AY288977.2) has been
confirmed to be a novel splice variant of CAB39L (calcium binding
protein 39-like). MLAA-34 was first discovered in M5 in an effort
to identify monocytic leukemia-associated antigens by serologic
analysis of a recombinant cDNA expression library (SEREX) that
reacted exclusively with sera from allogeneic leukemia patients but
not with normal donor sera (6,7). The
1671 kb gene is located on 13q14.2 and was initially cloned in our
laboratory from U937 cells (7).
CAB39L has three alternative transcripts and has been predicted to
encode a 337 aa protein. The three alternative transcripts of
CAB39L have been recognized to encode the same protein, differing
only in their 5′ untranslated regions [GenBank nos. BC010993 (1482
bp), BX647518 (2371 bp) and AY288977.2].
In our previous study, MLAA-34 and CAB39L were
identified with RNA interference (RNAi) in the U937 cell line as
novel anti-apoptotic factors that are closely related to
carcinogenesis or progression of M5 (7). Clinical research has shown that
MLAA-34 mRNA expression is upregulated in refractory/relapsed M5
patients compared with newly diagnosed, healthy donors and AML
patients in complete remission; high expression of MLAA-34 is more
prominent in the M5 subtype than in other AML patients; MLAA-34
overexpression has been found to be associated with unfavorable
clinical features at diagnosis and has been shown to be an
independent prognostic factor (8).
However, for MLAA-34, there are no exact reports regarding its
cellular localization and expression in manifold cell lines; the
anti-apoptotic mechanism of MLAA-34 remains unclear.
The purpose of this study was to conduct an in-depth
search for the expression and anti-apoptotic mechanism of MLAA-34
through the lentivirus-mediated overexpression in the U937 cell
line, and to then apply proteomics to identify its correlated
proteins or pathways that might perform functions important for the
apoptosis and proliferation of U937 cells.
Materials and methods
Cell culture
U937, HL60, K562, RPMI-8226, HepG2, Hep3B, MHCC97-H,
RC-K8, SGC-7901, Eca109, BGC823, MKN45, GES-1, BxPC-3, A375, T24,
HUVEC, BMSCs, LO2, HeLa, 293T, 293, RD, RT4, 5637, EJ, UM-UC-3,
2537, J82, Tsu-Prl, MAH, LiBr, Hut-78, HCT116+, FBL-3,
C6, astrocyte, 3T3-L1, NIH3T3, Vero and MDCK cell lines were all
maintained in our laboratory and cultured in RPMI-1640 or DMEM
supplemented with 10% fetal calf serum. The medium for cell lines
expressing the neomycin resistance gene was supplemented with 0.5
mg/ml G418. Human epithelial tissue, normal human peripheral blood
mononuclear cells (PBMCs), M5 patient and non-M5 acute leukemia
patient PBMCs were all obtained from over 30 cases of patients or
healthy young individuals. Mouse splenocytes were obtained from 30
mice.
Antibodies and reagents
CAB39L and MLAA-34 share the same open reading frame
(ORF), the CAB39L antibody was used in this report. Antibodies
specific for CAB39L (sc-100390), β-catenin (sc-133240), Rab-3D
(sc-26559), Rap-1B (sc-1481) and PGK1 (sc-130335) were purchased
from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). A
monoclonal mouse antibody against β-actin was obtained from
Sigma-Aldrich (St. Louis, MO, USA). The SAP kit and AP-Red kit were
provided by Zhongshan Co. Beijing, China (SAP-9102, ZLI-9042). The
lentivirus packaging system and enhanced infection solution (ENi.S)
were purchased from GeneChem Limited Company (Shanghai, China). The
SYBR Green PCR kit and SYBR Master Mixture were purchased from
Takara Bio, Inc. (Dalian, China). The Endo-free Plasmid Mini kit
was purchased from Qiagen, USA (12163). M-PER® Mammalian
Protein Extraction Reagent was purchased from Pierce, Rockford, IL,
USA (78503).
Western blot analysis
Cells were collected at a concentration of
2×107/ml. Following sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were
transferred to polyvinylidene fluoride membranes, which were
incubated with the primary antibody CAB39L (1:200). Western blot
analyses were performed according to standard methods. The protein
bands were visualized by applying SuperSignal West Pico
Chemiluminescent Substrate (34079; Pierce). The exposed film was
then analyzed using a densitometer.
Immunohistochemistry and
immunofluorescence
For analysis of the subcellular localization of
MLAA-34, U937 cells were washed with ice-cold PBS, blocked with 10%
normal goat serum and incubated with a primary antibody against
CAB39L at a dilution of 1:50 for 2 h at 37°C. Next, the cells were
washed again and incubated with the appropriate biotinylated
secondary antibody (goat anti-mouse IgG antibody) for 20 min at
37°C. Incubation with serum alkaline phosphatase (SAP; ALP) was
then performed at 37°C for 20 min, and the immunolabeling was
visualized with a mixture of AP-Red solution. Counterstaining with
hematoxylin was performed. For immunofluorescence, the cell samples
were incubated with the monoclonal antibody CAB39L (diluted 1:50)
and fluorescein isothiocyanate (FITC)-labeled or rhodamine-labeled
goat anti-mouse IgG as the primary and secondary antibodies,
respectively. The mounted cells were visualized with a fluorescent
microscope.
Construction and identification of the
MLAA-34 lentivirus vector and upregulated MLAA-34 stably
transfected cell line
The full-length MLAA-34 cDNA sequence was assembled
by searching the NCBI database and amplified by RT-PCR from U937
cells. First-strand cDNA synthesis was performed using a commercial
kit (Boehringer Mannheim, Milan, Italy). The restriction enzyme
site for AgeI (ACCGGT) was introduced into the 5′ and 3′ PCR
primers. To generate cDNA coding for full-length MLAA-34 by PCR,
the following primers were designed using plasmid MLAA-34 as the
template: MLAA-34-Age, I-F, GAGGATCCCCGGGTACCGGTCGCCACCATGAAAAAAATGCCTTTG
and MLAA-34-Age, I-R, TCACCATGGTGGCGACCGGAGGGGCCGTTTTCTTCAAG. The
PCR conditions consisted of 30 cycles, and the cycle parameters
were: 94°C for 5 min, then 30 cycles of 94°C for 30 sec, 55°C for
30 sec, 68°C for 1 min, followed by a final extension of 68°C for
10 min. The PCR product was purified using an Agarose Gel DNA
Purification kit (Takara Bio, Inc.). The two recovered products
were ligated using an In-Fusion kit (631774; Becton, Dickinson and
Co., USA). To confirm that the ligation was correct, MLAA-34-SEQF,
GACAGATAGGCACTCGGAG; Ubi-F, GGGTCAATATGTAATTTTCAGTG; and EGFP-N-R,
CGTCGCCGTCCAGCTCGACCAG primers were designed. The cycle parameters
were: 30 cycles of 94°C for 30 sec, 94°C for 30 sec, 60°C for 30
sec, 72°C for 50 sec, followed by a final extension of 72°C for 6
min. For detection of MLAA-34 expressed by recombinant lentivirus
in vitro, purified pGC-FU-MLAA-34 vectors were transfected
into 293T cells using Lipofectamine 2000 reagent (11668-019;
Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions. This vector was termed MLAA-34-Lentivirus, and the
vector without MLAA-34 cDNA was pGC-FU-GFP-LV. The titer of the
recombinant lentivirus was determined by real-time qPCR on 293T
cells. For identification of the recombinant MLAA-34 lentivirus
vector, the virus was added to targeted U937 cells at multiplicity
of infections (MOIs) of 10, 20, 50, 80, 100, 120 and 200 with ENi.S
and 5 μg/ml polybrene. MLAA-34-Lentivirus and pGC-FU-GFP-LV
transfected U937 cells were used as the test, and non-transfected
cells were used as the control. The expression level of MLAA-34 was
detected by western blot analysis and RT-PCR. The best MOI was
chosen.
Cells were grown in selective media (containing
G418) for two weeks, expanded and grown as independent clones for
at least two weeks. Resistant colonies were counted, and the
expression of GFP was confirmed by fluorescence microscopy, RT-PCR
and western blot analysis.
Fluorescence microscopy, MTT, flow
cytometry and DNA ladder
To determine the effect of upregulation of MLAA-34
by the MLAA-34-Lentivirus, non-transfected cells and cells
transfected with pGC-FU-GFP-LV and MLAA-34-Lentivirus were
examined. Cells were seeded in 96-well plates at a density of
1×104 cells/well. Cellular proliferation was measured
once per day during a seven-day period. In brief, 20 μl of sterile
MTT (Sigma) dye (5 mg/ml) was added to the cells, which were then
incubated for another 4 h at 37°C. Then, 150 μl of
dimethylsulfoxide was added to each well. The spectrophotometric
absorbance was measured at a wavelength of 490 nm on an enzyme
immunoassay analyzer.
Fixed cells were stained with 2.5 g/ml of DAPI
(4′,6-diamidino-2-phenylindole) solution to detect apoptotic
nuclei. Quantification of apoptosis was determined by counting the
number of apoptotic cells. The cells were stained using an Annexin
V-PE/7-AAD apoptosis detection kit (KGA1015; Nanjing KeyGen
Biotech. Co., Ltd.) according to the manufacturer’s instructions
and were analyzed by flow cytometry using a Beckman Coulter flow
cytometer.
For cell cycle analysis, the cells were fixed in 70%
ethanol and stained with propidium iodide (PI; Biosea Biotechnology
Co., Beijing, China) at a final concentration of 20 μg/ml in Triton
X-100 containing 10 mg/ml RNase. Following incubation, the samples
were analyzed on a flow cytometer.
Fragmented DNA was isolated using a DNA extraction
kit (C0008; Beyotime) according to the manufacturer’s instructions.
The eluants containing DNA pellets were electrophoresed on a 1%
agarose gel at 80 V for 1.5 h. The gel was examined and
photographed using an ultraviolet gel documentation system.
Co-immunoprecipitation (Co-IP) and
SDS-PAGE
Co-IP was performed using a Profound™ Mammalian
Co-IP kit (23605; Pierce). Transfected U937 cells
(2×107/ml) were washed, centrifuged and resuspended in
lysis buffer for incubation. The cell lysates were centrifuged to
remove the supernatant material, and the CAB39L antibody was
cross-linked to the antibody coupling resin. The lysed cell sample
was then applied to the antibody support to form immune complexes.
Then, unbound proteins were washed away three times. The samples
were then eluted, and coupling buffer was added to obtain the
immunoprecipitated protein. Finally, the Co-IP protein
concentrations were determined using a BCA Protein Assay kit
(23225; Pierce). The proteins were analyzed by SDS-PAGE, and the
gel was stained with Coomassie Blue.
Mass spectrometry analysis (MS, shotgun)
and protein identification
After separation by SDS-PAGE, discrete bands were
excised from and subjected to in-gel tryptic digestion. The
extracted peptides were analyzed using shotgun HPLC-ESI-MS
proteomics approach (LTQ; Thermo Finnigan, San Jose, CA, USA).
High-performance liquid chromatography (HPLC) separation was
performed with a capillary LC pump. The mobile phases used for the
reverse phase were i) 0.1% formic acid in water, pH 3.0; ii) 0.1%
formic acid in ACN. The collision energy was set automatically by
the LTQ system. Following acquisition of full scan mass spectrum,
three MS/MS scans were acquired for the next three most intense
ions using dynamic exclusion. Peptides and proteins were identified
using Bioworks Browser 3.1 software (Thermo Finnigan), which uses
the MS and MS/MS spectra of peptide ions to search against the NCBI
human protein database. The protein identification criteria that we
used were based on Delta CN (≥0.1) and Xcorr (one charge ≥1.9, two
charges ≥2.2, three charges ≥3.75). The protein identification
results were extracted from the SEQUEST out file with in-house
software (BuildSummary). The cellular localization, molecular
function and biologic process were determined using the gene
ontology annotation DAVID (http://david.abcc.ncifcrf.gov/). For pathway analysis,
the KEGG database was searched. To identify the corresponding
proteins in mixed protein obtained by Co-IP, western blot analysis
was performed as previously described.
Statistical analysis
The RT-PCR results were analyzed by the
self-contained software of iQ5 (Bio-Rad Co.). Statistical analyses
were performed using an analysis of variance (ANOVA). All results
are expressed as the means ± standard deviations from at least
three experiments. P<0.05 was considered to indicate
statistically significant differences.
Results
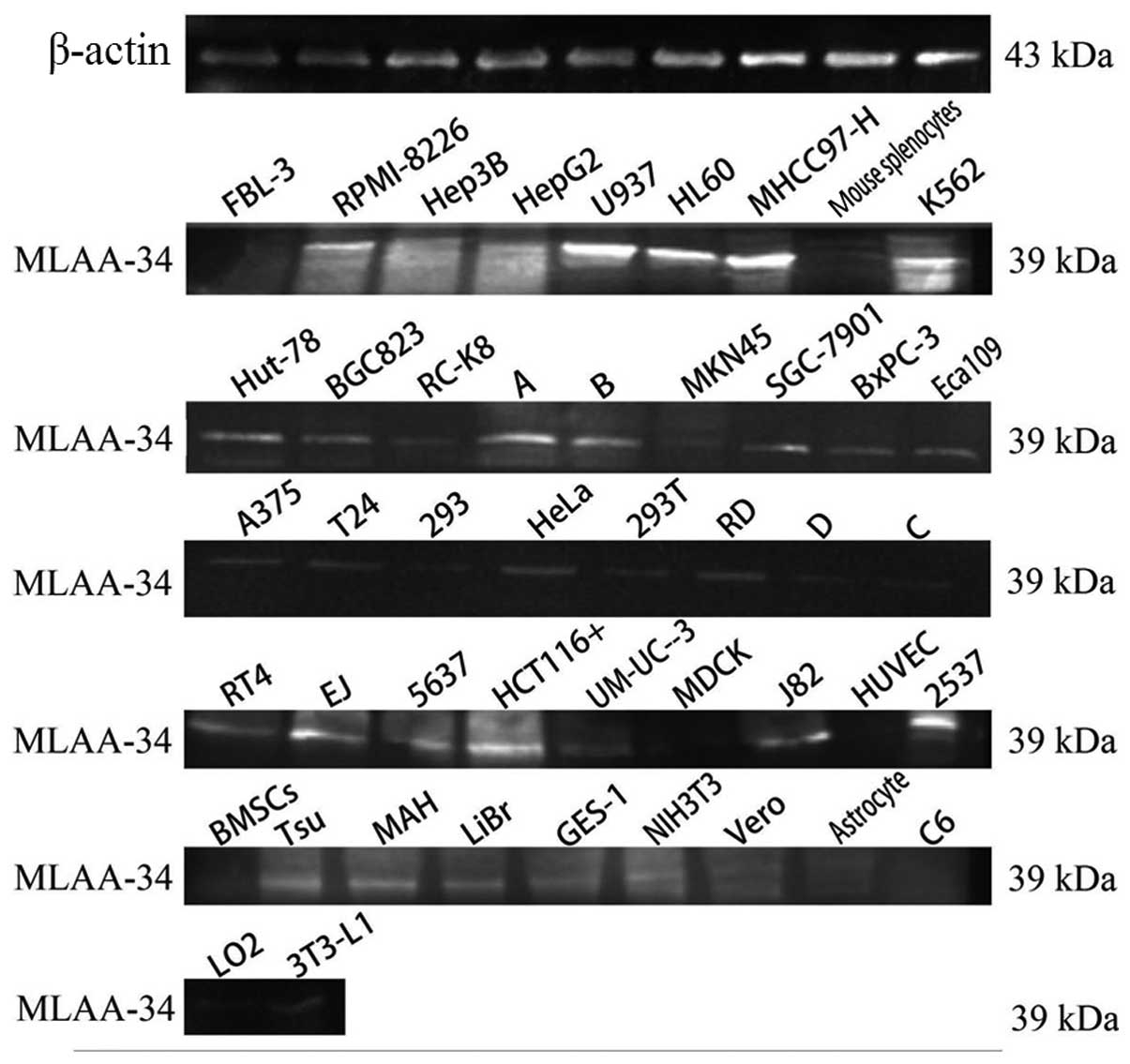
Expression of human MLAA-34 protein
With western blot analysis, a strong specific band
of ~39 kDa was observed in U937 and MHCC97-H cells, and reduced
expression was observed in other leukemia or lymphoma cell lines
and PBMCs from leukemia patients. Much fainter bands were observed
in solid tumor cell lines, and no expression was detected in normal
human cell lines or primary animal cells (Fig. 1).
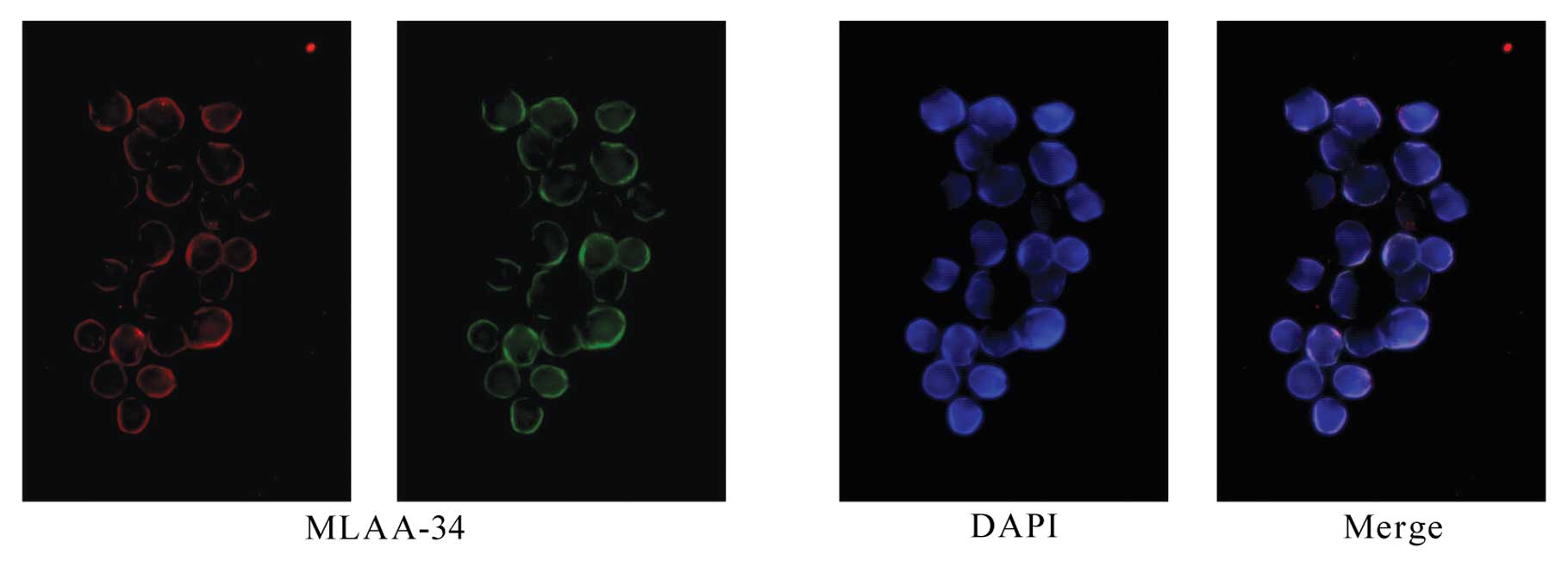
Identification and cellular localization
of MLAA-34
Immunohistochemical staining confirmed the presence
of MLAA-34 in U937 cells and the subcellular localization was
detected primarily in the cytomembrane and cytoplasm (Fig. 2).
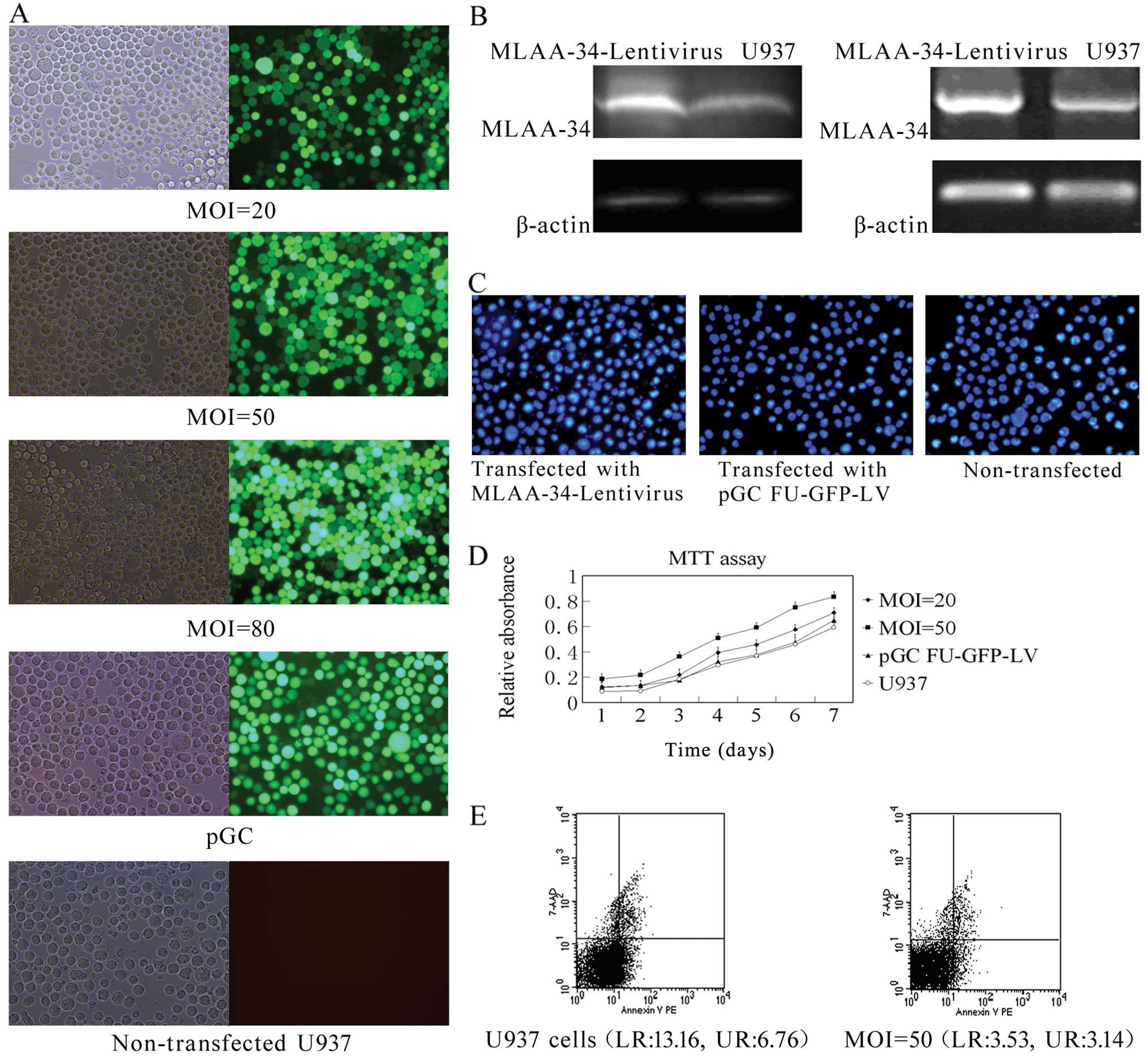
MLAA-34 is upregulated by the lentiviral
vector
A human MLAA-34 lentivirus gene transfer vector
encoding the green fluorescent protein (GFP) sequence was
constructed. The pGC-FU-MLAA-34-GFP plasmid has an insert of ~771
bp, which is in accord with the MLAA-34 cDNA [identities, 1009/1012
(99%)]. The pilot experiments showed that 293T cells could be
successfully infected by the packaged virus; the virus titer
reached higher than 2×108 TU/ml, indicating that a
high-titer lentiviral packaging platform was preliminarily
established. The pGC-FU-MLAA-34-GFP plasmid was confirmed by
western blot analysis. MLAA-34-Lentivirus and control pGC-FU-GFP-LV
virus were produced. After obtaining ideal U937 cells, we
transfected the cells with the MLAA-34-Lentivirus and pGC-FU-GFP-LV
viruses at different MOIs. The transfection efficiency was ~95% or
higher on Day 5 or later at the MOI of 50 (Fig. 3A). Five days after transfection, the
recombinant MLAA-34-Lentivirus caused a pronounced increase in the
expression of MLAA-34 compared with non-transfected U937 cells
(Fig. 3B).
Establishment of U937 cell line stably
overexpressing MLAA-34
In preliminary studies, 400 μg/ml of G418 were found
to maintain adequate selection pressure. The expression of GFP and
MLAA-34 were observed. After the cells had been frozen in liquid
nitrogen for six months and revived monthly, the U937 cells
expressed higher levels of MLAA-34 in ~400 μg/ml of G418, and ~95%
of the lentivirus-transfected U937 cells overexpressed MLAA-34.
These results suggested that the stably transfected U937 cell line
was successfully established by lentivirus and that the expression
of MLAA-34 can be long lasting even after passage.
Effect of upregulating MLAA-34 on
apoptosis and growth of U937 cells
Observations of morphology revealed increasing cell
shrinkage, nuclear condensation and fragmentation in
non-transfected and pGC-FU-GFP-LV transfected cells. By contrast,
cells transfected with MLAA-34-Lentivirus predominantly appeared
uniformly stained without condensation (Fig. 3C). These results further support the
findings that anti-apoptotic changes in the cell and nuclear
morphology are induced by MLAA-34 overexpression. MTT assays
suggested that the lentiviral overexpression of MLAA-34 induces
anti-apoptotic effects that result in a promotion effect on U937
cells; these data suggest that MLAA-34 might accelerate cell
proliferation (Fig. 3D). In
agreement with the anti-apoptotic effects of MLAA-34, cells
overexpressing MLAA-34 accumulated in the S-phase (~67.63% compared
with ~49.6% of cells in the S-phase in the control) and showed a
corresponding increase in cell numbers in the G2/M phase. The
percentages of early (lower right) and late apoptotic (upper right)
cells were markedly reduced in U937 cells after transfection with
MLAA-34-Lentivirus (Fig. 3E). These
results are in agreement with the DNA ladder assay and are even
more evident at the MOI=50, in which the cells transfected with
MLAA-34-Lentivirus showed a further increase. All of these results
suggest that MLAA-34 inhibits apoptosis in U937 cells.
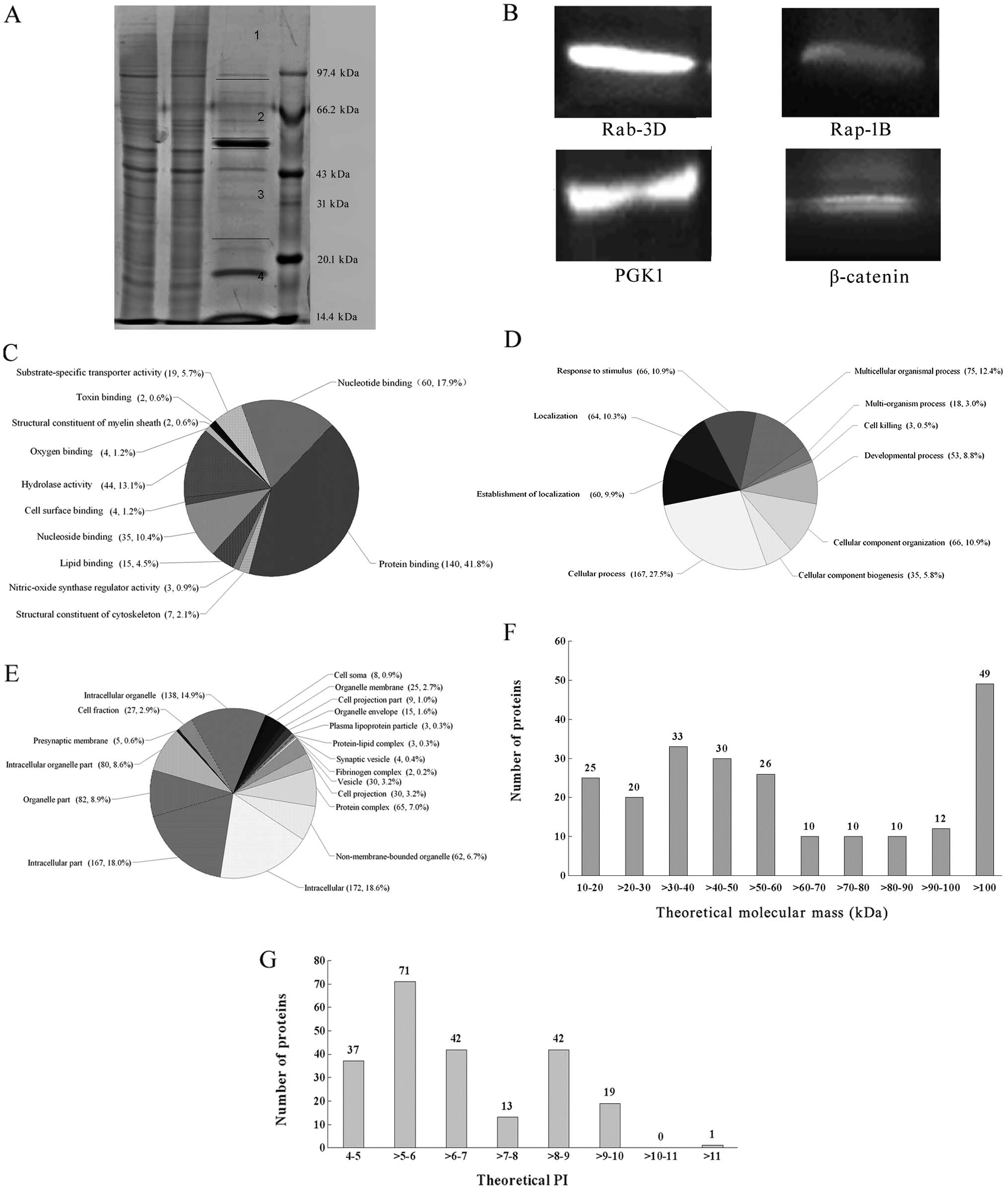
Co-IP, shotgun and western blot
analysis
Protein extracts with Co-IP were separated by
SDS-PAGE and the gel was cut into four pieces for shotgun ESI-MS
analysis (Fig. 4A). A total of 256
proteins were identified by the LC ESI-MS analysis and BIOWORKS in
the NCBI HUMAN protein databases, of which 225 (87.9%) proteins
were annotated by DAVID and the remaining 31 (12.1%) proteins have
no DAVID terms (Table I). The
expression of Rap-1B, Rab-3D, β-catenin and PGK1 was verified by
western blot analysis (Fig.
4B).
 | Table IThe 225 annotated proteins identified
by the shotgun approach. |
Table I
The 225 annotated proteins identified
by the shotgun approach.
| No. | Accession no. | Protein
description | Biological processes
(partly) | KEGG pathways
(partly) |
|---|
| 1 | IPI00022434 | ALB albumin | Apoptosis | |
| 2 | IPI00023598 | TUBB4 tubulin β-4
chain | | |
| 3 | IPI00013475 | TUBB2A tubulin β-2A
chain | | |
| 4 | IPI00180675 | TUBA1A tubulin α-1A
chain | | |
| 5 | IPI00387144 | TUBA1B tubulin α-1B
chain | | |
| 6 | IPI00013683 | TUBB3 tubulin β-3
chain | | |
| 7 | IPI00007752 | TUBB2C tubulin β-2C
chain | Apoptosis | |
| 8 | IPI00218343 | TUBA1C tubulin α-1C
chain | | |
| 9 | IPI00021439 | ACTB actin,
cytoplasmic 1 | | |
| 10 | IPI00646909 | TUBA8 tubulin α-8
chain | | |
| 11 | IPI00646779 | TUBB6 tubulin β-6
chain | | |
| 12 | IPI00257508 | DPYSL2
dihydropyrimidinase-related protein 2 | | |
| 13 | IPI00908469 | cDNA FLJ52712,
highly similar to Tubulin β-6 chain | | |
| 14 | IPI00410714 | HBA1; HBA2
hemoglobin, α-2; hemoglobin, α-1 | | |
| 15 | IPI00021428 | ACTA1 actin, α
skeletal muscle | | |
| 16 | IPI00026268 | GNB1 guanine
nucleotide-binding protein
G(I)/G(S)/G(T) subunit β-1 | Cell proliferation,
Ras protein signal transduction | Chemokine signaling
pathway, Attenuation of GPCR signaling, Erk1/Erk2 MAPK signaling
pathway, CXCR4 signaling pathway |
| 17 | IPI00220281 | GNAO1 isoform A-1
of Guanine nucleotide-binding protein G(o) subunit α | Regulation of
calcium ion transport, G-protein coupled receptor protein signaling
pathway | |
| 18 | IPI00021907 | MBP myelin basic
protein | | |
| 19 | IPI00024067 | CLTC clathrin heavy
chain 1 | | |
| 20 | IPI00216171 | ENO2 enolase 2 (γ,
neuronal) | | |
| 21 | IPI00465248 | ENO1 isoform
α-enolase of A-enolase | | |
| 22 | IPI00220706 | HBG1 hemoglobin
subunit γ-1 | | |
| 23 | IPI00219018 | GAPDH
glyceraldehyde-3-phosphate dehydrogenase | | |
| 24 | IPI00398700 | GNAO1 isoform A-2
of Guanine nucleotide-binding protein G(o) subunit α | Regulation of
calcium ion transport, G-protein coupled receptor protein signaling
pathway | |
| 25 | IPI00025363 | GFAP glial
fibrillary acidic protein | | |
| 26 | IPI00303476 | ATP5B ATP synthase,
H+ transporting, mitochondrial F1 complex, β
polypeptide | | |
| 27 | IPI00022977 | CKB creatine kinase
B-type | | |
| 28 | IPI00413140 | DNM1 dynamin 1 | | |
| 29 | IPI00154742 | IGL λ protein | | |
| 30 | IPI00022463 | TF
serotransferrin | | |
| 31 | IPI00220737 | NCAM1 neural cell
adhesion molecule 1 | Regulation of
calcium-mediated signaling | |
| 32 | IPI00022891 | SLC25A4 ADP/ATP
translocase 1 | | Calcium signaling
pathway |
| 33 | IPI00007188 | SLC25A5 ADP/ATP
translocase 2 | | Calcium signaling
pathway |
| 34 | IPI00009532 | ABAT
4-aminobutyrate aminotransferase, mitochondrial | | |
| 35 | IPI00012451 | GNB4 guanine
nucleotide-binding protein subunit β-4 | | Chemokine signaling
pathway |
| 36 | IPI00291006 | MDH2 malate
dehydrogenase 2, NAD (mitochondrial) | | |
| 37 | IPI00298497 | FGB fibrinogen β
chain | | |
| 38 | IPI00220993 | CNP 2′,3′-cyclic
nucleotide 3′ phosphodiesterase | | |
| 39 | IPI00027547 | DCD dermcidin | | |
| 40 | IPI00219446 | PEBP1
phosphatidylethanolamine-binding protein 1 | Regulation of
cAMP-mediated signaling, regulation of MAPKKK cascade | Calcium signaling
pathway |
| 41 | IPI00414123 | CRMP1 collapsin
response mediator protein 1 | | |
| 42 | IPI00217507 | NEFM neurofilament,
medium polypeptide | | |
| 43 | IPI00465439 | ALDOA aldolase A,
fructose-bisphosphate | | |
| 44 | IPI00029111 | DPYSL3
dihydropyrimidinase-like 3 | | |
| 45 | IPI00219813 | RTN1
reticulon-1 | | |
| 46 | IPI00001453 | INA
α-internexin | | |
| 47 | IPI00237671 | NEFL neurofilament
light polypeptide | Apoptosis | |
| 48 | IPI00743576 | ATP6V0A1 ATPase,
H+ transporting, lysosomal V0 subunit a1 | | |
| 49 | IPI00418262 | ALDOC
fructose-bisphosphate aldolase C | Apoptosis | |
| 50 | IPI00029751 | CNTN1
contactin-1 | Notch signaling
pathway | |
| 51 | IPI00549543 | NCDN
neurochondrin | | |
| 52 | IPI00024975 | KIF15 kinesin
family member 15 | | |
| 53 | IPI00027497 | GPI
glucose-6-phosphate isomerase | | |
| 54 | IPI00010154 | GDI1 GDP
dissociation inhibitor 1 | Small GTPase
mediated signal transduction | |
| 55 | IPI00554752 | PRKAR2B protein
kinase, cAMP-dependent, regulatory, type II, β | | Apoptosis, Insulin
signaling pathway |
| 56 | IPI00028888 | HNRNPD
heterogeneous nuclear ribonucleoprotein D0 | | |
| 57 | IPI00033025 | SEPT7 septin 7 | | |
| 58 | IPI00784156 | AP2B1
adaptor-related protein complex 2, β1 subunit | | |
| 59 | IPI00026272 | HIST1H2AB;
HIST1H2AE histone cluster 1, H2ae; histone cluster 1, H2ab | | |
| 60 | IPI00219661 | PLP1 proteolipid
protein 1 | | |
| 61 | IPI00015671 | TUBAL3 tubulin α
chain-like 3 | | |
| 62 | IPI00216298 | TXN
thioredoxin | Cell
proliferation | |
| 63 | IPI00215715 | CAMK2A
calcium/calmodulin-dependent protein kinase II α | Regulation of NF-κB
transcription factor activity | ErbB signaling
pathway, Calcium signaling pathway, Wnt signaling pathway |
| 64 | IPI00020926 | HOXA4 homeobox
A4 | | |
| 65 | IPI00022314 | SOD2 superoxide
dismutase 2, mitochondrial | Cell proliferation,
apoptosis | |
| 66 | IPI00024266 | MGST3 microsomal
glutathione S-transferase 3 | | |
| 67 | IPI00382470 | HSP90AA1 heat shock
protein 90 kDa α (cytosolic), class A member 1 isoform 1 | | NOD-like receptor
signaling pathway, pathways in cancer, Ahr signal transduction
pathway, AKT signaling pathway |
| 68 | IPI00019971 | STXBP2
syntaxin-binding protein 2 | | |
| 69 | IPI00289861 | ZCCHC11 zinc finger
CCHC domain-containing protein 11 | | |
| 70 | IPI00013508 | ACTN1
α-actinin-1 | | |
| 71 | IPI00007682 | ATP6V1A V-type
proton ATPase catalytic subunit A | | |
| 72 | IPI00003925 | PDHB pyruvate
dehydrogenase E1 component subunit β, mitochondrial | | |
| 73 | IPI00910290 | Aryl hydrocarbon
receptor nuclear translocator | Cell
proliferation | Pathways in cancer,
Ahr signal transduction pathway |
| 74 | IPI00022488 | HPX hemopexin | Regulation of
protein kinase cascade, interferon-γ-mediated signaling pathway,
regulation of JAK-STAT cascade | |
| 75 | IPI00023302 | SYN2
synapsin-2 | | |
| 76 | IPI00902614 | USP24 ubiquitin
carboxyl-terminal hydrolase 24 | | |
| 77 | IPI00220644 | PKM2 pyruvate
kinase isozymes M1/M2 | | |
| 78 | IPI00414676 | HSP90AB1 heat shock
protein HSP 90-β |
Interferon-γ-mediated signaling pathway,
type I interferon-mediated signaling pathway | NOD-like receptor
signaling pathway, pathways in cancer |
| 79 | IPI00647704 | IGHA1
immunoglobulin heavy constant α 1 | | |
| 80 | IPI00215747 | FABP7 fatty
acid-binding protein, brain | Cell
proliferation | PPAR signaling
pathway |
| 81 | IPI00026053 | CLDN11
claudin-11 | | |
| 82 | IPI00025447 | EEF1A1 elongation
factor 1-α | | |
| 83 | IPI00182944 | CAMK2B
calcium/calmodulin-dependent protein kinase type II β chain | | ErbB signaling
pathway, Calcium signaling pathway, Wnt signaling pathway |
| 84 | IPI00411486 | OPALIN opalin | | |
| 85 | IPI00299608 | PSMD1 proteasome
(prosome, macropain)
26S subunit, non-ATPase, 1 | | |
| 86 | IPI00299399 | S100B protein
S100-B | Cell
proliferation | Calcium signaling
pathway |
| 87 | IPI00175169 | ARFGAP1
ADP-ribosylation factor
GTPase-activating protein 1 | Small GTPase
mediated signal transduction, Ras protein signal transduction | |
| 88 | IPI00005614 | SPTBN1 spectrin β
chain, brain 1 | | |
| 89 | IPI00017597 | MAPRE3
microtubule-associated protein
RP/EB family member 3 | | |
| 90 | IPI00175092 | RNF149 ring finger
protein 149 | | |
| 91 | IPI00293613 | TBK1 TANK-binding
kinase 1 | Regulation of
protein kinase cascade, regulation of I-κB kinase/NF-κB
cascade | Toll-like receptor
signaling pathway, RIG-I-like receptor signaling pathway |
| 92 | IPI00015029 | PTGES3
prostaglandin E synthase 3 | | |
| 93 | IPI00169383 | PGK1
phosphoglycerate kinase 1 | | |
| 94 | IPI00015148 | RAP1B ras-related
protein Rap-1b | Cell proliferation,
small GTPase mediated signal transduction | MAPK signaling
pathway, Chemokine signaling pathway |
| 95 | IPI00028946 | RTN3
reticulon-3 | Apoptosis | |
| 96 | IPI00163849 | EPS15L1 epidermal
growth factor receptor substrate 15-like 1 | Calcium ion
binding | |
| 97 | IPI00645078 | UBA1 ubiquitin-like
modifier-activating enzyme 1 | | Ubiquitin mediated
proteolysis |
| 98 | IPI00005705 | PPP1CC γ-1 of
serine/threonine-protein phosphatase PP1-γ catalytic subunit | | Insulin signaling
pathway |
| 99 | IPI00159927 | NCAN neurocan core
protein | Calcium ion
binding | |
| 100 | IPI00003420 | MAPRE2
microtubule-associated protein, RP/EB family, member 2 | Cell
proliferation | |
| 101 | IPI00017566 | FBXL7
F-box/LRR-repeat protein 7 | | |
| 102 | IPI00027252 | PHB2
prohibitin-2 | | |
| 103 | IPI00015141 | CKMT2 creatine
kinase, sarcomeric mitochondrial | | |
| 104 | IPI00027770 | SYP
synaptophysin | | |
| 105 | IPI00290035 | PCDH15
protocadherin-15 | Calcium ion
binding | |
| 106 | IPI00027462 | S100A9 S100 calcium
binding protein A9 | Calcium ion
binding | |
| 107 | IPI00022462 | TFRC transferrin
receptor protein 1 | | |
| 108 | IPI00300020 | SLC1A2 excitatory
amino acid transporter 2 | | |
| 109 | IPI00019884 | ACTN2
α-actinin-2 | Apoptosis | |
| 110 | IPI00000875 | EEF1G cDNA
FLJ56389, highly similar to Elongation factor 1-γ | | |
| 111 | IPI00435928 | RASGRF1 Ras
protein-specific guanine nucleotide-releasing factor 1 | | Ras signaling
pathway |
| 112 | IPI00790581 | MPRIP protein | | |
| 113 | IPI00383660 | ZNF530 zinc finger
protein 530 | | |
| 114 | IPI00218896 | ADH1A alcohol
dehydrogenase 1A | | |
| 115 | IPI00300341 | TCEB1 transcription
elongation factor B polypeptide 1 | | Ubiquitin mediated
proteolysis, pathways in cancer |
| 116 | IPI00021891 | FGG Γ-B of
Fibrinogen γ chain | | |
| 117 | IPI00747180 | WDR52 WD repeat
protein 52 | | |
| 118 | IPI00642126 | KIAA1618 isoform 1
of protein ALO17 | | |
| 119 | IPI00164441 | UNC13A unc-13
homolog A | | |
| 120 | IPI00027820 | ESPN isoform 1 of
Espin | | |
| 121 | IPI00185659 | CCDC62 isoform 2 of
Coiled-coil domain-containing protein 60 | | |
| 122 | IPI00000816 | YWHAE 14-3-3
protein epsilon | Apoptosis | |
| 123 | IPI00160552 | TNR isoform 1 of
Tenascin-R | | |
| 124 | IPI00164347 | CNGB1 cyclic
nucleotide gated channel β 1 isoform b | | |
| 125 | IPI00166979 | KIAA1239
Leucine-rich repeat and WD repeat-containing protein KIAA1239 | | |
| 126 | IPI00217240 | WDR75 WD
repeat-containing protein 75 | | |
| 127 | IPI00017704 | COTL1
coactosin-like protein | | |
| 128 | IPI00008305 | HPCAL4
hippocalcin-like protein 4 | | |
| 129 | IPI00440493 | ATP5A1 ATP synthase
subunit α, mitochondrial | Cell
proliferation | |
| 130 | IPI00024547 | C2orf25 chromosome
2 open reading frame 25 | | |
| 131 | IPI00074962 | ANK2 isoform 4 of
Ankyrin-2 | | |
| 132 | IPI00395663 | ANKS1A ankyrin
repeat and SAM domain-containing protein 1A | | |
| 133 | IPI00029769 | HCK isoform p59-HCK
of Tyrosine-protein kinase HCK | | Chemokine signaling
pathway, GPCR signaling |
| 134 | IPI00216592 | HNRNPC isoform C1
of Heterogeneous nuclear ribonucleoproteins C1/C2 | | |
| 135 | IPI00000792 | CRYZ quinone
oxidoreductase | | |
| 136 | IPI00219806 | S100A7 S100 calcium
binding protein A7 | S100/CaBP-9k-type,
calcium binding | |
| 137 | IPI00216856 | ANKMY2 ankyrin
repeat and MYND domain-containing protein 2 | | |
| 138 | IPI00027434 | RHOC rho-related
GTP-binding protein RhoC | Small GTPase
mediated signal transduction, regulation of I-κB kinase/NF-κB
cascade | Ras signaling
pathway |
| 139 | IPI00396341 | C2orf67 chromosome
2 open reading frame 67 | | |
| 140 | IPI00015785 | CRB1 crumbs homolog
1 | Calcium ion
binding | |
| 141 | IPI00893234 | OBSL1 obscurin-like
1 | | |
| 142 | IPI00028277 | FTO isoform 1 of
Protein fto | | |
| 143 | IPI00060800 | LOC124220
uncharacterized protein UNQ773/PRO1567 | | |
| 144 | IPI00007765 | HSPA9 stress-70
protein, mitochondrial | Anti-apoptosis | |
| 145 | IPI00852669 | ZNF516 zinc finger
protein 516 | | |
| 146 | IPI00024994 | TULP4 tubby-related
protein 4 | | |
| 147 | IPI00010466 | PRKCB isoform B-I
of protein kinase C β type | | MAPK signaling
pathway, ErbB signaling pathway, Calcium signaling pathway,
Chemokine signaling pathway, Phosphatidylinositol signaling system,
Wnt signaling pathway, VEGF signaling pathway, pathways in
cancer |
| 148 | IPI00815811 | ZNF235 zinc finger
protein 235 | | |
| 149 | IPI00163187 | FSCN1 fascin
homolog 1 | Cell
proliferation | |
| 150 | IPI00793780 | TMCO5B
transmembrane and coiled-coil domain-containing protein 5B | | |
| 151 | IPI00011088 | CLDN12
claudin-12 | | |
| 152 | IPI00011986 | C5orf42 chromosome
5 open reading frame 42 | | |
| 153 | IPI00061780 | ITCH itchy E3
ubiquitin protein ligase homolog | Cell
proliferation | Ubiquitin mediated
proteolysis |
| 154 | IPI00152653 | DNAH5 dynein heavy
chain 5, axonemal | | |
| 155 | IPI00175416 |
PLCH11-phosphatidylinositol-4,5-bisphosphate
phosphodiesterase β-1 | Calcium ion
binding | |
| 156 | IPI00218352 | ESR1 estrogen
receptor1 | Estrogen receptor
signaling pathway | |
| 157 | IPI00791536 | MCF.2 cell line
derived transforming sequence-like 2 | Regulation of Rho
protein signal transduction, regulation of Ras protein signal
transduction, regulation of small GTPase mediated signal
transduction | |
| 158 | IPI00009619 | CADM3 isoform 2 of
cell adhesion molecule 3 | | |
| 159 | IPI00179330 | UBC; RPS27A; UBB
ubiquitin and ribosomal protein S27a precursor | Apoptosis | |
| 160 | IPI00217776 | ICK intestinal cell
(MAK-like) kinase | | |
| 161 | IPI00009439 | SYT1
synaptotagmin-1 | Calcium ion
binding | |
| 162 | IPI00784869 | DNAH10 isoform 1 of
Dynein heavy chain 10, axonemal | | |
| 163 | IPI00020265 | ANKRD20A1 ankyrin
repeat domain-containing protein 20A1 | | |
| 164 | IPI00007189 | CDC42 isoform 1 of
cell division control protein 42 homolog | | MAPK signaling
pathway, Chemokine signaling pathway, VEGF signaling pathway,
Pathways in cancer, Ras signaling pathway |
| 165 | IPI00032325 | CSTA
cystatin-A | | |
| 166 | IPI00032808 | RAB3D ras-related
protein Rab-3D | Small GTPase
mediated signal transduction | Ras signaling
pathway |
| 167 | IPI00216308 | VDAC1
voltage-dependent anion-selective channel protein 1 | Apoptosis | Calcium signaling
pathway |
| 168 | IPI00021841 | APOA1
apolipoprotein A-I | Cell proliferation,
small GTPase mediated signal transduction, Ras protein signal
transduction, Rho protein signal transduction, Cdc42 protein signal
transduction, G-protein coupled receptor protein signaling
pathway | PPAR signaling
pathway |
| 169 | IPI00645906 | CXorf39 isoform 1
of uncharacterized protein CXorf39 | | |
| 170 | IPI00220032 | CTNND2 isoform 2 of
Catenin δ-2 | | |
| 171 | IPI00002459 | ANXA6 Annexin VI
isoform 2 | Calcium ion
transport | |
| 172 | IPI00184119 | DNAJC6 isoform 2 of
putative tyrosine-protein phosphatase auxilin | | |
| 173 | IPI00152949 | TMEM168
transmembrane protein 168 | | |
| 174 | IPI00032402 | ATP8A1 isoform long
of probable phospholipid-transporting ATPase IA | | |
| 175 | IPI00008380 | PPP2CA
serine/threonine-protein phosphatase 2A catalytic subunit α
isoform | Apoptosis, MAPKKK
cascade, second-messenger-mediated signaling, regulation of
JAK-STAT cascade | Wnt signaling
pathway, TGF-β signaling pathway, AKT signaling pathway, Erk1/Erk2
MAPK signaling pathway |
| 176 | IPI00219217 | LDHB L-lactate
dehydrogenase B chain | | |
| 177 | IPI00394855 | C12orf63 chromosome
12 open reading frame 63 | | |
| 178 | IPI00025366 | CS citrate
synthase, mitochondrial | | |
| 179 | IPI00218570 | PGAM2
phosphoglycerate mutase 2 | | |
| 180 | IPI00303484 | OR52K2 olfactory
receptor 52K2 | G-protein coupled
receptor protein signaling pathway | |
| 181 | IPI00005565 | DGKQ diacylglycerol
kinase θ | G-protein coupled
receptor protein signaling pathway, activation of protein kinase C
activity by G-protein coupled receptor protein signaling
pathway |
Phosphatidylinositol signaling system |
| 182 | IPI00168218 | DOK7 isoform 2 of
protein Dok-7 | | |
| 183 | IPI00154645 | TBC1D15 isoform 1
of TBC1 domain family member 15 | Rab protein signal
transduction, Ras protein signal transduction, small GTPase
mediated signal transduction | |
| 184 | IPI00386494 | SPPL2B isoform 1 of
signal peptide peptidase-like 2B | | |
| 185 | IPI00465436 | CAT catalase | Apoptosis,
regulation of protein kinase cascade, regulation of
phosphoinositide 3-kinase cascade, regulation of NF-κB
transcription factor activity | |
| 186 | IPI00007612 | KCNJ1 isoform 1 of
ATP-sensitive inward rectifier potassium channel 1 | | |
| 187 | IPI00020153 | BSN protein
bassoon | | |
| 188 | IPI00298547 | PARK7 protein
DJ-1 | Small GTPase
mediated signal transduction, Ras protein signal transduction | |
| 189 | IPI00456969 | DYNC1H1 cytoplasmic
dynein 1 heavy chain 1 | | |
| 190 | IPI00030144 | PPIAL4C; PPIAL4A;
PPIAL4G; PPIAL4B Peptidylprolyl cis-trans isomerase A-like 4B | | |
| 191 | IPI00376119 | PRKACB isoform 2 of
cAMP-dependent protein kinase catalytic subunit β | G-protein coupled
receptor protein signaling pathway, second-messenger-mediated
signaling, cAMP-mediated signaling | MAPK signaling
pathway, Calcium signaling pathway, Chemokine signaling pathway,
Apoptosis, Wnt signaling pathway, Hedgehog signaling pathway,
Insulin signaling pathway |
| 192 | IPI00025753 | DSG1
desmoglein-1 | Calcium ion
binding | |
| 193 | IPI00292934 | USP53 inactive
ubiquitin carboxyl-terminal hydrolase 53 | | |
| 194 | IPI00024684 | MX2
interferon-induced GTP-binding protein Mx2 | | |
| 195 | IPI00384998 | NFASC isoform 7 of
Neurofascin | | |
| 196 | IPI00217494 | SMG7 smg-7 homolog,
nonsense mediated
mRNA decay factor (C. elegans) | | |
| 197 | IPI00171594 | DACT1 dapper
homolog 1 | Wnt receptor
signaling pathway | |
| 198 | IPI00006612 | SNAP91 isoform 1 of
Clathrin coat assembly protein AP180 | | |
| 199 | IPI00291922 | PSMA5 proteasome
subunit α type-5 | | |
| 200 | IPI00056040 | NRN1L neuritin-like
protein | | |
| 201 | IPI00013421 | GPM6B isoform 1 of
neuronal membrane glycoprotein M6-b | | |
| 202 | IPI00738216 | KIAA0947 isoform 1
of uncharacterized protein KIAA0947 | | |
| 203 | IPI00022133 | EPB41L2 4.1G
protein | | |
| 204 | IPI00853219 | RAPGEF2 rap guanine
nucleotide exchange factor 2 | Small GTPase
mediated signal transduction, cAMP-mediated signaling | MAPK signaling
pathway |
| 205 | IPI00375609 | JAKMIP3 janus
kinase and microtubule-interacting protein 3 | | |
| 206 | IPI00178185 | BICD2 isoform 1 of
protein bicaudal D homolog 2 | | |
| 207 | IPI00006146 | SAA2 serum amyloid
A2 | | |
| 208 | IPI00014843 | LRRC16A isoform 1
of Leucine-rich repeat-containing protein 16A | | |
| 209 | IPI00183368 | SMG1
phosphatidylinositol 3-kinase-related kinase (C.
elegans) | | |
| 210 | IPI00019038 | LYZ lysozyme C | | |
| 211 | IPI00397801 | FLG2
filaggrin-2 | Calcium ion
binding | |
| 212 | IPI00186290 | EEF2 elongation
factor 2 | | |
| 213 | IPI00783097 | GARS glycyl-tRNA
synthetase | | |
| 214 | IPI00029468 | ACTR1A
α-centractin | | |
| 215 | IPI00010845 | NDUFS8 NADH
dehydrogenase (ubiquinone) iron-sulfur protein 8,
mitochondrial | | |
| 216 | IPI00256861 | MACF1
microtubule-actin crosslinking factor 1 | Wnt receptor
signaling pathway, calcium ion binding | |
| 217 | IPI00017292 | CTNNB1 Catenin
β-1 | Apoptosis, cell
proliferation, regulation of MAPKKK cascade | Wnt signaling
pathway, pathways in cancer |
| 218 | IPI00005966 | NDUFA12 13 kDa
differentiation-associated protein variant (Fragment) | | |
| 219 | IPI00022229 | APOB apolipoprotein
B-100 | | |
| 220 | IPI00307259 | DNAJC13 dnaJ
homolog subfamily C member 13 | | |
| 221 | IPI00216085 | COX6B1 cytochrome
c oxidase subunit VIb isoform 1 | | |
| 222 | IPI00022774 | VCP
valosin-containing protein | Apoptosis,
ER-nuclear signaling pathway | |
| 223 | IPI00479640 | C1orf113 chromosome
1 open reading frame 113 | | |
| 224 | IPI00033019 | KCNB1 potassium
voltage-gated channel subfamily B member 1 | | |
| 225 | IPI00455876 | RING1 isoform 2 of
E3 ubiquitin-protein ligase RING1 | | |
Classification of the 225 annotated proteins in
terms of molecular function, biological process and cellular
localization was performed according to the DAVID. Molecular
function was clustered and the protein binding (140, 41.8%) and
nucleotide binding (60, 17.9%) groups were the majority (Fig. 4C). For biological processes,
annotated proteins are particularly involved in the cell process
(167, 27.5%) and the multicellular organismal process (75, 12.4%)
(Fig. 4D). Most (172, 18.6%) of the
annotated proteins were localized in the intracellular (Fig. 4E). Distribution of molecular mass
and isoelectric points (PI) of the annotated proteins was analyzed.
Molecular mass ranged between 10.19 and 620.42 kDa in size, most of
them were between 10 and 60 kDa (Fig.
4F). PI of the proteins ranged between 4.35 and 11.05 with the
most PIs between four and ten (Fig.
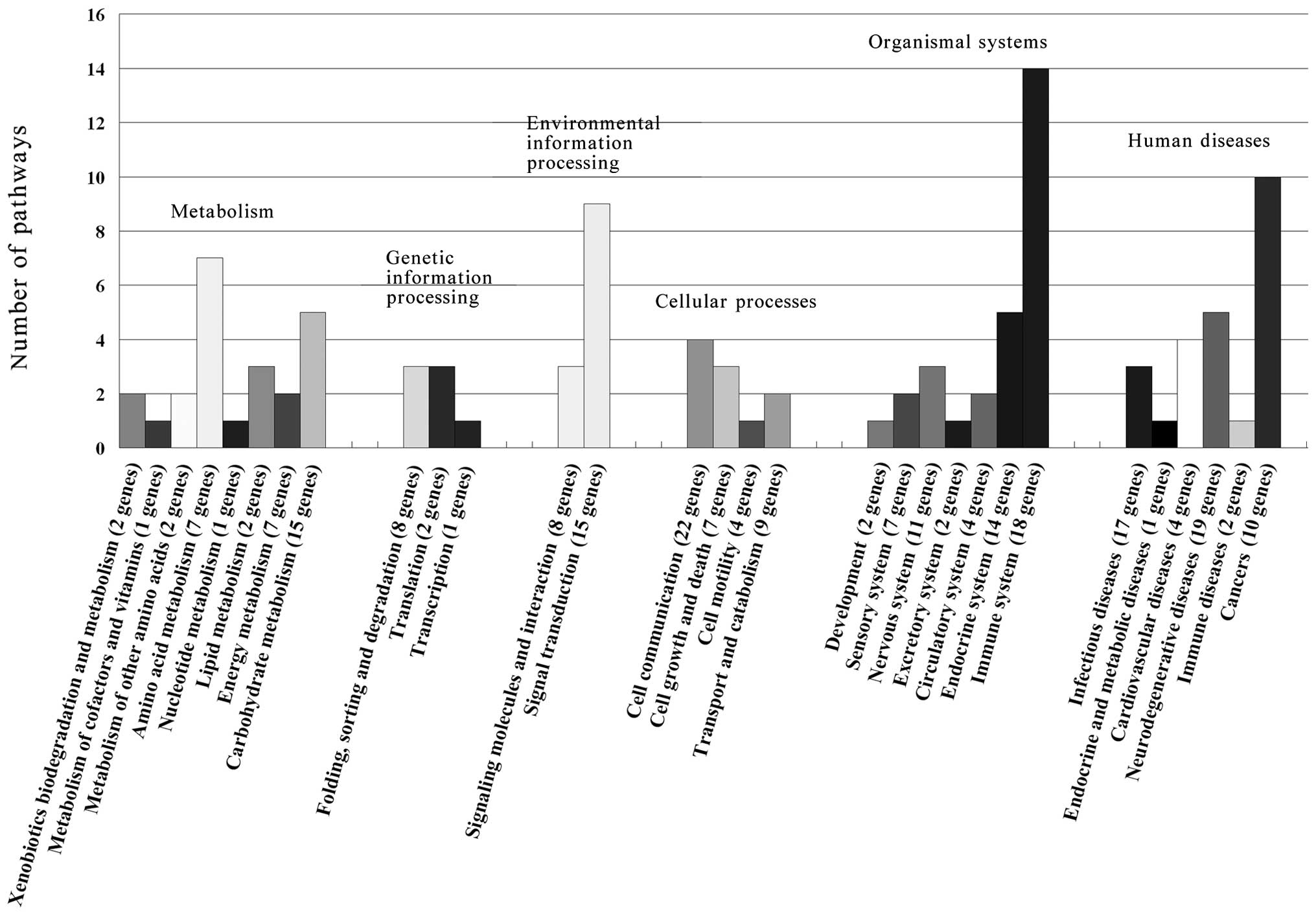
4G). To uncover the signaling pathways of the 225 annotated
proteins, the protein sequences were searched against the KEGG
reference pathway database. The pathways were ascribed to
metabolism, genetic information processing, environmental
information processing, cellular processes, organismal systems and
human diseases (Fig. 5). Among
them, the immune system, cancer and signal transduction were more
than others. On the other hand, the specific expressed proteins
related pathways displayed more differences and 71 proteins were
involved in cell apoptosis or proliferation biological processes
and KEGG pathways (Table I).
Discussion
In this study, to evaluate the function of MLAA-34
in M5 cells, we used the well-characterized cell line U937. In our
previous research, we reported that the MLAA-34 protein is probably
a cytoplasmic protein predicted by the amino acid sequence analysis
of the encoded protein (7). Here,
we verified that MLAA-34 is localized in the cytoplasm and cell
membrane. Western blot analysis showed that the expression of
MLAA-34 differed between different cell types and was observed to
be stronger in U937. Although U937 cells are generally difficult to
transfect, the U937 cells were transfected with MLAA-34-Lentivirus
and pGC-FU-GFP-LV. A stably transfected U937 cell line was
successfully established and expressed MLAA-34 at a high level,
which aided in the study exploring the effect of MLAA-34 on M5 and
will be critical for further research using U937 cells and animal
models. In addition, an analysis of the cell morphology, apoptosis,
proliferation and cell cycle revealed that the overexpression of
MLAA-34 markedly inhibited apoptosis of U937 cells. These results
suggested that MLAA-34 maybe a novel anti-apoptotic factor of M5,
which is consistent with the RNAi in our previous study.
The proteins that interact with MLAA-34 or CAB39L
remain unclear. To analyze complex mixtures of proteins, shotgun is
considered the most powerful (9,10).
Using the MLAA-34 protein as bait, 256 proteins were identified and
225 of them have DAVID terms. Among these proteins, 71 proteins
correlated with cell apoptosis or proliferation biological
processes and KEGG pathways. Twenty-eight proteins are involved in
cell apoptosis or proliferation; nine proteins are associated with
the calcium signaling pathway and seven proteins participate in the
chemokine signaling pathway; 17 proteins are concerned with the Ras
signaling transduction pathway and 8 proteins are concerned with
Wnt signaling pathway. The Ras, Wnt, calcium and chemokine
signaling pathways may be involved in anti-apoptosis with MLAA-34
in U937 cells. As is known, the Ras family plays an important role
in the molecular pathogenesis of myeloid leukemia, and Ras
mutations have been preferentially associated with monocytic
subtypes in AML (11). The Ras and
Wnt signaling pathways are known to be key anti-apoptosis pathways
in AML-M5 (12). Understanding the
molecular genetics of leukemia has led to an appreciation that
particular molecular abnormalities give rise to specific subtypes
of the disease. For example, in myeloid leukemogenesis, PML-RAR-α
and BCR-ABL are defining features of acute promyelocytic leukemia
and chronic myeloid leukemia, respectively (13). In this case, MLAA-34 may either play
an important role in leukemogenesis or play a dual role in
subsequent differentiation, as in the case of PML/RAR. The results
suggest that MLAA-34 might be an important agent for subtype
diagnosis in AML. However, an understanding of how these identified
proteins or pathways interact with MLAA-34 requires further
study.
In addition to the typical pathways such as pathways
in cancer and apoptosis, there were several notable pathways such
as the GPCR signaling, the insulin signaling pathway, the ErbB
signaling pathway, the NOD-like receptor signaling pathway, the Ahr
signal transduction pathway, the AKT signaling pathway, the
Toll-like receptor signaling pathway, the RIG-I-like receptor
signaling pathway, the ubiquitin mediated proteolysis, the hedgehog
signaling pathway, the phosphatidylinositol signaling system, the
PPAR signaling pathway, the VEGF signaling pathway and the TGF-β
signaling pathway worthy of further validation (Table I). Otherwise, there are some
proteins mainly involved in tumorigenesis concerned with MLAA-34 as
discussed below. PGK1 is secreted by tumor cells and may play a
role in inhibiting tumor angiogenesis (14). GAPDH has been shown to be
upregulated in several types of cancer and downregulated by
chemotherapeutic drugs, and could be considered a potential target
to observe the effects of bisphosphonates on cancer cells (15). In addition, GAPDH was the best
control gene in the apoptosis pattern on the myeloid cell lines
(16). CRMP1 is a suppressor of
tumor cell invasion of the local stroma and might be a functional
modulator of the Wnt signaling pathway in vivo(17,18).
As the trigger of TBK-1 pathway, TBK1 is important for tumor
angiogenesis and tumor-associated microvascular inflammation and
expressed at significant levels in many solid tumors (19,20). A
recent study has demonstrated that SEPT7 could function in
gliomagenesis and in the suppression of glioma cell proliferation
(21).
Markedly, some p53 or caspase-related proteins were
also identified, such as CLTC, PPP2CA, SOD2, PARK7, HSPA9, TXN,
ESR1 and YWHAE. CLTC associates with p53 not only in nuclei but
also in cytosol, and co-localizes with p53 at the plasma membrane
in human cancer cells (22). CLTC
expression enhances p53-dependent transactivation (23). As a downstream mediator of the
antiproliferative effects of PPP2CA, p53 plays an important role in
PPP2CA-directed cell cycle arrest and apoptosis (24). The SOD2 growth-retarding functions
are at least partially due to triggering of a p53-dependent
cellular senescence program (25).
DJ-1 (PARK7) bound to p53 in vitro and in vivo and
they were found colocalized. DJ-1 positively regulates p53 through
Topors-mediated sumoylation (26).
Previous studies indicated that HSPA9 could bind to p53 and
sequesters it in the cytoplasm, thus providing a mechanism of
inactivation of wild-type p53 and contributing to human
carcinogenesis (27,28). Additional studies have shown that
TXN induces p53 DNA binding activity in vitro and enhances
p53-dependent expression of its target gene p21 and DNA repair
genes (29). Additional studies
also indicated that caspases could be activated by TXN due to its
disulfide reducing properties (30). ESR1 might activate caspases-8, −9
and −3 and induce tumor cell apoptosis, it also showed the
downregulation of β-catenin signaling implicating the suppression
of proliferation and metastasis of tumor cells (31,32).
The cleavage of YWHAE by caspase-3 during apoptosis might
contribute to cell death by preventing the association of YWHAE
with Bad (33). The key event
during apoptosis that is common to all pathways is the activation
of caspases. P53 is a well-known tumor suppressor gene, and
mutational inactivation of p53 function or deletion of the gene
increases susceptibility to cancer (34–37).
On the basis of these findings, we will further study the
interaction between MLAA-34 and caspases or p53 to investigate the
anti-apoptotic mechanisms of MLAA-34 in U937 cells.
To our knowledge, this is the first report showing
the cellular localization and expression of MLAA-34 in U937 cells.
We have demonstrated for the first time that the overexpression of
MLAA-34 by lentivirus can significantly suppress the apoptosis of
U937 cells, and a cell line stably overexpressing MLAA-34 was
successfully established. Another key finding of this study is the
information from proteomics evidence that MLAA-34 may be a
tumor-correlated gene, and this is the first time it is revealed
that the preliminary framework of proteins and pathways interlink
with MLAA-34 in U937. Furthermore, it will be essential to
integrate data from many different sources to obtain an accurate
understanding of MLAA-34 protein networks.
Gene therapy remains the most promising, if not the
only, approach to treating genetic diseases. An example of this is
the use of rituximab for the treatment of lymphoma and other types
of cancer. Rituximab is a mouse/human chimeric IgG(1)-κ monoclonal antibody that targets the
CD20 antigen found on the surface of malignant and normal B
lymphocytes (38). Most cellular
processes are performed by multiprotein complexes. The
identification and analysis of their components provides insight
into how the ensemble of expressed proteins (the proteome) is
organized into functional units (39). Nevertheless, for a viable clinical
approach, extensive research is needed in the future to regulate
the expression of the target gene and improve its safety.
In conclusion, our current results provide new
evidence that MLAA-34 may be a novel anti-apoptotic factor in
vitro, and the data presented here show a strong correlation
between anti-apoptosis with the upregulation of MLAA-34. In
addition, preliminary proteomic analysis suggests that a number of
genes belonging to different signaling pathways may be involved in
apoptosis in U937 cells in association with MLAA-34, which would
disclose a novel cross-link between MLAA-34 and the Ras, Wnt,
calcium and chemokine signaling pathways. Findings of the present
study will lead to a better understanding of the mechanisms
involved in M5, and MLAA-34 may serve as a potential novel marker
for the early diagnosis and gene therapy of M5.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China under award nos. 30971284, 81000219 and
18110021.
References
|
1
|
Ossenkoppele GJ, Graveland WJ, Sonneveld
P, Daenen SM, Biesma DH, Verdonck LF, Schaafsma MR, Westveer PH,
Peters GJ, Noordhuis P, Muus P, Selleslag D, van der Holt B,
Delforge M, Lowenberg B and Verhoef GE: The value of fludarabine in
addition to ARA-C and G-CSF in the treatment of patients with
high-risk myelodysplastic syndromes and AML in elderly patients.
Blood. 103:2908–2913. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Faderl S, Gandhi V, O’Brien S, Bonate P,
Cortes J, Estey E, Beran M, Wierda W, Garcia-Manero G, Ferrajoli A,
Estrov Z, Giles FJ, Du M, Kwari M, Keating M, Plunkett W and
Kantarjian H: Results of a phase 1–2 study of clofarabine in
combination with cytarabine (ara-C) in relapsed and refractory
acute leukemias. Blood. 105:940–947. 2005.
|
|
3
|
Fiedler W, Serve H, Döhner H, Schwittay M,
Ottmann OG, O’Farrell AM, Bello CL, Allred R, Manning WC,
Cherrington JM, Louie SG, Hong W, Brega NM, Massimini G, Scigalla
P, Berdel WE and Hossfeld DK: A phase 1 study of SU11248 in the
treatment of patients with refractory or resistant acute myeloid
leukemia (AML) or not amenable to conventional therapy for the
disease. Blood. 105:986–993. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niitsu N, Yamamoto-Yamaguchi Y, Kasukabe
T, Okabe-Kado J, Umeda M and Honma Y: Antileukemic efficacy of
2-deoxycoformycin in monocytic leukemia cells. Blood. 96:1512–1516.
2000.PubMed/NCBI
|
|
5
|
Tallman MS, Kim HT, Paietta E, Bennett JM,
Dewald G, Cassileth PA, Wiernik PH and Rowe JM: Acute monocytic
leukemia (French-American-British classification M5) does not have
a worse prognosis than other subtypes of acute myeloid leukemia: a
report from the Eastern Cooperative Oncology Group. J Clin Oncol.
22:1276–1286. 2004. View Article : Google Scholar
|
|
6
|
Chen G, Zhang W, Cao X, Li F, Liu X and
Yao L: Serological identification of immunogenic antigens in acute
monocytic leukemia. Leuk Res. 29:503–509. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang PY, Zhang WG, He AL, Wang JL and Li
WB: Identification and functional characterization of the novel
acute monocytic leukemia associated antigen MLAA-34. Cancer Immunol
Immunother. 58:281–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao J, He A, Zhang W, Meng X and Gu L:
Quantitative assessment of MLAA-34 expression in diagnosis and
prognosis of acute monocytic leukemia. Cancer Immunol Immunother.
60:587–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu B, McClatchy DB, Kim JY and Yates JR
III: Strategies for shotgun identification of integral membrane
proteins by tandem mass spectrometry. Proteomics. 8:3947–3955.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He P, He HZ, Dai J, Wang Y, Sheng QH, Zhou
LP, Zhang ZS, Sun YL, Liu F, Wang K, Zhang JS, Wang HX, Song ZM,
Zhang HR, Zeng R and Zhao X: The human plasma proteome: analysis of
Chinese serum using shotgun strategy. Proteomics. 5:3442–3453.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bowen DT, Frew ME, Hills R, Gale RE,
Wheatley K, Groves MJ, Langabeer SE, Kottaridis PD, Moorman AV,
Burnett AK and Linch DC: RAS mutation in acute myeloid leukemia is
associated with distinct cytogenetic subgroups but does not
influence outcome in patients younger than 60 years. Blood.
106:2113–2119. 2005.PubMed/NCBI
|
|
12
|
Morgan MA, Dolp O and Reuter CW:
Cell-cycle-dependent activation of mitogen-activated protein kinase
kinase (MEK-1/2) in myeloid leukemia cell lines and induction of
growth inhibition and apoptosis by inhibitors of RAS signaling.
Blood. 97:1823–1834. 2001. View Article : Google Scholar
|
|
13
|
Pearn L, Fisher J, Burnett AK and Darley
RL: The role of PKC and PDK1 in monocyte lineage specification by
Ras. Blood. 109:4461–4469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lay AJ, Jiang XM, Kisker O, Flynn E,
Underwood A, Condron R and Hogg PJ: Phosphoglycerate kinase acts in
tumour angiogenesis as a disulphide reductase. Nature. 408:869–873.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valenti MT, Bertoldo F, Dalle Carbonare L,
Azzarello G, Zenari S, Zanatta M, Balducci E, Vinante O and Lo
Cascio V: The effect of bisphosphonates on gene expression: GAPDH
as a housekeeping or a new target gene? BMC Cancer. 6:492006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ullmannová V and Haskovec C: The use of
housekeeping genes (HKG) as an internal control for the detection
of gene expression by quantitative real-time RT-PCR. Folia Biol
(Praha). 49:211–216. 2003.PubMed/NCBI
|
|
17
|
Pan SH, Chao YC, Hung PF, Chen HY, Yang
SC, Chang YL, Wu CT, Chang CC, Wang WL, Chan WK, Wu YY, Che TF,
Wang LK, Lin CY, Lee YC, Kuo ML, Lee CH, Chen JJ, Hong TM and Yang
PC: The ability of LCRMP-1 to promote cancer invasion by enhancing
filopodia formation is antagonized by CRMP-1. J Clin Invest.
121:3189–3205. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stelzl U, Worm U, Lalowski M, Haenig C,
Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A,
Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S,
Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W,
Lehrach H and Wanker EE: A human protein-protein interaction
network: a resource for annotating the proteome. Cell. 122:957–968.
2005.PubMed/NCBI
|
|
19
|
Korherr C, Gille H, Schäfer R,
Koenig-Hoffmann K, Dixelius J, Egland KA, Pastan I and Brinkmann U:
Identification of proangiogenic genes and pathways by
high-throughput functional genomics: TBK1 and the IRF3 pathway.
Proc Natl Acad Sci USA. 103:4240–4245. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Czabanka M, Korherr C, Brinkmann U and
Vajkoczy P: Influence of TBK-1 on tumor angiogenesis and
microvascular inflammation. Front Biosci. 13:7243–7249. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia ZF, Huang Q, Kang CS, Yang WD, Wang
GX, Yu SZ, Jiang H and Pu PY: Overexpression of septin 7 suppresses
glioma cell growth. J Neurooncol. 98:329–340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Endo Y, Sugiyama A, Li SA, Ohmori K, Ohata
H, Yoshida Y, Shibuya M, Takei K, Enari M and Taya Y: Regulation of
clathrin-mediated endocytosis by p53. Genes Cells. 13:375–386.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Enari M, Ohmori K, Kitabayashi I and Taya
Y: Requirement of clathrin heavy chain for p53-mediated
transcription. Genes Dev. 20:1087–1099. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ofek P, Ben-Meir D, Kariv-Inbal Z, Oren M
and Lavi S: Cell cycle regulation and p53 activation by protein
phosphatase 2C alpha. J Biol Chem. 278:14299–14305. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Behrend L, Mohr A, Dick T and Zwacka RM:
Manganese superoxide dismutase induces p53-dependent senescence in
colorectal cancer cells. Mol Cell Biol. 25:7758–7769. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shinbo Y, Taira T, Niki T, Iguchi-Ariga SM
and Ariga H: DJ-1 restores p53 transcription activity inhibited by
Topors/p53BP3. Int J Oncol. 26:641–648. 2005.PubMed/NCBI
|
|
27
|
Grover A, Priyandoko D, Gao R, Shandilya
A, Widodo N, Bisaria VS, Kaul SC, Wadhwa R and Sundar D: Withanone
binds to mortalin and abrogates mortalin-p53 complex: Computational
and experimental evidence. Int J Biochem Cell Biol. 44:496–504.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wadhwa R, Yaguchi T, Hasan MK, Mitsui Y,
Reddel RR and Kaul SC: Hsp70 family member, mot-2/mthsp70/GRP75,
binds to the cytoplasmic sequestration domain of the p53 protein.
Exp Cell Res. 274:246–253. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ueno M, Masutani H, Arai RJ, Yamauchi A,
Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J and Nikaido T:
Thioredoxin-dependent redox regulation of p53-mediated p21
activation. J Biol Chem. 274:35809–35815. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ueda S, Nakamura H, Masutani H, Sasada T,
Yonehara S, Takabayashi A, Yamaoka Y and Yodoi J: Redox regulation
of caspase-3(-like) protease activity: regulatory roles of
thioredoxin and cytochrome c. J Immunol. 161:6689–6695.
1998.PubMed/NCBI
|
|
31
|
Hsu HH, Cheng SF, Chen LM, Liu JY, Chu CH,
Weng YJ, Li ZY, Lin CS, Lee SD, Kuo WW and Huang CY: Over-expressed
estrogen receptor-α upregulates hTNF-α gene expression and
downregulates β-catenin signaling activity to induce the apoptosis
and inhibit proliferation of LoVo colon cancer cells. Mol Cell
Biochem. 289:101–109. 2006.
|
|
32
|
Kouzmenko AP, Takeyama K, Ito S, Furutani
T, Sawatsubashi S, Maki A, Suzuki E, Kawasaki Y, Akiyama T, Tabata
T and Kato S: Wnt/β-catenin and estrogen signaling converge in
vivo. J Biol Chem. 279:40255–40258. 2004.
|
|
33
|
Won J, Kim DY, La M, Kim D, Meadows GG and
Joe CO: Cleavage of 14–3–3 protein by caspase-3 facilitates bad
interaction with Bcl-x(L) during apoptosis. J Biol Chem.
278:19347–19351. 2003.
|
|
34
|
Vousden KH and Prives C: P53 and
prognosis: new insights and further complexity. Cell. 120:7–10.
2005.PubMed/NCBI
|
|
35
|
Vousden KH and Lane DP: p53 in health and
disease. Nat Rev Mol Cell Biol. 8:275–283. 2007. View Article : Google Scholar
|
|
36
|
Pietsch EC, Sykes SM, McMahon SB and
Murphy ME: The p53 family and programmed cell death. Oncogene.
27:6507–6521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar
|
|
38
|
Plosker GL and Figgitt DP: Rituximab: a
review of its use in non-Hodgkin’s lymphoma and chronic lymphocytic
leukaemia. Drugs. 63:803–843. 2003.
|
|
39
|
Gavin AC, Bösche M, Krause R, Grandi P,
Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM,
Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A,
Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S,
Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth
E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B,
Kuster B, Neubauer G and Superti-Furga G: Functional organization
of the yeast proteome by systematic analysis of protein complexes.
Nature. 415:141–147. 2002. View
Article : Google Scholar : PubMed/NCBI
|