Introduction
Cancer is the major cause of mortality in human
populations worldwide, and human hepatocellular carcinoma is one of
the most lethal types of cancers (1,2).
Typical treatment approaches to human hepatocellular carcinoma
include hepatic resection, chemotherapy, percutaneous ablation and
transcatheter arterial chemoembolization and transplantation, yet
patient outcomes are not satisfactory (3,4).
Currently, investigators are focusing on new agents and novel
targets for human hepatocellular carcinoma treatment (5–7).
Caspases are important proteases in cells. Following
apoptotic stimuli, caspases can stimulate intracellular cascades
and activate downstream caspase members (8,9).
Several apoptotic stimuli have been reported that include extrinsic
pathways (receptor-ligand interaction) and intrinsic pathways
(mitochondrial-involved) (10–12).
In the intrinsic apoptosis pathway, caspase-9 acts as a major
initiator caspase, while in the extrinsic pathway, caspase-8 is a
major initiator caspase (16–18).
Endoplasmic reticulum (ER) stress induces apoptotic
cell death (13–15). Recent studies have identified ER as
a third pathway implicated in apoptosis. ER has several biological
functions including protein folding, protein trafficking and
regulation of the intracellular calcium concentration in apoptosis
(15,19,20).
When ER disrupts the biological function, the unfolded protein
response is triggered and this response occurs through the
activation of ER stress sensor proteins, including
inositol-requiring enzyme 1 (IRE1), GADD153 and activating
transcription factor 6 (ATF-6) (10,11,21).
The ubiquitin-proteasome system plays an important role in the
degradation of unfolded proteins (22,23).
The continued increase of unfolded proteins in the ER lumen
disrupts Ca2+ homeostasis in the ER and ultimately leads
to apoptosis. The major initiator caspase is caspase-4 in human
cells or caspase-12 in murine cells (24–26).
In our previous study, we designed and synthesized a
series of 2-phenyl-4-quinolone compounds as novel antitumor agents
(27–30).
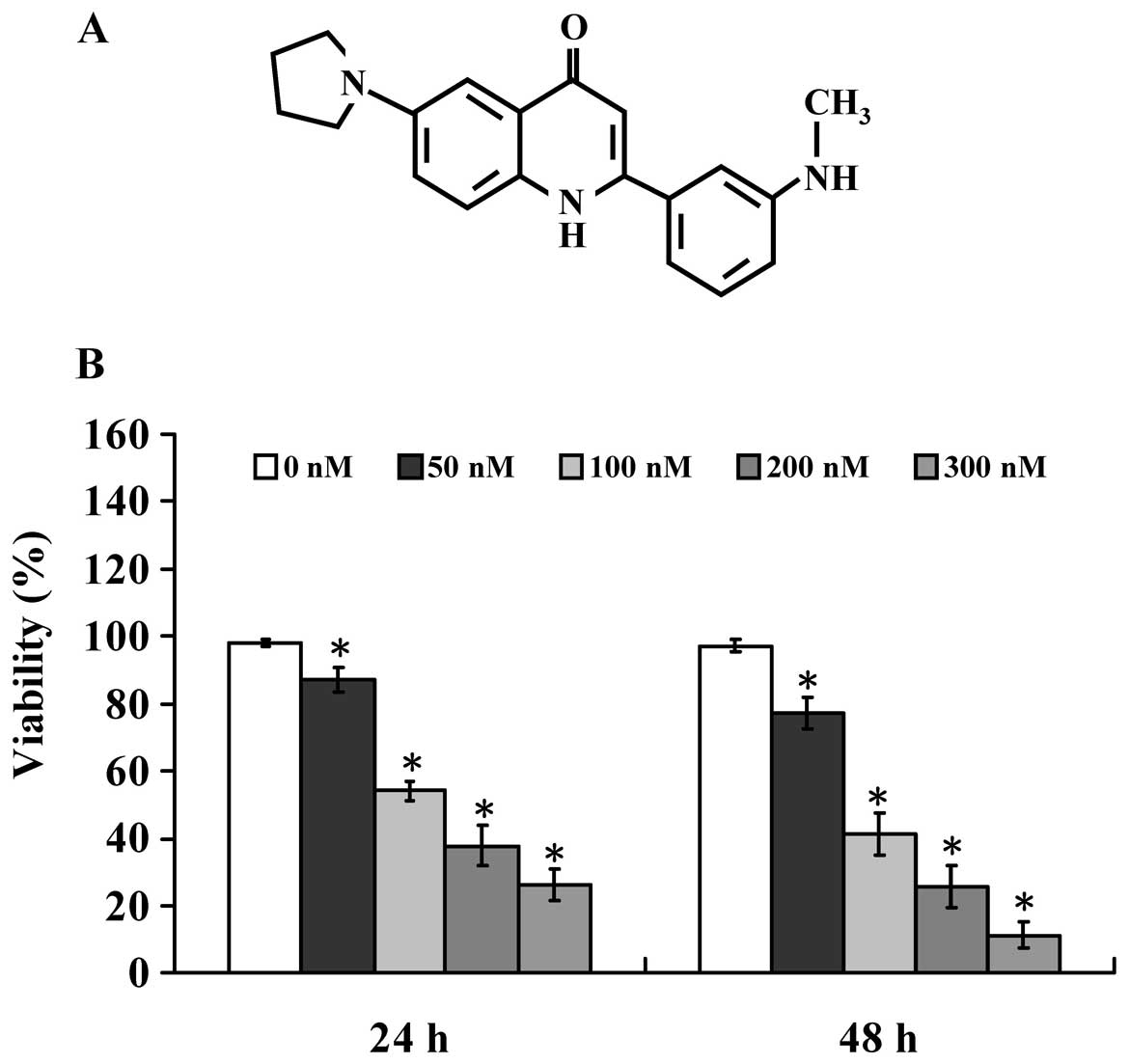
2-(3-(Methylamino)phenyl)-6-(pyrrolidin-1-yl)quinolin-4-one (Smh-3)
(Fig. 1A) is a candidate exhibiting
the most potential for antitumor activities. We demonstrated that
Smh-3 induces G2/M phase arrest and
mitochondrial-dependent apoptotic cell death through inhibition of
CDK1 and AKT activity in HL-60 human leukemia cells (31). However, neither the cytotoxic
effects of Smh-3 on human hepatocellular carcinoma cells, nor the
molecular mechanisms underlying its anticancer activity have been
investigated. Therefore, this study investigated the molecular
mechanisms of the antitumor effects of Smh-3 on Hep3B cells in
vitro.
Materials and methods
Materials, chemicals and reagents
MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide],
potassium phosphate, trypan blue, propidium iodide (PI), Triton
X-100, Tris-HCl and ribonuclease-A were obtained from Sigma-Aldrich
Corp. (St. Louis, MO). 3′-Dihexyloxacarbocyanine iodide
(DiOC6), RPMI-1640 medium, L-glutamine, fetal bovine
serum (FBS), Trypsin-EDTA, penicillin, nitrocellulose membrane and
the iBlot Dry Blotting system were obtained from Invitrogen Life
Technologies (Carlsbad, CA). Caspase-4 activity substrate
(Ac-LEVD-pNA) was purchased from BioVision (Mountain View, CA) and
caspase-3 and -9 activity assay kits were purchased from R&D
Systems (Minneapolis, MN). Primary antibodies (anti-caspase-4,
anti-GADD153 and anti-β-actin) and second antibodies for western
blotting were obtained from Santa Cruz Biotechnology (Santa Cruz,
CA).
Cell culture
The human hepatocellular carcinoma Hep3B cell line
was obtained from the Food Industry Research and Development
Institute (Hsinchu, Taiwan). The Hep3B cells were incubated in 5%
CO2 at 37°C in DMEM medium with 2 mM L-glutamine,
supplemented with 10% heat-inactivated FBS and 1%
antibiotic/antimycotic (100 units/ml penicillin and 100 μg/ml
streptomycin) (32).
Determination of cell morphology and the
percentage of viable cells
For analysis of cell morphological changes, cells
treated with Smh-3 (100 nM) in the well were examined and
photographed under a phase-contrast microscope at a magnification
of ×400. The quantitative analysis of cell viability was performed
by MTT assay. Cells (1×104 cells/well) on 96-well plates
were exposed to Smh-3 (0, 50, 100, 200 and 300 nM) and 0.1% DMSO as
a vehicle control. After a 24- and 48-h incubation, 100 ml MTT (0.5
mg/ml) solution was added to each well, and the plate was incubated
at 37°C for 4 h. Then, 0.04 N HCl in isopropanol was added, and the
absorbance at 570 nm was measured for each well. All results were
representative of 3 independent experiments (33,34).
DNA content and cell cycle distribution
analysis
Hep3B cells were incubated with 0, 50, 100, 200 and
300 nM of Smh-3 for 24 h. For determination of cell cycle phase and
apoptosis, cells were fixed gently in 70% ethanol at −20°C
overnight, and then re-suspended in PBS containing 40 μg/ml PI, 0.1
mg/ml RNase and 0.1% Triton X-100 in a dark room. Cell cycle
distribution and apoptotic nuclei were determined by flow cytometry
(31,35,36).
CDK1 kinase assay
CDK1 kinase activity was analyzed according to the
protocol outlined for the CDK1 kinase assay kit (Medical &
Biological Laboratories International, Nagoya, Japan). In brief,
the ability of the cell extract prepared from each treatment to
phosphorylate its specific substrate, MV peptide, was measured as
previously described (31,33).
Caspase activity assay
Hep3B cells were incubated with 0, 50, 100, 200 and
300 nM of Smh-3 for 24 h. Cells were lysed in lysis buffer [50 mM
Tris-HCl (pH 7.4), 1 mM EDTA, 10 mM EGTA, 10 mM digitonin and 2 mM
DTT]. Approximately 50 μg of cytosol proteins was incubated with
caspase-4 (BioVision), caspase-9 and caspase-3-specific substrates
(R&D System) for 1 h at 37°C. The caspase activity was
determined by measuring OD405 as previously described
(31,33,37).
Assay of intracellular Ca2+
levels
Hep3B cells were treated with 0, 50, 100, 200 and
300 nM of Smh-3 for 24 h. Cells were harvested, washed twice and
re-suspended in 3 mg/ml of Fluo-3/AM (Calbiochem; La Jolla, CA) at
37°C for 30 min and analyzed by flow cytometry (Becton-Dickinson
FACSCalibur) (38,39).
Determination of mitochondrial membrane
potential (ΔΨm)
Hep3B cells were treated with 0, 50, 100, 200 and
300 nM of Smh-3. The cells were harvested and washed twice,
resuspended in DiOC6 (4 mmol/l) and incubated for 30 min
before being analyzed by flow cytometry (Becton-Dickinson
FACSCalibur) (38,39).
Western blot assay
Hep3B cells were placed into 75-T flask. Cells in
each well were treated without and with 0, 50, 100, 200 and 300 nM
of Smh-3 for 24 h. Cells were collected and total protein from each
treatment was extracted and placed into buffer (PRO-PREP™ protein
extraction solution, Korea) and centrifuged at 12,000 rpm for 10
min at 4°C. The quantitated total protein from each treatment was
determined by Bradford assay. Proteins from each treatment were
resolved on an SDS polyacrylamide gel through electrophoresis
(SDS-PAGE) and transferred to nitrocellulose membranes. The
membranes were incubated with a blocking buffer of 5% non-fat dry
milk in Tris-buffered saline containing Tween-20 for 1 h at room
temperature and then incubated with the specific primary antibodies
(anti-GADD153 and anti-caspase-4). The membranes were washed and
then treated by appropriate horseradish peroxidase (HRP)-conjugated
secondary antibodies and visualized using an ECL detection kit (GE
Healthcare, Princeton, NJ) (33,40).
cDNA microarray analysis
Hep3B cells were treated with or without 100 nM of
Smh-3 for 24 h. Then cells from each treatment were harvested, and
the total RNA was extracted using the Qiagen RNeasy Mini kit
(Qiagen, Inc., Valencia, CA, USA). The isolated total RNA was used
for cDNA synthesis and labeling and microarray hybridization. The
fluorescence-labeled cDNA were then hybridized to their complements
on the chip (Affymetrix GeneChip Human Gene 1.0 ST array,
Affymetrix, Santa Clara, CA, USA). Finally the resulting localized
concentrations of fluorescent molecules were detected and
quantitated (Asia Bio-Innovations Corp.). The resulting data were
analyzed using Expression Console software (Affymetrix) with
default RMA parameters. Genes regulated by citosol were determined
to have a 1.5-fold change in expression (41).
Statistical analysis
Significance of the mean values between the
Smh-3-treated group and control group was obtained using the
Student’s t-test. Data were expressed as the means ± SD. P<0.05
was considered to indicate a statistically significant difference
(33,40).
Results
Smh-3 decreases the percentage of Hep3B
viable cells
To investigate the effect of Smh-3 on cell
proliferation, Hep3B cells were treated with 0, 50, 100, 200 and
300 nM of Smh-3 for 48 h. The cell viability following each
treatment was analyzed by MTT assay. As shown in Fig. 1B, Smh-3 inhibited Hep3B cell growth
in a dose- and time-dependent manner. The half maximal inhibitory
concentration IC50 following a 48-h treatment of Smh-3
was 68.26±3.24 nM.
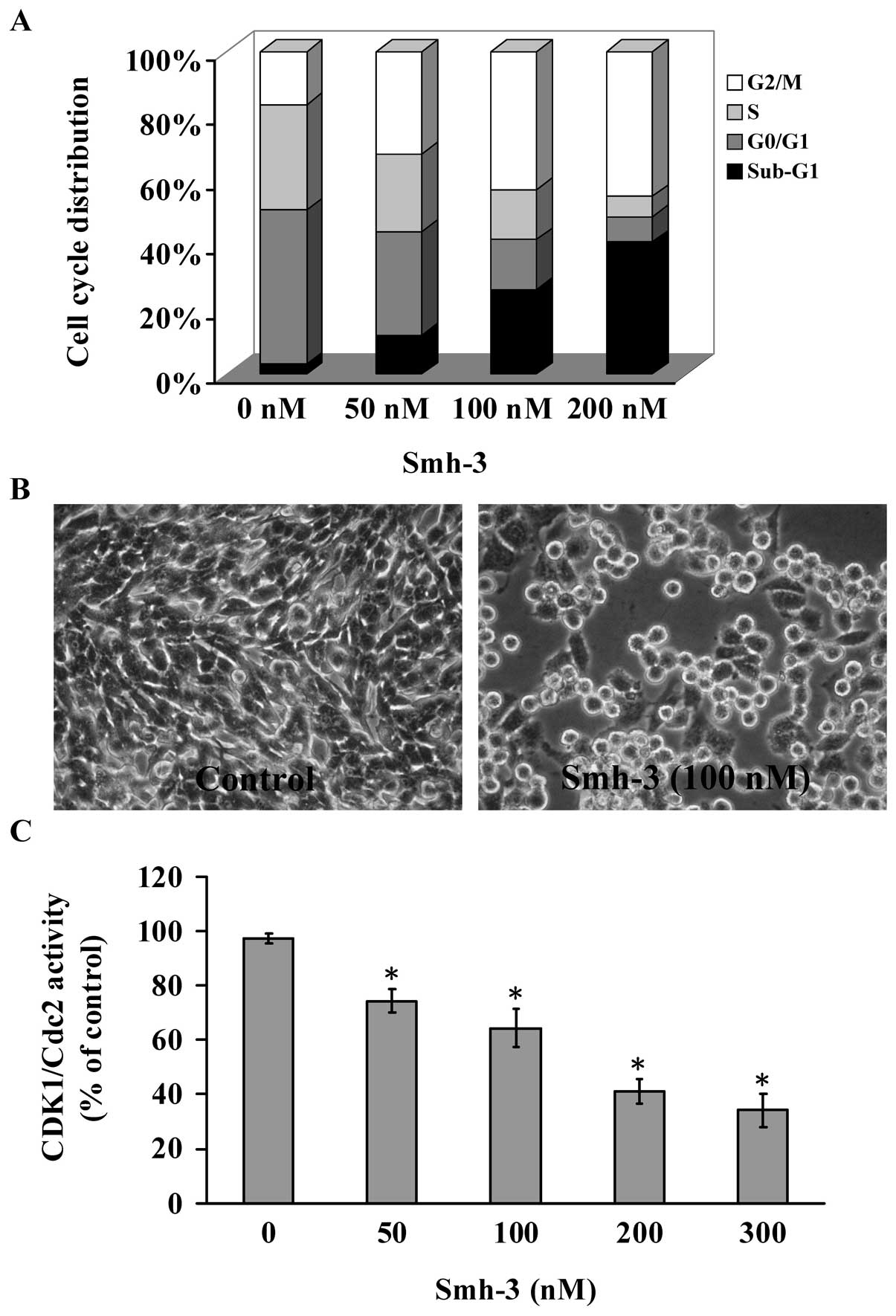
Smh-3 induces G2/M arrest and
decreases CDK1 activity and apoptosis in Hep3B cells
Smh-3 induced cell morphological changes and
decreased the cell numbers of Hep3B cells (Fig. 2B). Mitotic and apoptotic cells
appeared smaller, round and blunt in size following exposure to
Smh-3. To investigate the cell cycle distribution of Hep3B cells
following Smh-3 treatment, cells were stained with propidium iodide
(PI). Flow cytometry revealed that Smh-3 treatment (0, 50, 100 and
200 nM) of Hep3B cells significantly increased the G2/M
cell population at 48 h (Fig. 2A).
Furthermore, Smh-3 treatment increased the sub-G1 cell
population at 48 h in a concentration-dependent manner. These data
suggest that Smh-3 effectively induces G2/M arrest and
promotes cell death. We examined the CDK1 activity in Smh-3-treated
Hep3B cells. Treatment with 0, 50, 100, 200 and 300 nM Smh-3 caused
a significant decrease in CDK1 activity (Fig. 2C). Our results suggest that the
downregulation of CDK1 activity plays an important role in
G2/M phase arrest in Smh-3-treated Hep3B cells.
cDNA microarray analysis
The microarray analysis indicated 192 genes were
upregulated and 278 genes were downregulated in Hep3B cells
following treatment with Smh-3. Moreover, the mRNA descriptions of
the genes are listed in Table I.
DNA microarray assay revealed that many differentially expressed
genes related to angiogenesis, autophagy, calcium-mediated ER
stress signaling, cell adhesion, cell cycle and mitosis, cell
migration, cytoskeleton organization, DNA damage and repair,
mitochondrial-mediated apoptosis and cell signaling pathways were
present in the Hep3B cells following Smh-3 treatment.
 | Table IGenes exhibiting more than 1.5-fold
changes in mRNA levels in Hep-3B cells following a 24-h treatment
with Smh-3 as identified using DNA microarray. |
Table I
Genes exhibiting more than 1.5-fold
changes in mRNA levels in Hep-3B cells following a 24-h treatment
with Smh-3 as identified using DNA microarray.
| Fold-change | Gene symbol | mRNA
description |
|---|
| Biological process:
angiogenesis |
| 1.73 | RBPJ | Homo sapiens
mRNA for H-2K binding factor-2 |
| 1.52 | GPI | Homo sapiens
glucose phosphate isomerase |
| −1.64 | SH2D2A | Homo sapiens
SH2 domain containing 2A |
| −1.74 | RNH1 | Homo sapiens
ribonuclease/angiogenin inhibitor 1 |
| −1.79 | RTN4 | Homo sapiens
reticulon 4 |
| −1.87 | VEGFA | Homo sapiens
vascular endothelial growth factor A |
| Biological process:
apoptosis and anti-apoptosis |
| 1.73 | PERP | Homo sapiens
PERP, TP53 apoptosis effector |
| 1.61 | BTG1 | Homo sapiens
B-cell translocation gene 1 |
| 1.59 | DBH | Homo sapiens
dopamine β-hydroxylase |
| 1.58 | PARP11 | Homo sapiens
poly(ADP-ribose)polymerase family, member 11 |
| 1.58 | SEMA3A | Homo sapiens
sema domain |
| −1.51 | ABR | Homo sapiens
active BCR-related gene |
| −1.51 | BLOC1S2 | Homo sapiens
biogenesis of lysosomal organelles complex-1 |
| −1.51 | BCL2L1 | Homo sapiens
BCL2-like 1 |
| −1.52 | CARD17 | Homo sapiens
caspase recruitment domain family, member 17 |
| −1.53 | YARS | Homo sapiens
tyrosyl-tRNA synthetase |
| −1.54 | USP17L2 | Homo sapiens
ubiquitin specific peptidase 17-like 2 |
| −1.56 | APH1B | Homo sapiens
anterior pharynx defective 1 homolog B |
| −1.62 | IHH | Homo sapiens
Indian hedgehog homolog |
| −1.75 | CIDEB | Homo sapiens
cell death-inducing DFFA-like effector b |
| −1.85 | BCL6 | Homo sapiens
B-cell CLL/lymphoma 6, transcript variant 1 |
| Biological process:
autophagy |
| 1.97 | ATG12 | Homo sapiens
ATG12 autophagy related 12 homolog |
| 1.85 | CLEC4F | Homo sapiens
C-type lectin domain family 4, member F |
| 1.64 | ACP2 | Homo sapiens
acid phosphatase 2, lysosomal |
| 1.55 | HPS1 | Homo sapiens
Hermansky-Pudlak syndrome 1 |
| 1.53 | NEU1 | Homo sapiens
sialidase 1 |
| 1.51 | GABARAP | Homo sapiens
GABA receptor-associated protein |
| −1.62 | CHIT1 | Homo sapiens
chitinase 1 |
| −1.68 | VPS53 | Homo sapiens
vacuolar protein sorting 53 homolog |
| −2.12 | SYT3 | Homo sapiens
synaptotagmin III |
| Biological process:
calcium-mediated signaling |
| 1.68 | CCL3 | Homo sapiens
chemokine ligand 3 |
| 1.66 | STC1 | Homo sapiens
stanniocalcin 1 |
| 1.53 | CHRNA10 | Homo sapiens
cholinergic receptor |
| 1.53 | GRIN1 | Homo sapiens
glutamate receptor |
| 1.50 | ETNK2 | Homo sapiens
ethanolamine kinase 2 |
| −1.50 | CX3CL1 | Homo sapiens
chemokine ligand 1 |
| −1.52 | PLCG2 | Homo sapiens
phospholipase C, γ 2 |
| −1.55 | NMUR1 | Homo sapiens
neuromedin U receptor 1 |
| −1.64 | CALCB | Homo sapiens
calcitonin-related polypeptide β |
| −2.43 | KCNA5 | Homo sapiens
potassium voltage-gated channel |
| Biological process:
cell adhesion |
| 2.50 | PVRL2 | Homo sapiens
poliovirus receptor-related 2 |
| 1.71 | SYMPK | Homo sapiens
symplekin |
| 1.68 | SIGLEC5 | Homo sapiens
sialic acid binding Ig-like lectin 5 |
| 1.63 | LPXN | Homo sapiens
leupaxin |
| Biological process:
cell adhesion |
| 1.63 | CDH6 | Homo sapiens
cadherin 6 |
| 1.56 | GPR56 | Homo sapiens
G protein-coupled receptor 56 |
| −1.50 | PVRL4 | Homo sapiens
poliovirus receptor-related 4 |
| −1.51 | SIGLEC14 | Homo sapiens
sialic acid binding Ig-like lectin 14 |
| −1.51 | CPXM2 | Homo sapiens
carboxypeptidase X, member 2 |
| −1.52 | MMRN1 | Homo sapiens
multimerin 1 |
| −1.53 | NCAM1 | Homo sapiens
neural cell adhesion molecule 1 |
| −1.54 | PCDHB4 | Homo sapiens
protocadherin β 4 |
| −1.54 | PCDHB14 | Homo sapiens
protocadherin β 14 |
| −1.58 | PCDHB13 | Homo sapiens
protocadherin β 13 |
| −1.58 | PXN | Homo sapiens
paxillin |
| −1.61 | HSPB11 | Homo sapiens
heat shock protein family B |
| −1.61 | ITGA7 | Homo sapiens
integrin, α 7 |
| −1.62 | SIRPG | Homo sapiens
signal-regulatory protein γ |
| −1.62 | NINJ2 | Homo sapiens
ninjurin 2 |
| −1.62 | NEO1 | Homo sapiens
neogenin homolog 1 |
| −1.68 | CYR61 | Homo sapiens
cysteine-rich, angiogenic inducer, 61 |
| Biological process:
cell cycle and mitosis |
| 2.13 | CNNM3 | Homo sapiens
cyclin M3 |
| 1.84 | RAB11B | Homo sapiens
RAB11B, member RAS oncogene family |
| 1.80 | CDKN3 | Homo sapiens
cyclin-dependent kinase inhibitor 3 |
| 1.64 | 40789 | Homo sapiens
septin 3 |
| 1.59 | MGC16703 | Homo sapiens
tubulin, α pseudogene |
| 1.55 | KRT18 | Homo sapiens
keratin 18 |
| 1.52 | ARL8B | Homo sapiens
ADP-ribosylation factor-like 8B |
| 1.52 | RPRM | Homo sapiens
reprimo, TP53 dependent G2 arrest mediator candidate |
| −1.50 | HAUS6 | Homo sapiens
HAUS augmin-like complex, subunit 6 |
| −1.52 | HSPA8 | Homo sapiens
heat shock 70 kDa protein 8 |
| −1.55 | ACLY | Homo sapiens
ATP citrate lyase |
| −1.57 | CCNG1 | Homo sapiens
cyclin G1 |
| −1.62 | CHEK2 | Homo sapiens
CHK2 checkpoint homolog |
| −1.63 | TAF1 | Homo sapiens
TAF1 RNA polymerase II |
| −1.73 | PHF16 | Homo sapiens
PHD finger protein 16 |
| −1.87 | DUSP1 | Homo sapiens
dual specificity phosphatase 1 |
| −1.89 | PISD | Homo sapiens
THAP domain containing, apoptosis associated protein 1 |
| −1.96 | CLPX | Homo sapiens
non-SMC condensin I complex, subunit H |
| −2.10 | SLC25A26 | Homo sapiens
septin 2 |
| Biological process:
cell migration |
| −1.54 | PLXNB1 | Homo sapiens
plexin B1 |
| −1.56 | CLIC4 | Homo sapiens
chloride intracellular channel 4 |
| −1.66 | RNF20 | Homo sapiens
ring finger protein 20 |
| −1.72 | S100A2 | Homo sapiens
S100 calcium binding protein A2 |
| −1.73 | SEMA3C | Homo sapiens
sema domain, immunoglobulin domain, short basic domain |
| Biological process:
cytoskeleton organization and mitosis |
| 2.19 | TPM3 | Homo sapiens
tropomyosin 3 |
| 1.63 | CHD2 | Homo sapiens
chromodomain helicase DNA binding protein 2 |
| 1.59 | CORO2A | Homo sapiens
coronin, actin binding protein, 2A |
| 1.59 | ARHGAP26 | Homo sapiens
Rho GTPase activating protein 26 |
| 1.59 | DSTN | Homo sapiens
destrin (actin depolymerizing factor) |
| 1.57 | ACTB | Homo sapiens
actin |
| Biological process:
cytoskeleton organization and mitosis |
| 1.57 | ACTG1 | Homo sapiens
actin, γ 1 |
| 1.56 | TUBA1B | Homo sapiens
tubulin, α 1b |
| 1.51 | TUBB2C | Homo sapiens
tubulin, β 2C |
| 1.50 | KIF21A | Homo sapiens
kinesin family member 21A |
| −1.51 | KIF19 | Homo sapiens
kinesin family member 19 |
| −1.51 | MC1R | Homo sapiens
melanocortin 1 receptor |
| −1.51 | RUVBL1 | Homo sapiens
RuvB-like 1 |
| −1.53 | NDE1 | Homo sapiens
nudE nuclear distribution gene E homolog 1 |
| −1.58 | C2CD3 | Homo sapiens
C2 calcium-dependent domain containing 3 |
| −1.59 | NXF5 | Homo sapiens
nuclear RNA export factor 5 |
| −1.70 | BICD2 | Homo sapiens
bicaudal D homolog 2 |
| −1.76 | CYP1A1 | Homo sapiens
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| −1.88 | MAP1B | Homo sapiens
microtubule-associated protein 1B |
| −1.95 | KRT1 | Homo sapiens
keratin 1 |
| Biological process:
DNA damage and repair |
| 2.53 | NPM1 | Homo sapiens
nucleophosmin |
| 1.93 | DDIT3 | Homo sapiens
DNA-damage-inducible transcript 3 |
| 1.80 | BRCC3 | Homo sapiens
BRCA1/BRCA2-containing complex, subunit 3 |
| 1.56 | FTO | Homo sapiens
fat mass and obesity associated |
| 1.52 | HIPK1 | Homo sapiens
homeodomain interacting protein kinase 1 |
| −1.54 | MRPS11 | Homo sapiens
mitochondrial ribosomal protein S11 |
| −1.54 | GNL1 | Homo sapiens
guanine nucleotide binding protein-like 1 |
| −1.57 | NAA10 | Homo sapiens
N(α)-acetyltransferase 10, NatA catalytic subunit |
| −1.58 | MORF4L2 | Homo sapiens
mortality factor 4 like 2 |
| −1.59 | ATRX | Homo sapiens
α thalassemia/mental retardation syndrome X-linked |
| −1.59 | RUVBL2 | Homo sapiens
RuvB-like 2 |
| −1.63 | NCOA6 | Homo sapiens
nuclear receptor coactivator 6 |
| −1.72 | POLB | Homo sapiens
polymerase, β |
| −1.83 | ATMIN | Homo sapiens
ATM interactor |
| −2.03 | CIRBP | Homo sapiens
cold inducible RNA binding protein |
| −2.78 | SGK3 | Homo sapiens
serum/glucocorticoid regulated kinase family, member 3 |
| Biological process:
DNA transcription and replication |
| 1.96 | GLI1 | Homo sapiens
GLI family zinc finger 1 |
| 1.58 | CDC7 | Homo sapiens
cell division cycle 7 homolog |
| 1.53 | SET | Homo sapiens
SET nuclear oncogene |
| −1.55 | MCM6 | Homo sapiens
minichromosome maintenance complex component 6 |
| −1.59 | THRA | Homo sapiens
thyroid hormone receptor, α |
| −1.67 | NOBOX | Homo sapiens
NOBOX oogenesis homeobox |
| Biological process:
ER function and ER stress |
| 1.78 | NECAB1 | Homo sapiens
N-terminal EF-hand calcium binding protein1 |
| 1.77 | EFCAB4B | Homo sapiens
EF-hand calcium binding domain 4B |
| 1.71 | CHRNA7 | Homo sapiens
cholinergic receptor, nicotinic, α 7 |
| 1.65 | RYR1 | Homo sapiens
ryanodine receptor 1 |
| 1.63 | GRIN2A | Homo sapiens
glutamate receptor |
| 1.61 | SNAP25 | Homo sapiens
synaptosomal-associated protein |
| 1.53 | ALG13 | Homo sapiens
asparagine-linked glycosylation 13 homolog |
| 1.53 | MLEC | Homo sapiens
malectin |
| 1.51 | CYP2B6 | Homo sapiens
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| −1.50 | UBQLN4 | Homo sapiens
ubiquilin 4 |
| −1.50 | FITM1 | Homo sapiens
fat storage-inducing transmembrane protein 1 |
| Biological process:
ER function and ER stress |
| −1.55 | SEC22B | Homo sapiens
SEC22 vesicle trafficking protein homolog B |
| −1.55 | DERL1 | Homo sapiens
Der1-like domain family, member 1 |
| −1.63 | FOXRED2 | Homo sapiens
FAD-dependent oxidoreductase domain containing 2 |
| −1.63 | CYP4A22 | Homo sapiens
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| −1.69 | PLA2G2A | Homo sapiens
phospholipase A2, group IIA |
| −1.72 | HSP90B2P | Homo sapiens
heat shock protein 94b |
| −2.36 | NOX4 | Homo sapiens
NADPH oxidase 4 |
| −4.84 | S100A7 | Homo sapiens
S100 calcium binding protein A7 |
| Biological process:
mitochondrial function |
| 1.91 | ACAD8 | Homo sapiens
5-methyltetrahydrofolate-homocysteine methyltransferase
reductase |
| 1.74 | ACOT2 | Homo sapiens
ATP synthase, H+ transporting, mitochondrial F0
complex |
| 1.73 | SPNS1 | Homo sapiens
MpV17 mitochondrial inner membrane protein |
| 1.72 | NMRAL1 | Homo sapiens
NmrA-like family domain containing 1, mitochondrial |
| 1.69 | ACSS2 | Homo sapiens
NADH dehydrogenase 1 β subcomplex, 4, 15 kDa |
| 1.68 | SAT2 | Homo sapiens
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| 1.56 | MTRR | Homo sapiens
cytochrome b-561 |
| 1.54 | ATP5J | Homo sapiens
potassium inwardly-rectifying channel |
| 1.52 | MPV17 | Homo sapiens
pyruvate dehydrogenase phosphatase regulatory subunit |
| 1.50 | LONP1 | Homo sapiens
solute carrier family 3, member 1 |
| −1.50 | NDUFB4 | Homo sapiens
thymidine phosphorylase |
| −1.54 | CYP2D6 | Homo sapiens
dynamin 1-like |
| −1.55 | CYB561 | Homo sapiens
solute carrier family 25, member 30 |
| −1.57 | KCNJ11 | Homo sapiens
MOCO sulphurase C-terminal domain containing 2 |
| −1.58 | PDPR | Homo sapiens
cytochrome c oxidase subunit VIb polypeptide 1 |
| −1.59 | SLC3A1 | Homo sapiens
phosphatidylserine decarboxylase |
| −1.62 | TYMP | Homo sapiens
ClpX caseinolytic peptidase X homolog |
| −1.62 | DNM1L | Homo sapiens
solute carrier family 25, member 26 |
| −1.64 | SLC25A30 | Homo sapiens
yrdC domain containing |
| −1.65 | MOSC2 | Homo sapiens
surfeit 1 |
| −2.10 | COX6B1 | Homo sapiens
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| Biological process:
cell signal transduction |
| 2.54 | ZCCHC8 | Homo sapiens
Rho GTPase activating protein 12 |
| 2.18 | DHX35 | Homo sapiens
cannabinoid receptor 2 |
| 2.12 | DGCR14 | Homo sapiens
guanine nucleotide binding protein |
| 2.08 | RPS13 | Homo sapiens
calcitonin-related polypeptide α |
| 1.97 | SNRPG | Homo sapiens
opsin 4 |
| 1.83 | RBMY1A1 | Homo sapiens
growth factor receptor-bound protein 10 |
| 1.83 | ARHGAP12 | Homo sapiens
ribosomal protein S6 kinase, 90 kDa, polypeptide 3 |
| 1.80 | CNR2 | Homo sapiens
prolactin |
| 1.78 | GNAO1 | Homo sapiens
ras homolog gene family, member G |
| 1.77 | CALCA | Homo sapiens
leukocyte-associated immunoglobulin-like receptor 2 |
| 1.75 | OPN4 | Homo sapiens
CD79b molecule, immunoglobulin-associated β |
| 1.74 | GRB10 | Homo sapiens
leukocyte immunoglobulin-like receptor |
| 1.74 | RPS6KA3 | Homo sapiens
CD28 molecule |
| 1.69 | PRL | Homo sapiens
A kinase anchor protein 3 |
| 1.69 | RHOG | Homo sapiens
RAB6C |
| 1.68 | LAIR2 | Homo sapiens
RAB41 |
| 1.66 | CD79B | Homo sapiens
RAB15 |
| 1.66 | LILRB2 | Homo sapiens
ras homolog gene family, member V |
| 1.65 | CD28 | Homo sapiens
RAB, member of RAS oncogene family-like 2A |
| Biological process:
cell signal transduction |
| 1.65 | AKAP3 | Homo sapiens
RAB, member of RAS oncogene family-like 2B |
| 1.65 | RAB6C | Homo sapiens
RAB43, member RAS oncogene family |
| 1.63 | RAB41 | Homo sapiens
leukemia inhibitory factor |
| 1.63 | RAB15 | Homo sapiens
CD81 molecule |
| 1.62 | RHOV | Homo sapiens
CD74 molecule |
| 1.59 | RABL2A | Homo
sapienss transforming growth factor, β 3 |
| 1.59 | RABL2B | Homo sapiens
sema domain |
| 1.58 | RAB43 | Homo sapiens
acylglycerol kinase |
| 1.56 | LIF | Homo sapiens
pleckstrin and Sec7 domain containing |
| 1.56 | CD81 | Homo sapiens
fibroblast growth factor binding protein 1 |
| 1.54 | CD74 | Homo sapiens
olfactory receptor |
| 1.53 | TGFB3 | Homo sapiens
vomeronasal 1 receptor 3 |
| 1.53 | SEMA4C | Homo sapiens
olfactory receptor |
| 1.52 | AGK | Homo sapiens
olfactory receptor |
| 1.52 | PSD | Homo sapiens
olfactory receptor |
| 1.51 | FGFBP1 | Homo sapiens
olfactory receptor |
| 1.51 | OR51A4 | Homo sapiens
taste receptor |
| 1.51 | VN1R3 | Homo sapiens
olfactory receptor |
| −1.50 | OR5M10 | Homo sapiens
olfactory receptor |
| −1.51 | OR52N1 | Homo sapiens
neuromedin B receptor |
| −1.51 | OR1B1 | Homo sapiens
G protein-coupled receptor 120 |
| −1.52 | OR3A3 | Homo sapiens
olfactory receptor |
| −1.52 | TAS2R1 | Homo sapiens
olfactory receptor |
| −1.53 | OR3A1 | Homo sapiens
olfactory receptor |
| −1.53 | OR1D5 | Homo sapiens
olfactory receptor |
| −1.53 | NMBR | Homo sapiens
MAS-related GPR, member X2 |
| −1.54 | GPR120 | Homo sapiens
G protein-coupled receptor 119 |
| −1.54 | OR10C1 | Homo sapiens
olfactory receptor |
| −1.54 | OR10H5 | Homo sapiens
G protein-coupled receptor 109B |
| −1.55 | OR10G4 | Homo sapiens
G protein-coupled receptor 83 |
| −1.56 | OR4D5 | Homo sapiens
G protein-coupled receptor 39 |
| −1.56 | MRGPRX2 | Homo sapiens
olfactory receptor |
| −1.56 | GPR119 | Homo sapiens
olfactory receptor |
| −1.57 | OR7C2 | Homo sapiens
insulin receptor substrate 1 |
| −1.57 | GPR109B | Homo sapiens
insulin-like 3 |
| −1.58 | GPR83 | Homo sapiens
FYN binding protein |
| −1.58 | GPR39 | Homo sapiens
dual specificity phosphatase 16 |
| −1.59 | OR2L13 | Homo sapiens
hypocretin receptor 1 |
| −1.62 | OR52A1 | Homo sapiens
secretogranin V |
| −1.63 | IRS1 | Homo sapiens
GRB2-related adaptor protein |
| −1.63 | INSL3 | Homo sapiens
regulatory factor X-associated ankyrin-containing protein |
| −1.66 | FYB | Homo sapiens
ubiquitin specific peptidase 8 |
| −1.69 | DUSP16 | Homo sapiens
syndecan binding protein |
| −1.72 | HCRTR1 | Homo sapiens
discoidin domain receptor tyrosine kinase 2 |
| −1.72 | SCG5 | Homo sapiens
AXL receptor tyrosine kinase |
| −1.74 | GRAP | Homo sapiens
adenylate cyclase activating polypeptide 1 |
| −1.75 | RFXANK | Homo sapiens
EPH receptor A8 |
| Biological process:
cell signal transduction |
| −1.75 | USP8 | Homo sapiens
TBC1D30 mRNA for TBC1 domain family, member 30 |
| −1.75 | SDCBP | Homo sapiens
Rho guanine nucleotide exchange factor |
| −1.77 | DDR2 | Homo sapiens
RAS p21 protein activator 4 |
| −1.78 | AXL | Homo sapiens
RAS-like, family 12 |
| −1.78 | ADCYAP1 | Homo sapiens
mediator complex subunit 13 |
| −1.82 | EPHA8 | Homo sapiens
mitogen-activated protein kinase kinase 7 |
| −1.92 | TBC1D30 | Homo sapiens
dishevelled, dsh homolog 3 |
| −2.28 | ARHGEF10 | Homo sapiens
LIM domain binding 1 |
| −3.04 | RASA4 | Homo sapiens
coiled-coil domain containing 88C |
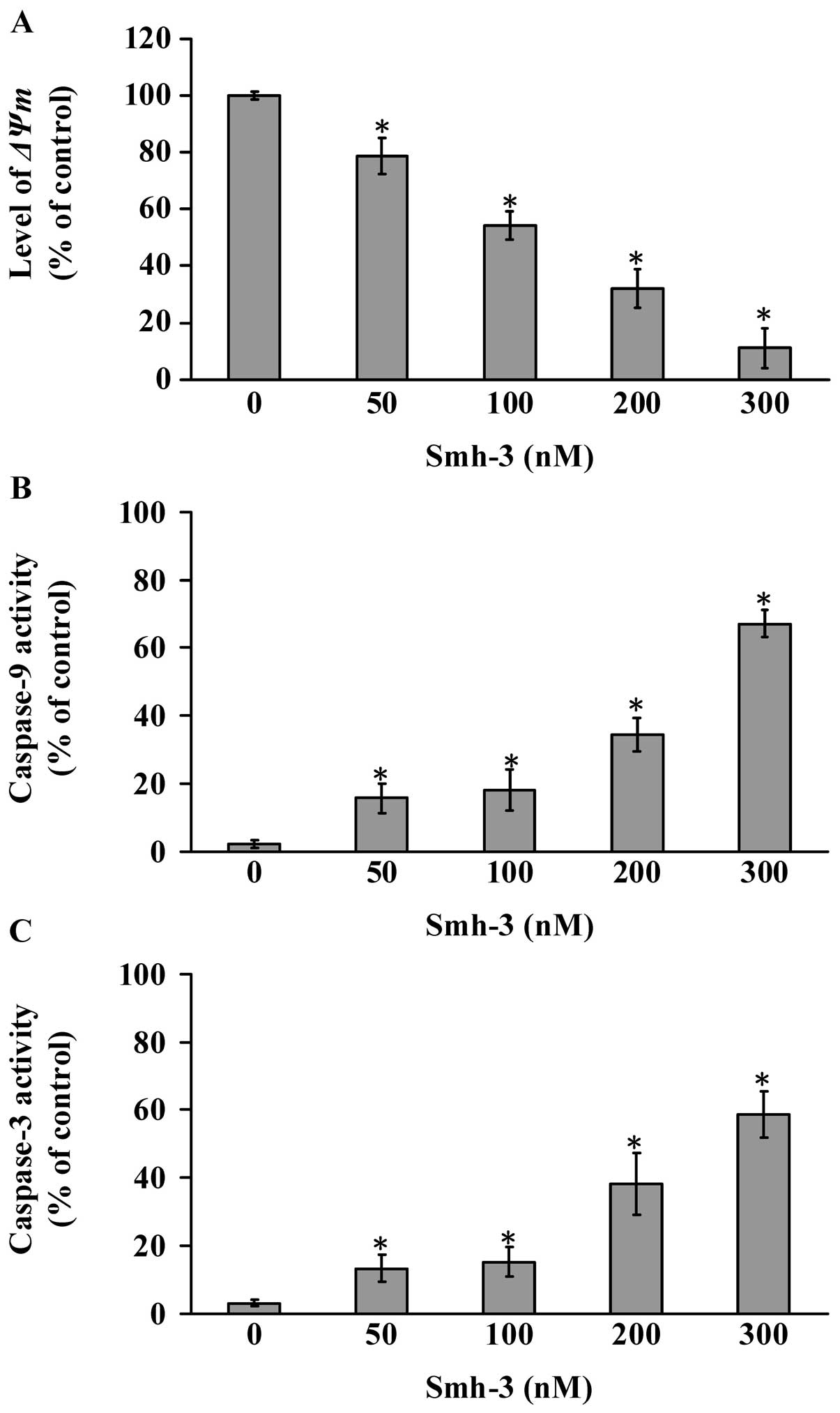
Effects of Smh-3 on ER stress in Hep3B
cells
Our previous research demonstrated that Smh-3 acts
against HL-60 leukemia cells in vitro via G2/M
phase arrest, downregulation of AKT activity and induction of
mitochondrial-dependent apoptotic pathways (31). To confirm the possibility that the
Smh-3-induced apoptosis could be related to contributions from the
ER stress signal pathways, Hep3B cells were treated with 0, 50,
100, 200 and 300 nM of Smh-3 for 24 h. The intracellular
Ca2+ release, caspase-4 activity and the levels of ER
stress-associated proteins were examined. The quantities and
results are shown in Fig. 3.
Results from the flow cytometric assay indicated that Smh-3 induced
the production of intracellular Ca2+ release (Fig. 3A). To evaluate whether or not
Smh-3-induced apoptosis is involved in activation of caspase-4, we
determined the caspase-4 activity using caspase colorimetric
analysis. Smh-3 at concentrations of 0, 50, 100, 200 and 300 nM
stimulated caspase-4 activity in a concentration-dependent manner
Fig. 3B. It was reported that
GADD153 is a hallmark of ER stress (42,43).
Smh-3-treated Hep3B cells were harvested for western blot analysis
of the expression levels of the ER stress pathway-related GADD153
and caspase-4 proteins. Smh-3 promoted the protein levels of
GADD153 and caspase-4 (Fig. 3C).
Based on these results, we suggest that Smh-3-induced apoptosis in
Hep3B cells may be mediated through the ER stress-dependent
apoptotic signaling pathway.
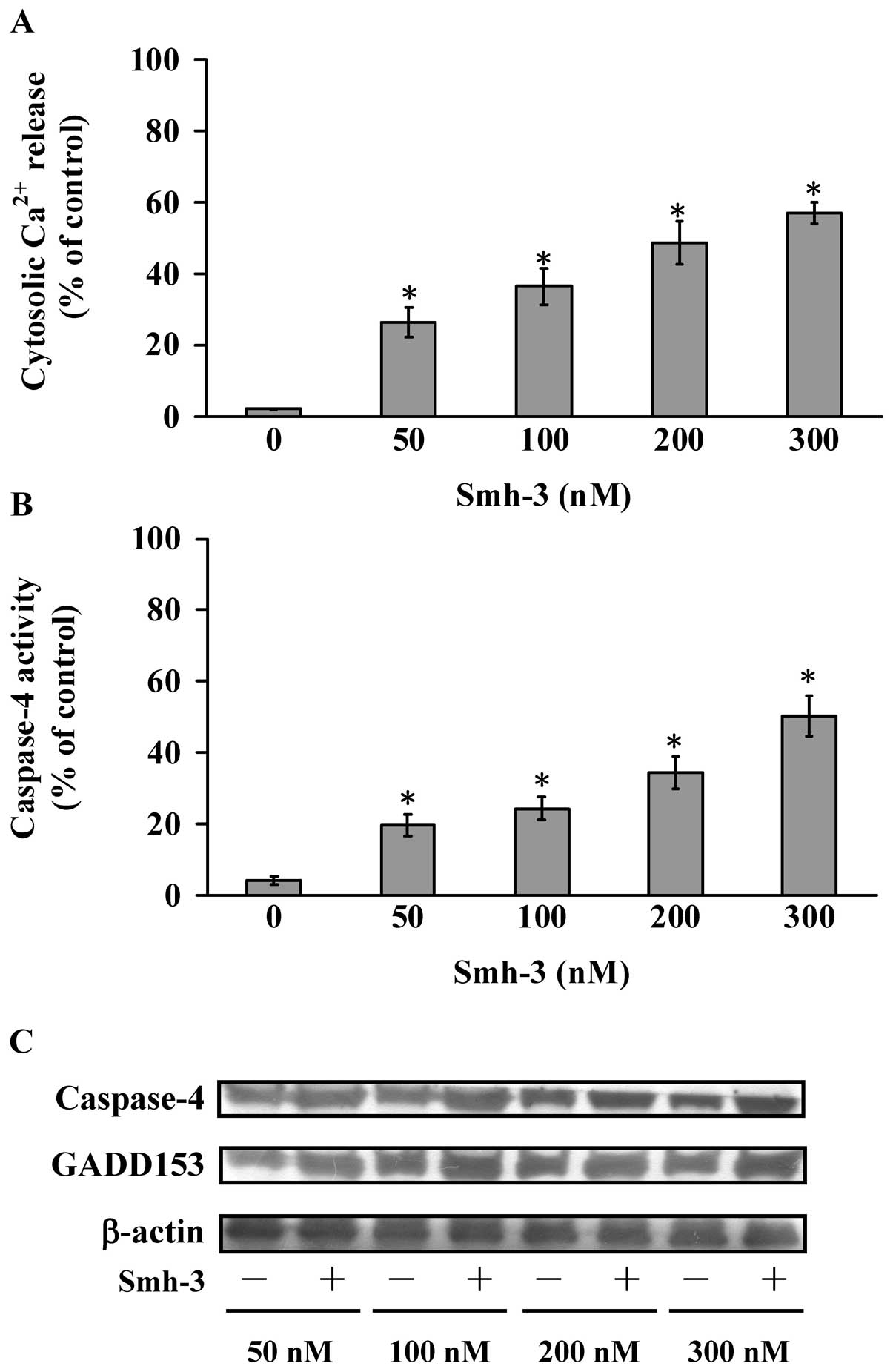 | Figure 3Effects of Smh-3 on ER stress in
Hep3B cells. (A) Cells were treated with 0, 50, 100, 200 and 300 nM
of Smh-3 for 24 h and the intracellular Ca2+ levels were
measured using flow cytometric analysis. (B) Cells were treated
with 0, 50, 100, 200 and 300 nM of Smh-3 for 24 h and the whole
cell lysates were subjected to caspase-4 activity assay. Each data
point is the means ± SD of 3 experiments. *P<0.05.
(C) Cells were treated with 0, 50, 100, 200 and 300 nM of Smh-3 for
24 h, and whole cell lysate were prepared and the caspase-4 and
GADD153 protein levels were estimated by western blotting as
described in Materials and methods. |
Effects of Smh-3 on the loss of ΔΨm
level, caspase-9 and caspase-3 activities in Hep3B cells
To confirm the possibility that Smh-3-induced
apoptosis is related to contributions from the mitochondrial signal
pathways, Hep3B cells were treated with 0, 50, 100, 200 and 300 nM
of Smh-3 for 24 h, and changes in ΔΨm, caspase-9 and caspase-3
activities were examined; the quantities and results are shown in
Fig. 4. Results from the flow
cytometric assay indicated that Smh-3 decreased the level of ΔΨm
(Fig. 4A). To evaluate whether or
not Smh-3-induced apoptosis is involved in activation of caspase-9
and caspase-3, we detected the caspase-9 and caspase-3 activities
using caspase colorimetric analysis. Concentrations of 0, 50, 100,
200 and 300 nM of Smh-3 stimulated caspase-9 activity (Fig. 4B) and caspase-3 activity (Fig. 4C) in a concentration-dependent
manner.
Discussion
There are no reports concerning the effects of Smh-3
on apoptosis and associated gene expression in Hep3B cells. The
present study is the first to show that Smh-3 induces a cytotoxic
effect which includes induction of G2/M phase arrest and
apoptosis and changes in the expression of associated gene in Hep3B
cells. In previous studies, we showed that Smh-3 triggered
apoptosis in HL-60 human leukemia cells (31); however, the involvement of ER stress
in Smh-3-induced apoptosis in cancer cells is still unclear. Our
results demonstrated that Smh-3 treatment increased the protein
levels of caspase-4 and GADD153 in Hep3B cells (Fig. 3B and C). Our novel findings suggest
that these events demonstrate that the ER stress apoptotic pathway
is involved in the Smh-3 effects in vitro. It has been
reported that cellular organelles such as mitochondria, ER,
lysosomes and Golgi apparatus are also major targets of apoptotic
initiation (44,45). Many chemotherapy agents that
strongly affect the function of the ER are identified as strong
inducers of GADD153. This suggests that increased intracellular
Ca2+ induces mitochondrial swelling (39,46).
Following mitochondrial permeabilization, cytochrome c,
Apaf-1, procaspase-9, Endo G and AIF are released into the cytosol,
activating caspase-3 via caspase-9 (47,48).
Smh-3 induced the activation of caspase-9 and caspase-3 after a
48-h treatment (Fig. 4B and C),
suggesting that Smh-3 possibly activates the mitochondrial
signaling pathway. Our results demonstrated that Smh-3 decreased
the ΔΨm after a 24 h treatment with Smh-3 (Fig. 4A), and then promoted caspase-9 and
caspase-3 activities in Hep3B cells (Fig. 4B and C). In addition, caspase-8
activity exhibited no significant increase in the Smh-3-treated
Hep3B cells (data not shown). Based on the above evidence,
Smh-3-stimulated apoptotic cell death is involved in the crosstalk
between the ER and mitochondria.
Based on the change in gene expression profile in
Smh-3-treated Hep3B cells by DNA microarray, we found that cellular
and molecular responses to Smh-3 treatment are multifaceted and are
likely to be mediated via a variety of regulatory pathways. Smh-3
regulated the expression of important genes that control cell
growth, angiogenesis, autophagy, calcium-mediated ER stress
signaling, cell adhesion, cell cycle and mitosis, cell migration,
cytoskeleton organization, DNA damage and repair,
mitochondrial-mediated apoptosis, transcription and translation and
cell signaling pathways (Table I).
Regulation of these genes may be responsible for inhibiting the
proliferation of Hep3B cells. It was reported that cyclins
associate with cyclin-dependent protein kinases (CDKs) and CDK
inhibitor (CKI) to control the process of the cell cycle. The CDK
inhibitor (CKI) has been demonstrated to arrest the cell cycle and
inhibit the cell growth of cancer cells (33,40,46).
From a gene expression profile, we found that Smh-3 altered the
expression of cyclin and cyclin-dependent kinase inhibitors and
cytoskeleton organization genes including CNNM3, CDKN3, RPRM,
CCNG1, ACTB, ACTG1, TUBA1B, TUBB2C and MAP1B, suggesting a change
in cyclin, cyclin-dependent kinase inhibitors, and microtubule
interaction which could finally lead to cell cycle G2/M
arrest (Fig. 2B).
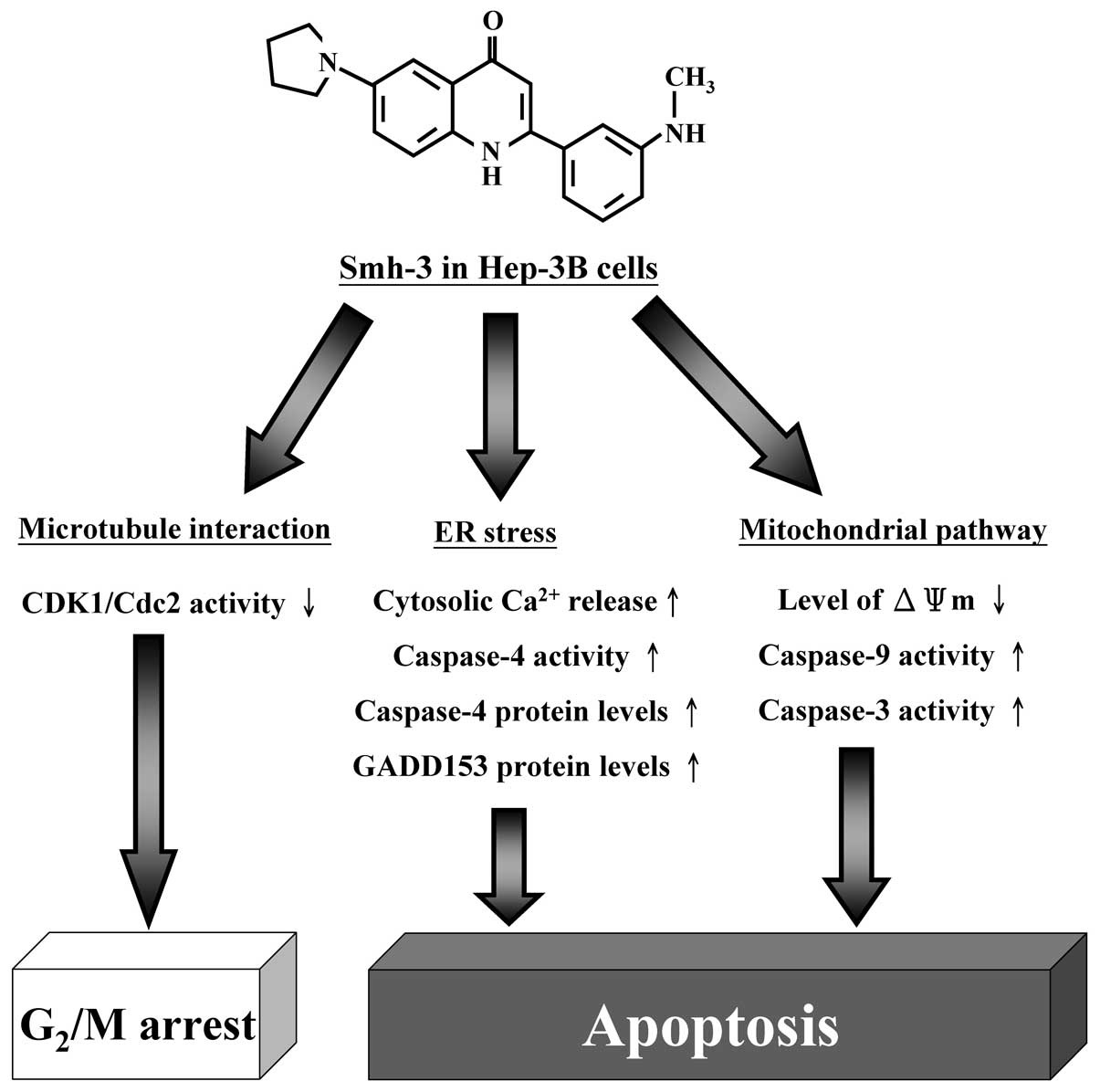
In conclusion, the molecular signaling pathways
involved in Smh-3 effects on Hep-3B cells are summarized in
Fig. 5. Based on these data,
further detailed investigations including anti-metastasis,
anti-angiogenesis and autophagy induction studies are required in
order to establish cause and effect relationships between these
altered genes and the outcome of human hepatocellular carcinoma
patients.
Acknowledgements
The study was supported by research grants from the
National Science Council of P.R. China awarded to S.-C.K. (NSC
100-2320-B-039-001) and Taiwan Department of Health, China Medical
University Hospital Cancer Research Center of Excellence
(DOH101-TD-C-111-005).
References
|
1
|
Moriguchi M, Takayama T, Higaki T, et al:
Early cancer-related death after resection of hepatocellular
carcinoma. Surgery. 151:232–237. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen TH, Chen CJ, Yen MF, et al:
Ultrasound screening and risk factors for death from hepatocellular
carcinoma in a high risk group in Taiwan. Int J Cancer. 98:257–261.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ueda H, Fukuchi H and Tanaka C: Toxicity
and efficacy of hepatic arterial infusion chemotherapy for advanced
hepatocellular carcinoma (Review). Oncol Lett. 3:259–263.
2012.PubMed/NCBI
|
|
4
|
Blum HE: Hepatocellular carcinoma: HCC.
Hepat Mon. 11:69–70. 2011.
|
|
5
|
Abou-Alfa GK: New agents in hepatocellular
carcinoma. Clin Adv Hematol Oncol. 6:423–424. 2008.
|
|
6
|
Marquardt JU, Galle PR and Teufel A:
Hepatocellular carcinoma: molecular pathogenesis and novel targets
for therapy. Dtsch Med Wochenschr. 137:855–860. 2012.(In
German).
|
|
7
|
Hoshida Y, Toffanin S, Lachenmayer A,
Villanueva A, Minguez B and Llovet JM: Molecular classification and
novel targets in hepatocellular carcinoma: recent advancements.
Semin Liver Dis. 30:35–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chai F, Truong-Tran AQ, Ho LH and Zalewski
PD: Regulation of caspase activation and apoptosis by cellular zinc
fluxes and zinc deprivation: a review. Immunol Cell Biol.
77:272–278. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crawford ED, Seaman JE, Barber AE II, et
al: Conservation of caspase substrates across metazoans suggests
hierarchical importance of signaling pathways over specific targets
and cleavage site motifs in apoptosis. Cell Death Differ. Aug
24–2012.(Epub ahead of print).
|
|
10
|
Oyadomari S and Mori M: Roles of
CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ.
11:381–389. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nozaki S, Sledge GW Jr and Nakshatri H:
Repression of GADD153/CHOP by NF-kappaB: a possible cellular
defense against endoplasmic reticulum stress-induced cell death.
Oncogene. 20:2178–2185. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McCullough KD, Martindale JL, Klotz LO, Aw
TY and Holbrook NJ: Gadd153 sensitizes cells to endoplasmic
reticulum stress by down-regulating Bcl2 and perturbing the
cellular redox state. Mol Cell Biol. 21:1249–1259. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hogstrand K, Hejll E, Sander B, Rozell B,
Larsson LG and Grandien A: Inhibition of the intrinsic but not the
extrinsic apoptosis pathway accelerates and drives MYC-driven
tumorigenesis towards acute myeloid leukemia. PLoS One.
7:e313662012. View Article : Google Scholar
|
|
14
|
Poellinger L and Lendahl U: Modulating
Notch signaling by pathway-intrinsic and pathway-extrinsic
mechanisms. Curr Opin Genet Dev. 18:449–454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harding HP and Ron D: Endoplasmic
reticulum stress and the development of diabetes: a review.
Diabetes. 51(Suppl 3): S455–S461. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muhlethaler-Mottet A, Bourloud KB,
Auderset K, Joseph JM and Gross N: Drug-mediated sensitization to
TRAIL-induced apoptosis in caspase-8-complemented neuroblastoma
cells proceeds via activation of intrinsic and extrinsic pathways
and caspase-dependent cleavage of XIAP, Bcl-xL and RIP. Oncogene.
23:5415–5425. 2004. View Article : Google Scholar
|
|
17
|
Chong ZZ, Kang JQ and Maiese K:
Erythropoietin fosters both intrinsic and extrinsic neuronal
protection through modulation of microglia, Akt1, Bad, and
caspase-mediated pathways. Br J Pharmacol. 138:1107–1118. 2003.
View Article : Google Scholar
|
|
18
|
Perrin FE, Boisset G, Lathuiliere A and
Kato AC: Cell death pathways differ in several mouse models with
motoneurone disease: analysis of pure motoneurone populations at a
presymptomatic age. J Neurochem. 98:1959–1972. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim MK, Kim HS, Lee IK and Park KG:
Endoplasmic reticulum stress and insulin biosynthesis: a review.
Exp Diabetes Res. 2012:5094372012.PubMed/NCBI
|
|
20
|
Gregor MF and Hotamisligil GS: Thematic
review series: Adipocyte Biology. Adipocyte stress: the endoplasmic
reticulum and metabolic disease. J Lipid Res. 48:1905–1914. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chakrabarti A, Chen AW and Varner JD: A
review of the mammalian unfolded protein response. Biotechnol
Bioeng. 108:2777–2793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shore GC, Papa FR and Oakes SA: Signaling
cell death from the endoplasmic reticulum stress response. Curr
Opin Cell Biol. 23:143–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Endo H, Murata K, Mukai M, Ishikawa O and
Inoue M: Activation of insulin-like growth factor signaling induces
apoptotic cell death under prolonged hypoxia by enhancing
endoplasmic reticulum stress response. Cancer Res. 67:8095–8103.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Obeng EA and Boise LH: Caspase-12 and
caspase-4 are not required for caspase-dependent endoplasmic
reticulum stress-induced apoptosis. J Biol Chem. 280:29578–29587.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Song R, Shen Y, et al: Targeting
sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2 by
curcumin induces ER stress-associated apoptosis for treating human
liposarcoma. Mol Cancer Ther. 10:461–471. 2011.PubMed/NCBI
|
|
26
|
Wu Z, Liang F, Hong B, et al: An
endoplasmic reticulum-bound Ca(2+)/Mn(2+) pump, ECA1, supports
plant growth and confers tolerance to Mn(2+) stress. Plant Physiol.
130:128–137. 2002.
|
|
27
|
Lai YY, Huang LJ, Lee KH, et al: Synthesis
and biological relationships of 3′,6-substituted
2-phenyl-4-quinolone-3-carboxylic acid derivatives as antimitotic
agents. Bioorg Med Chem. 13:265–275. 2005.
|
|
28
|
Lee HZ, Lin WC, Yeh FT and Wu CH:
2-Phenyl-4-quinolone prevents serotonin-induced increases in
endothelial permeability to albumin. Eur J Pharmacol. 354:205–213.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee HZ: Inhibitory effect of
2-phenyl-4-quinolone on serotonin-mediated changes in the
morphology and permeability of endothelial monolayers. Eur J
Pharmacol. 335:245–254. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang JP, Hsu MF, Raung SL and Kuo SC:
Suppressive effect of 2-phenyl-4-quinolone (YT-1) on hind-paw edema
and cutaneous vascular plasma extravasation in mice. Naunyn
Schmiedebergs Arch Pharmacol. 349:324–330. 1994.PubMed/NCBI
|
|
31
|
Huang SM, Yang JS, Tsai SC, et al: The
novel synthesized
2-(3-(methylamino)phenyl)-6-(pyrrolidin-1-yl)quinolin-4-one (Smh-3)
compound induces G2/M phase arrest and mitochondrial-dependent
apoptotic cell death through inhibition of CDK1 and AKT activity in
HL-60 human leukemia cells. Int J Oncol. 38:1357–1364. 2011.
View Article : Google Scholar
|
|
32
|
Ni CH, Chen PY, Lu HF, et al:
Chrysophanol-induced necrotic-like cell death through an impaired
mitochondrial ATP synthesis in Hep3B human liver cancer cells. Arch
Pharm Res. 35:887–895. 2012. View Article : Google Scholar
|
|
33
|
Tsai SC, Yang JS, Peng SF, et al: Bufalin
increases sensitivity to AKT/mTOR-induced autophagic cell death in
SK-HEP-1 human hepatocellular carcinoma cells. Int J Oncol.
41:1431–1442. 2012.PubMed/NCBI
|
|
34
|
Hour MJ, Tsai SC, Wu HC, et al: Antitumor
effects of the novel quinazolinone MJ-33: inhibition of metastasis
through the MAPK, AKT, NF-kappaB and AP-1 signaling pathways in
DU145 human prostate cancer cells. Int J Oncol. 41:1513–1519.
2012.
|
|
35
|
Wu PP, Chung HW, Liu KC, et al: Diallyl
sulfide induces cell cycle arrest and apoptosis in HeLa human
cervical cancer cells through the p53, caspase- and
mitochondria-dependent pathways. Int J Oncol. 38:1605–1613.
2011.
|
|
36
|
Huang WW, Ko SW, Tsai HY, et al:
Cantharidin induces G2/M phase arrest and apoptosis in human
colorectal cancer colo 205 cells through inhibition of CDK1
activity and caspase-dependent signaling pathways. Int J Oncol.
38:1067–1073. 2011.
|
|
37
|
Yaguchi T, Saito M, Yasuda Y and Nishizaki
T: Caspase-4 activation in association with decreased adenosine
deaminase activity may be a factor for gastric ulcer. Digestion.
81:62–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu SH, Hang LW, Yang JS, et al: Curcumin
induces apoptosis in human non-small cell lung cancer NCI-H460
cells through ER stress and caspase cascade- and
mitochondria-dependent pathways. Anticancer Res. 30:2125–2133.
2010.PubMed/NCBI
|
|
39
|
Huang WW, Chiu YJ, Fan MJ, et al:
Kaempferol induced apoptosis via endoplasmic reticulum stress and
mitochondria-dependent pathway in human osteosarcoma U-2 OS cells.
Mol Nutr Food Res. 54:1585–1595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang WW, Yang JS, Lin MW, et al:
Cucurbitacin E induces G(2)/M phase arrest through STAT3/p53/p21
signaling and provokes apoptosis via Fas/CD95 and
mitochondria-dependent pathways in human bladder cancer T24 cells.
Evid Based Complement Alternat Med. 2012:9527622012. View Article : Google Scholar
|
|
41
|
Wu RC, Yu CS, Liu KC, et al: Citosol
(thiamylal sodium) triggers apoptosis and affects gene expressions
of murine leukemia RAW 264.7 cells. Hum Exp Toxicol. 31:771–779.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lin JP, Yang JS, Chang NW, et al: GADD153
mediates berberine-induced apoptosis in human cervical cancer Ca
ski cells. Anticancer Res. 27:3379–3386. 2007.PubMed/NCBI
|
|
43
|
Lu HF, Hsueh SC, Ho YT, et al: ROS
mediates baicalin-induced apoptosis in human promyelocytic leukemia
HL-60 cells through the expression of the Gadd153 and
mitochondrial-dependent pathway. Anticancer Res. 27:117–125.
2007.
|
|
44
|
Chambers KT, Unverferth JA, Weber SM, Wek
RC, Urano F and Corbett JA: The role of nitric oxide and the
unfolded protein response in cytokine-induced beta-cell death.
Diabetes. 57:124–132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim R, Emi M, Tanabe K and Murakami S:
Role of the unfolded protein response in cell death. Apoptosis.
11:5–13. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lu CC, Yang JS, Chiang JH, et al: Novel
quinazolinone MJ-29 triggers endoplasmic reticulum stress and
intrinsic apoptosis in murine leukemia WEHI-3 cells and inhibits
leukemic mice. PLoS One. 7:e368312012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schattenberg JM, Schuchmann M and Galle
PR: Cell death and hepatocarcinogenesis: dysregulation of apoptosis
signaling pathways. J Gastroenterol Hepatol. 26(Suppl 1): 213–219.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rosello A, Warnes G and Meier UC: Cell
death pathways and autophagy in the central nervous system and its
involvement in neurodegeneration, immunity and central nervous
system infection: to die or not to die - that is the question. Clin
Exp Immunol. 168:52–57. 2012. View Article : Google Scholar : PubMed/NCBI
|