Introduction
The incidence of tumours has risen over the past
half-century, resulting in cancer becoming one of the most lethal
human diseases. This threat to humanity is great and continues to
increase, and the urgency with which humans address this challenge
is also increasing. Rapid developments in science technology over
the past century have resulted in an independent medical
discipline, oncology, which has been further divided into a number
of branches.
With the rapid development of the oncology field,
several questions have been raised: What are the primary research
centres conducting research in this field? What are the major
topics of current oncology research? Which are the most important
academic communities? The answers to these questions are essential
for developing effective measures to address the updated status of
oncology research and practices. We used two information
techniques, document overview and knowledge domain visualisation
(KDViz) (1,2), to answer the above questions and
create a comprehensive picture of the field.
To visualise the intellectual structure of
scientific fields, two main scientometric methods can be employed:
document co-citation analysis and co-authorship analysis (3–8).
Document co-citation analysis has been used in various theoretical
and empirical studies, including research concerning anaesthesia
(3), environmental science
(4), knowledge management (5), ubiquitous computing (6), mapping scientometrics (7) and stem cells (8). Co-authorship analysis has been applied
to various research fields, including those involving digital
libraries (2) and hypertext
(7). Chen and Liu (2) revealed the co-authorship pattern of
scientometrics using the data from Science Citation Index (SCI).
These two methods are used to explore research fronts and hotspots
and the relationships between collaborators in selected scientific
fields. These above studies demonstrate their practical value and
advantages over document overview, but they are rarely used in
medical research.
In the present study, we used KDViz for co-citation
and co-authorship analysis to reveal the main research centres,
topics and academic communities of oncology to probe the
intellectual structure of oncology. Our study used a novel method
and a unique perspective. We expect that it will reveal an overview
of the development in this field from 2001 to 2010. The research
fronts are identified based on indicators computed by CiteSpace
without the intervention of domain experts or prior working
knowledge of the topic. This approach makes the analysis repeatable
with new data and verifiable by different analysts.
Materials and methods
Materials
Based on 185 oncology journals listed in the 2010
annual JCR report from the Web of Science database, we chose the
top 10% by impact factor (IF) [19 journals (A)] and total citations
[19 journals (B)]; 8 journals were in both groups A and B. We
assigned 30 journals to be our data.
After retrieving 30 journals from the SCI database
with the literature format ‘article’ and ‘review’, and removing
those with the format ‘news’, ‘meetings abstract’, ‘letter’ and
other non-original studies, we named the files ‘download*. txt’ and
downloaded them in their fully recorded format with references. In
total, we retrieved 143,152 articles from 2001 to 2010, including
the authors, title, keywords, abstract and citations as sample data
for our study. The last date on which we retrieved data was
November 1, 2011.
Methods
Knowledge domain visualisation
KDViz is a computer-supported information processing
technology that can reveal the developmental process and structure
of scientific knowledge in graphical form by analysing information
such as authors and publications and defining their relationships.
These relationships are expressed in two- or three-dimensional
knowledge landscapes to effectively describe large amounts of data
and outline the structure and evolution of a scientific field
(1,2).
Chen from Drexel University designed the
visualisation software CiteSpace II, which is written in Java and
can be used to analyse multiple-perspective co-citation networks to
identify and display new trends in scientific development based on
a large number of scientific studies (9). Knowledge maps drawn by CiteSpace II
are not only able to predict future trends in research but can also
aid in understanding the current forefronts (6,9–11).
Chen also used CiteSpace II to draw knowledge maps on large-scale
biological populations that are now extinct (1981–2004), terrorism
(1990–2003)(12) and emerging
trends in regenerative medicine (13).
In this study, CiteSpace II was used for the
co-citation analysis of network graphs (Figs. 1 and 3).
Co-citation analysis
Co-citation analysis is the most influential
citation analysis method and can be used not only to reveal the
developmental status and changes in the structure of a scientific
field but also to study the research fronts and domains and provide
support for those making critical decisions in the science and
technology field. Co-citation analyses include document co-citation
analysis, author co-citation analysis and institution co-citation
analysis. In this study, we introduced document co-citation
analysis, and the principles of the aforementioned analyses are
similar.
Document co-citation analysis (DCA) studies a
network of co-cited references (14). The fundamental assumption is that
co-citation clusters reveal the underlying intellectual structure.
The notion that cited documents are concept symbols was introduced
by Small (14). He found a high
degree of uniformity in how specific concepts and specific
references to documents (cited documents) are associated in the
chemistry literature. These cited documents serve as symbols for
scientific ideas, methods and experiments. The idea is further
extended to clusters of noun phrases that are extracted from
document citations. From the concept symbol perspective, the study
of a co-citation network focuses on interpreting the nature of a
cluster of cited documents and the interrelationships between the
clusters (10). The principle of
document co-citation analysis is as follows. If two documents are
cited together in one or more articles, we say that the two
documents are co-cited. Higher co-citation frequencies indicate
closer links between the documents. Thus, based on the document
citation relationship, we can analyse the affiliations between
documents. If the documents are divided into clusters and classes,
we can analyse the fronts of the current research according to the
contents of documents (15).
Social network analysis (SNA)
SNA focuses on ties between social entities; it
involves the mapping and measurement of relationships between
components in a system (16).
Graphs are used to detect and interpret patterns of social ties.
The vertices in the graphs represent social actors, and the edges
represent the social interactions between the actors. This
representation allows us to apply graph theory, a branch of
mathematics, to the analysis of what would otherwise be an
inherently elusive and poorly understood problem: the tangled web
of our social interactions. In the present study, SNA helps reveal
the connection between authors in the oncology field between 2001
and 2010.
The co-authorship network is an important type of
social network and has been widely used to detect the structure of
scientific collaborations and status of individual researchers.
Analysing collaboration through co-authorship is advantageous as it
is inexpensive and practical (17).
In the past few years, many studies have shown that the rates of
research collaboration, as measured by co-authorship, have greatly
increased. Gossart and Ozman (18)
analysed the collaboration patterns of Turkish social sciences and
humanities (SSH) by examining co-authored papers. Lu and Feng
(19) proposed a new method called
‘extensity centrality’ to analyse the importance of authors in
co-authorship networks. Yu et al(20) analysed research groups from the
co-authorship network in the oncology field in China. Morel et
al(21) used co-authorship
network analysis to generate valuable information on the strategic
planning, implementation and monitoring of the program launched by
the Brazilian Government to fund scientific research.
Results and Discussion
Data were analysed to generate the rich visual maps
shown in Figs. 1–3.
Primary research bodies
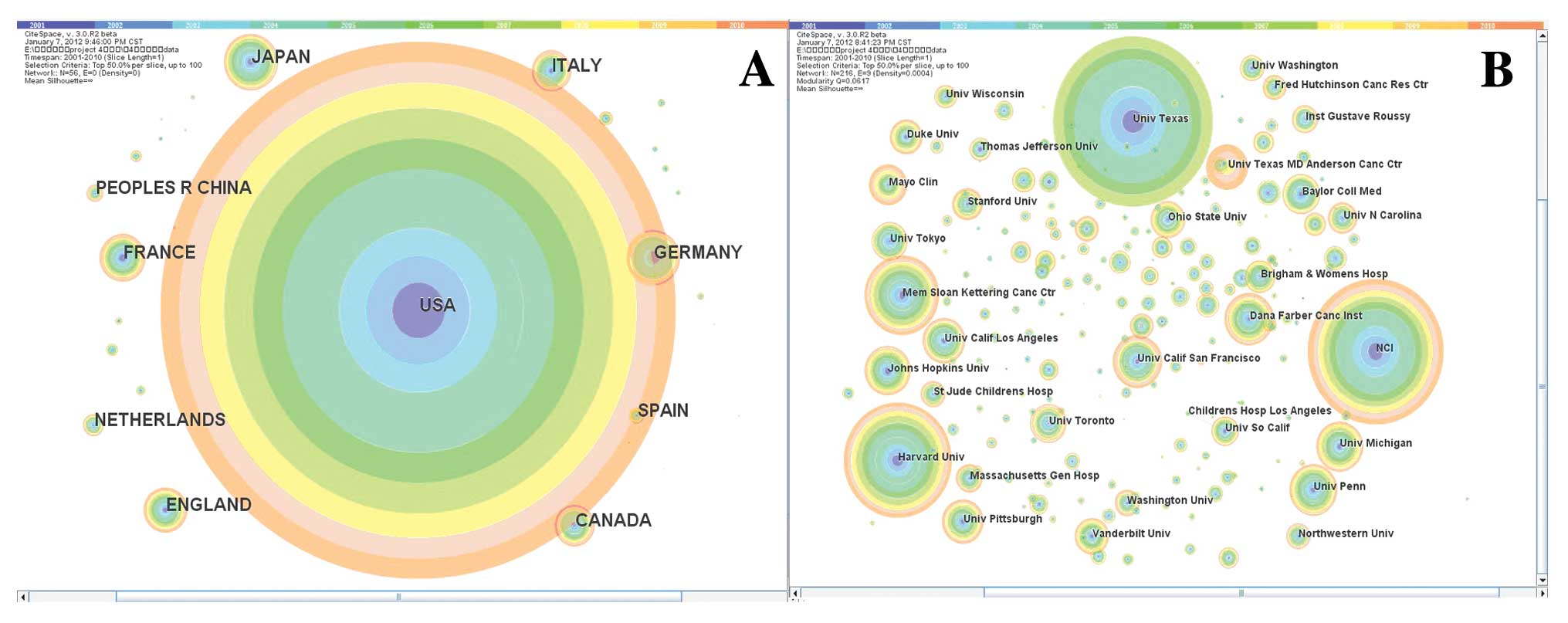
Core countries
Citation tree rings represent the citation history
of a country/institution. The colour of a citation ring denotes the
time slice of corresponding citations. The thickness of a ring is
proportional to the number of citations in a given time slice.
Larger rings indicate a higher frequency of citations for the
publications. Fig. 1A shows the top
20 most-cited countries, which are also listed in Table I.
 | Table IThe most-cited countries and their
citation frequencies in oncology (top 20, 2001–2010). |
Table I
The most-cited countries and their
citation frequencies in oncology (top 20, 2001–2010).
| Serial no. | Country | Citation
frequency |
|---|
| 1 | USA | 16,463 |
| 2 | Japan | 1809 |
| 3 | Germany | 1756 |
| 4 | France | 1523 |
| 5 | England | 1454 |
| 6 | Canada | 1334 |
| 7 | Italy | 1326 |
| 8 | The
Netherlands | 719 |
| 9 | P.R. China | 583 |
| 10 | Spain | 507 |
| 11 | Australia | 499 |
| 12 | Sweden | 415 |
| 13 | Republic of
Korea | 387 |
| 14 | Switzerland | 337 |
| 15 | Israel | 283 |
| 16 | Belgium | 277 |
| 17 | Taiwan | 276 |
| 18 | Scotland | 255 |
| 19 | Austria | 203 |
| 20 | Denmark | 201 |
In Fig. 1A, the
citation frequency of the United States is 16,463, which accounts
for 53.8% of the total citation frequency of the top 20 countries.
The United States is not only the most-cited country, but its ring
is evenly distributed, indicating that the United States has always
been the leader of the international field of oncology research.
The sole developing country in the top 10 is China, which ranks
9th. We did not find connection lines between the United States and
China among the top 20 countries. Therefore, we can say that they
are not co-cited, indicating that they have distinct research
directions.
Core institutions
Fig. 1B shows that
the contemporary oncology studies are primarily from the University
of Texas, the NCI, Harvard University and the Memorial Sloan
Kettering Cancer Center. Regarding both the quantity of issued
studies and the citation frequency, the University of Texas ranks
1st, the NCI ranks 2nd and Harvard University ranks 3th. The rings
of the NCI, Harvard University and other institutions are basically
evenly distributed, indicating that their research strengths
continued to steadily increase over the past 10 years. Fig. 1B shows the top 20 most-cited
institutions and their citation frequency, which are also listed in
Table II.
 | Table IIThe most cited institutions and their
citation frequencies in oncology (top 20, 2001–2010). |
Table II
The most cited institutions and their
citation frequencies in oncology (top 20, 2001–2010).
| Serial no. | Institution | Citation
frequency |
|---|
| 1 | University of
Texas | 1177 |
| 2 | National Cancer
Institute | 1007 |
| 3 | Harvard
University | 802 |
| 4 | Memorial
Sloan-Kettering Cancer Center | 555 |
| 5 | University of
California, San Francisco | 368 |
| 6 | Dana-Farber Cancer
Institute | 365 |
| 7 | University of
Pennsylvania | 362 |
| 8 | University of
Michigan | 353 |
| 9 | Johns Hopkins
University | 350 |
| 10 | University of
California, Los Angeles | 324 |
| 11 | University of
Pittsburgh | 317 |
| 12 | The University of
Texas: MD Anderson Cancer Center | 312 |
| 13 | Mayo Clinic | 287 |
| 14 | Baylor College of
Medicine | 285 |
| 15 | University of
Toronto | 273 |
| 16 | University of
Tokyo | 271 |
| 17 | Ohio State
University | 262 |
| 18 | Vanderbilt
University | 258 |
| 19 | Duke
University | 249 |
| 20 | Brigham and Women’s
Hospital | 236 |
Core academic communities
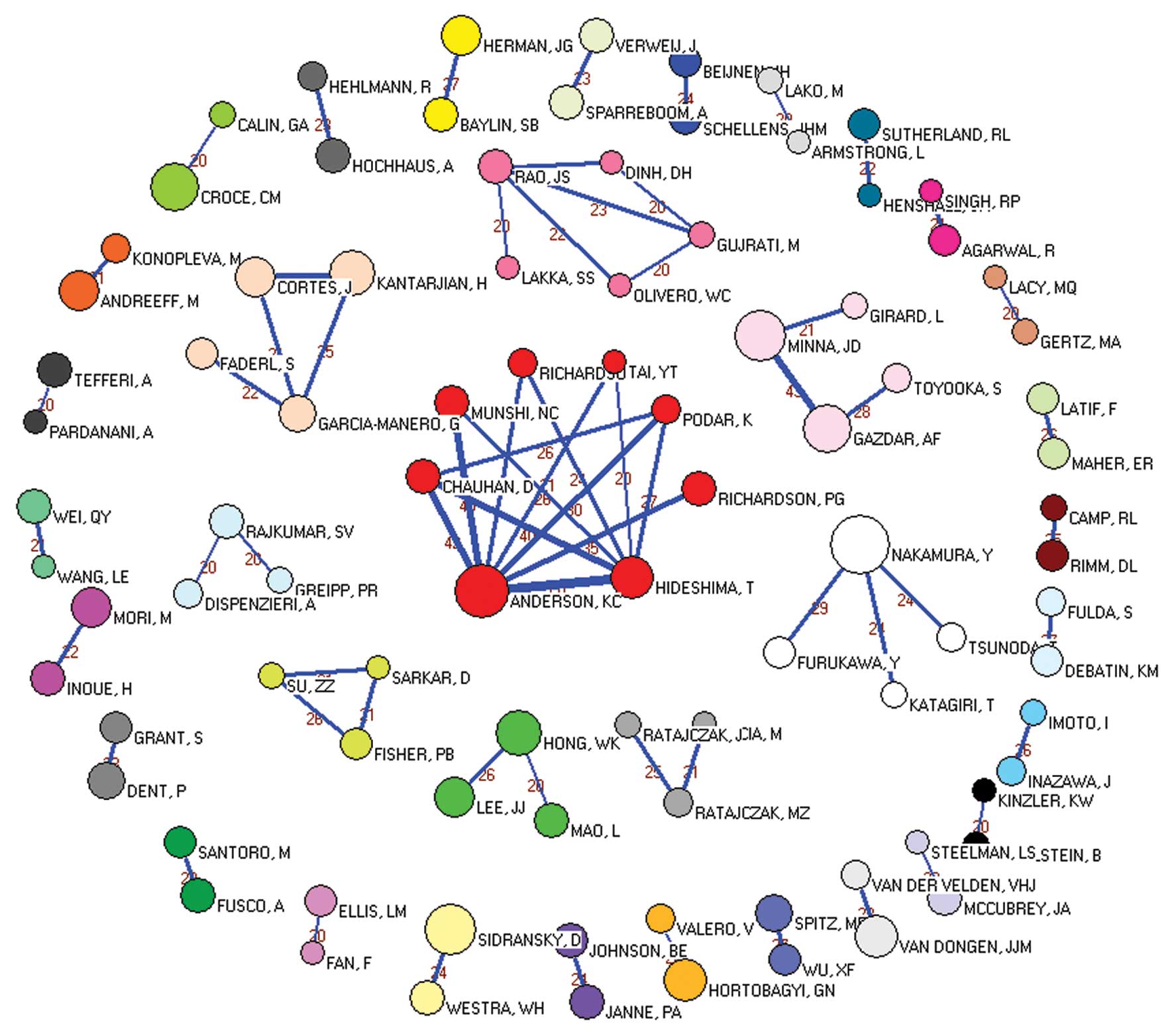
We performed co-authorship analysis using SNA and
Pajek was to develop the network graphs (Fig. 2).
As shown in Fig. 2,
our dataset was composed of 118,871 authors and was submitted to
Pajek. To obtain a clear picture, we deleted the lines with values
less than 20, i.e., we removed the authors whose cooperation
instances were below 20. As shown in Fig. 2, the most collaborative authors were
divided into 36 relatively dense components, including 91 vertices,
showing that there were 36 close collaborative communities
performing oncology research composed of 91 authors (Table III). The largest vertex was found
in the white cluster and represents the author with the highest
productivity, Nakamura Y, who published 117 studies from 2001 to
2010. The largest academic community (the biggest component) is the
red cluster in the middle, which contains 8 authors who are mainly
from the Harvard University School of Medicine, the Division of
Hematology and Oncology of the Dana Farber Cancer Institute, and
the Jerome Lipper Multiple Myeloma Center of Boston. Their research
is focused on multiple myeloma. The 5 authors in the second
component are all affiliated with the Department of Neurosurgery of
the University of Illinois College of Medicine at Peoria, and their
research focuses primarily on angiogenesis. The 4 authors in the
third component are all from the Department of Leukemia of the MD
Anderson Cancer Center at the University of Texas and perform
research primarily on acute lymphocytic leukaemia (ALL).
Furthermore, two vertices connect in the peripheral lap, which
indicates that two authors in these components have cooperated at
least 20 times; we consider these laboratories to be cohesive
subgroups.
 | Table IIIComponents for the most collaborative
authors in Oncology Research (2001–2010). |
Table III
Components for the most collaborative
authors in Oncology Research (2001–2010).
| Cluster | Freq | Freq % | CumFreq | CumFreq % | Representative |
|---|
| 1 | 8 | 8.7912 | 12 | 13.1868 | Hideshima T |
| 2 | 5 | 5.4945 | 58 | 63.7363 | Rao JS |
| 3 | 4 | 4.3956 | 18 | 19.7802 | Gazdar AF |
| 4 | 4 | 4.3956 | 22 | 24.1758 | Tsunoda T |
| 5 | 4 | 4.3956 | 75 | 82.4176 | Faderl S |
| 6 | 3 | 3.2967 | 53 | 58.2418 | Lee JJ |
| 7 | 3 | 3.2967 | 63 | 69.2308 | Sarkar D |
| 8 | 3 | 3.2967 | 80 | 87.9121 | Rajkumar SV |
| 9 | 3 | 3.2967 | 87 | 95.6044 | Kucia M |
| 10 | 2 | 2.1978 | 2 | 2.1978 | Baylin SB |
| 11 | 2 | 2.1978 | 4 | 4.3956 | Calin GA |
| 12 | 2 | 2.1978 | 14 | 15.3846 | Schellens JHM |
| 13 | 2 | 2.1978 | 24 | 26.3736 | Andreeff M |
| 14 | 2 | 2.1978 | 26 | 28.5714 | Janne PA |
| 15 | 2 | 2.1978 | 28 | 30.7692 | Spitz MR |
| 16 | 2 | 2.1978 | 30 | 32.967 | Wang LE |
| 17 | 2 | 2.1978 | 32 | 35.1648 | Fusco A |
| 18 | 2 | 2.1978 | 34 | 37.3626 | Grant S |
| 19 | 2 | 2.1978 | 36 | 39.5604 | Kinzler KW |
| 20 | 2 | 2.1978 | 38 | 41.7582 | Rimm DL |
| 21 | 2 | 2.1978 | 40 | 43.956 | Maher ER |
| 22 | 2 | 2.1978 | 42 | 46.1538 | Sidransky D |
| 23 | 2 | 2.1978 | 44 | 48.3516 | Inoue H |
| 24 | 2 | 2.1978 | 46 | 50.5495 | Henshall SM |
| 25 | 2 | 2.1978 | 48 | 52.7473 | Valero V |
| 26 | 2 | 2.1978 | 50 | 54.9451 | Singh RP |
| 27 | 2 | 2.1978 | 60 | 65.9341 | Debatin KM |
| 28 | 2 | 2.1978 | 65 | 71.4286 | Ellis LM |
| 29 | 2 | 2.1978 | 67 | 73.6264 | Sparreboom A |
| 30 | 2 | 2.1978 | 69 | 75.8242 | McCubrey JA |
| 31 | 2 | 2.1978 | 71 | 78.022 | Imoto I |
| 32 | 2 | 2.1978 | 77 | 84.6154 | Gertz MA |
| 33 | 2 | 2.1978 | 82 | 90.1099 | Armstrong L |
| 34 | 2 | 2.1978 | 84 | 92.3077 | Hochhaus A |
| 35 | 2 | 2.1978 | 89 | 97.8022 | Tefferi A |
| 36 | 2 | 2.1978 | 91 | 100 | Van Der Velden
VHJ |
Therefore, all of the aforementioned authors and
institutions have played an important role in forming and
connecting the collaborative oncology research network. The
oncology research strength is basically balanced between the
communities over the 10 years studied in this study. Only one
academic community had greater impact and features (the largest red
cluster) than the other communities. We also found that researchers
preferred to collaborate within their own institution.
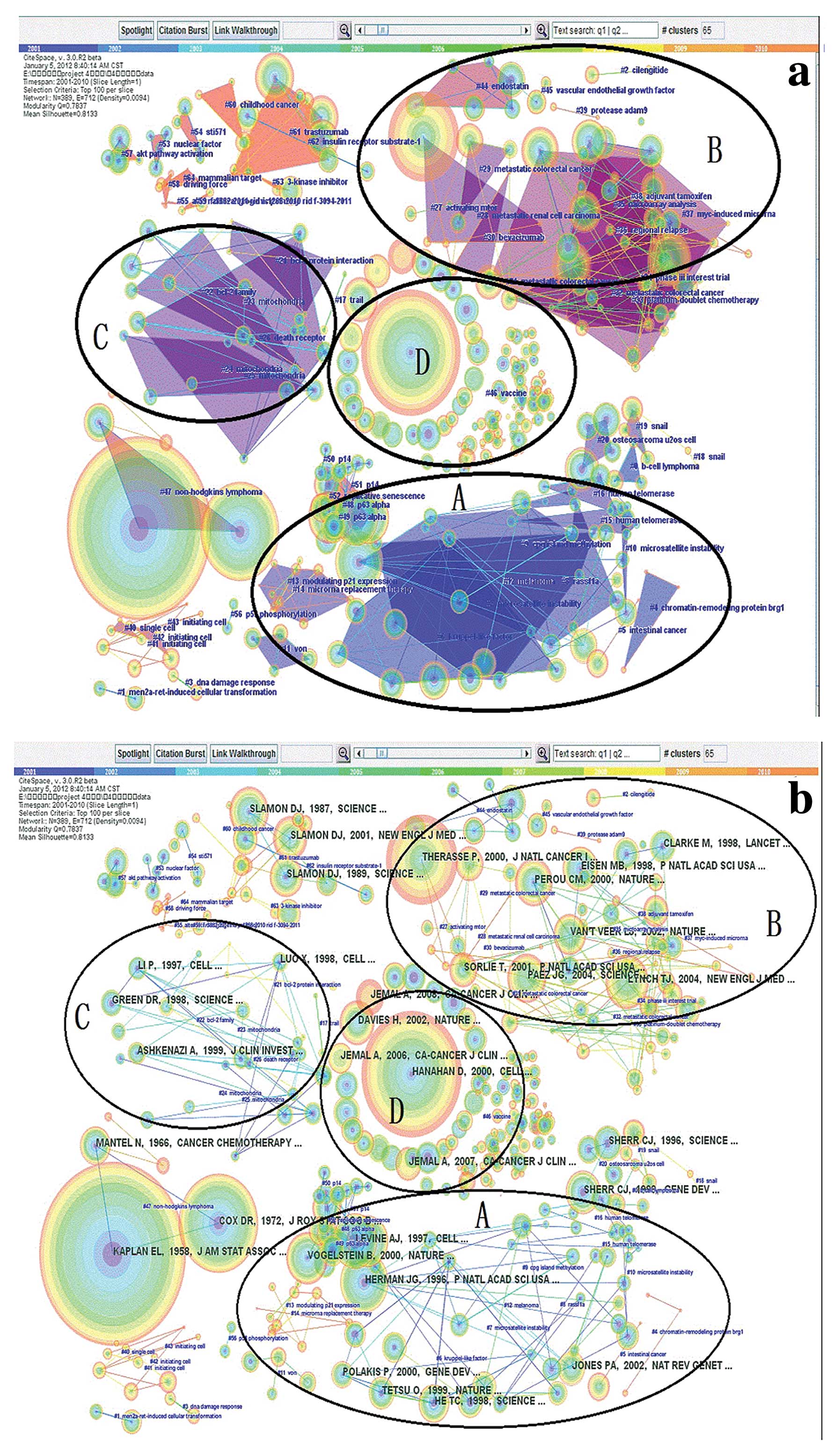
Oncology research fronts
Fig. 3 and Tables IV and V show the results of our document
co-citation analysis using Citespace II.
 | Table IVThe 4 largest DCA clusters of the
oncology research network (2001–2010). |
Table IV
The 4 largest DCA clusters of the
oncology research network (2001–2010).
| Field | C# | Size | Silhouette | Top terms by tf*
idf | Top terms by LLR
(P=0.0001) | Related
clusters |
|---|
| A | #9 | 100 | 0.754 | (7.04) kruppel-like
factor; (16.49) CpG island methylation; (15.51) p63α; (15.51) p53
mutant: (15.45) microsatellite instability; (15.44) promoter
methylation; (13.46) microRNA replacement therapy; (15.06) rassf1a;
(14.36) human telomerase; (14.36) aberrant promoter methylation;
(12.31) osteosarcoma U2OS cells; (10.78) p14; (9.73) Snail | (203.73) p53;
(117.03) microsatellite instability; (103.63) p14; (97.86) rassf1a;
(97.46) microRNA; (89.71) hypermethylation; (52.91) methylation;
(27.41) snail | #6, #48, #49, #7,
#14, #15, #16, #20, #50, #19 |
| B | #31 | 69 | 0.822 | (17.04) metastatic
colorectal cancer; (16.98) metastatic breast cancer; (16.48)
gefitinib; (15.67) endostatin; (14.4) adjuvant tamoxifen; (13.46)
regional relapse; (11.16) microarray analysis; (10.62) Myc-induced
microRNA; (10.62) platinum-doublet chemotherapy; (10.57)
bevacizumab; (6.94) vascular endothelial growth factor | (94.55) tamoxifen;
(76.03) gefitinib; (54.37) endostatin; (53.48) tumor growth;
(49.74) bevacizumab; (36.21) metastatic colorectal cancer; (14.37)
vascular endothelial growth factor | #29, #34, #44, #38,
#36, #35, #37, #33, #30, #45 |
| C | #23 | 45 | 0.346 | (12.45)
mitochondria; (12.14) death receptor; (11.87) adenine nucleotide
translocator; (8.73) Bcl-2 protein interaction; (8.73) Bcl-2 family
interaction; (11.42) tumour growth | (99.55) Trail;
(94.12) factor-related apoptosis-inducing ligand; (41.98)
drug-induced apoptosis; (41.09) adenine nucleotide translocator;
(38.37) apoptosis; (25.41) Bcl-2 protein | #24, #25, #26, #21,
#22, #17 |
| D | #46 | 41 | 0 | (16.49) vaccine;
(15.86) dendritic cells; (14.94) multiple myeloma; (14.7)
cyclooxygenase-2; (14.7) chemokine | (147.93) vaccine;
(87.73) patient; (79.36) prostate cancer; (68.49) novel | |
 | Table VThe most affective authors and their
most-cited study in oncology research fronts (2001–2010). |
Table V
The most affective authors and their
most-cited study in oncology research fronts (2001–2010).
| Serial no. | Research front | Representative
author | The author’s
most-cited study | Frequency |
|---|
| 1 | The mechanisms of
abnormal oncogene expression | Herman JG | DNA methylation
changes in hematologic malignancies: biologic and clinical
implications (Leukemia, 1997) (23) | 337 |
| Levine AJ | A single nucleotide
polymorphism in the MDM2 promoter attenuates the p53 tumor
suppressor pathway and accelerates tumor formation in humans (Cell,
2004) (29) | 316 |
| He TC | PPAR delta is an
APC-regulated target of non-steroidal anti-inflammatory drugs
(Cell, 1999) (46) | 218 |
| Serrano M | Cellular senescence
in cancer and aging (Cell, 2007) (47) | 208 |
| Jones PA | The fundamental
role of epigenetic events in cancer (Nat Rev Genet, 2002) (24) | 195 |
| 2 | Tumor metastasis
and angiogenesis | Therasse P | New guidelines to
evaluate the response to treatment in solid Tumors (J Natl Cancer
Inst, 2000) (48) | 498 |
| Lynch TJ | Activating
mutations in the epidermal growth factor receptor underlying
responsiveness of non-small cell lung cancer to gefitinib (N Engl J
Med, 2004) (34) | 307 |
| Paez JG | EGFR mutations in
lung cancer: correlation with clinical response to gefitinib
therapy (Science, 2004) (33) | 277 |
| Hurwitz H | Bevacizumab plus
irinotecan, fluorouracil, and leucovorin for metastatic colorectal
cancer (N Engl J Med, 2004) (31) | 194 |
| Pao W | EGF receptor gene
mutations are common in lung cancers from ‘never smokers’ and are
associated with sensitivity of tumors to gefitinib and erlotinib
(Proc Natl Acad Sci USA, 2004) (49) | 141 |
| 3 | The relationship
between cancer cells and apoptosis | Green DR | The pathophysiology
of mitochondrial cell death (Science, 2004) (39) | 152 |
| Walczak H | Tumoricidal
activity of tumor necrosis factor related apoptosis-inducing ligand
in vivo (Nat Med, 1999) (41) | 134 |
| Lip GY | Evidence of
platelet activation in hypertension (J Hum Hypertens, 1997)
(50) | 134 |
| LI HL | Solution structure
of BID, an intracellular amplifier of apoptotic signaling (Cell,
1999) (51) | 134 |
| Ashkenazi A | Safety and
antitumor activity of recombinant soluble Apo2 ligand (J Clin
Invest, 1999) (42) | 131 |
| 4 | Tumor vaccines | Hanahan D | The hallmarks of
cancer (Cell, 2000) (52) | 695 |
| Davies H | Mutations of the
BRAF gene in human cancer (Nature, 2002) (53) | 251 |
| Jemal A | Cancer statistics,
2008 (CA Cancer J Clin 2008) (54) | 195 |
| Lengauer C | 14-3-3 Sigma is
required to prevent mitotic catastrophe after DNA damage (Nature,
1999) (55) | 185 |
| Sambrook J | Dominant negative
ATM mutations in breast cancer families (J Natl Cancer Inst, 2002)
(56) | 179 |
Table IV shows the
automatically chosen cluster labels of the 4 largest DCA clusters,
along with their size, silhouette value and background colour shown
in Fig. 3a. The top-ranked title
terms according to LLR and tf* idf were selected as cluster labels.
The largest cluster (‘CpG island methylation’, ‘p63’, ‘p53’ and
‘microsatellite instability’ (#9 and its related clusters) had 100
members and a reasonably high silhouette value of 0.754, which
suggests a homogenous structure. The second largest cluster (#31
and its related clusters) had 69 members and a silhouette value of
0.822, which was the highest among the four co-citation networks,
and was labelled ‘metastatic colorectal cancer’, ‘endostatin’ and
‘vascular endothelial growth factor’. The third largest cluster
(#23 and its related clusters) had 45 members and was labelled
‘mitochondria’ and ‘death receptor’. The fourth largest cluster
(#46) was labelled ‘vaccine’ and had the lowest silhouette value of
0, indicating a heterogeneous structure since there were no links
found in this cluster (Fig. 3).
Other candidate labels for the clusters included
‘trastuzumab’ (#61) and ‘akt pathway activation’ (#57), confirming
that these clusters are subclusters (Fig. 3).
As shown in Fig. 3,
after removing a few small clusters that did not have relevant
themes, we determined the top 5 representative scholars and their
most influential studies in the four major research fronts
(Table V).
Research front 1
The mechanisms of abnormal oncogene expression
(
Fig. 3, Field A)
In our interpretation of Fig. 3, we noted that the most important
field of the four major fronts was Field A, which primarily
involves the study of the mechanisms of abnormal oncogene
expression. Field A includes the study of microsatellite
instability (#7 and #10), DNA methylation (#9), the role of
transcription factors in oncogene expression, tumour-suppressor
genes (#48-#52) and telomerase (#15 and #16).
Microsatellite instability in a human colon cancer
cell line was reported by Liu et al(22). They demonstrated that inhibiting the
expression of the mismatch-repair (MMR) gene is caused by the
inactivation of the two alleles of the gene. Their research is a
milestone in revealing the mechanism of tumorigenesis.
Knudon’s two-hit model is now considered a mechanism
of tumorigenesis, and some loci of microsatellite polymorphisms are
used to examine microsatellite instability. In addition to
microsatellite instability in autosomes, various researchers are
focusing on the relationship between mitochondrial DNA
microsatellite instability and cancer.
Another important area of research involves DNA
methylation (#9). MLH1, one of the mismatch-repair genes, is often
hypermethylated within its 5′ region in the apparently normal
colonic epithelium of patients with microsatellite instability. In
another example, during the progression of leukaemia, the
hypermethylation state of several gene promoters was observed to be
abnormal. This result suggests that epigenetic abnormalities could
be used to monitor disease activity during therapy (23). Based on the opinion of Jones and
Baylin (24), in neoplasia, the
chromatin structure and DNA methylation patterns including the
specific component differ from normal cells. These epigenetic
changes, particularly regarding aberrant promoter hypermethylation,
affect gene expression in tumour progression. Many clinical trials
have found that DNA methylation plays an important role in the
development of cervical cancer, ovarian cancer and endometrial
cancer. Therefore, DNA methylation is a candidate epigenetic marker
that can be used to diagnose tumours at an early stage and provide
prognostic evaluation.
Various transcription factors are also involved in
the abnormal expression of oncogenes. Kruppel-like transcription
factors (#6) that contain zinc finger domains play a dual role in
inhibiting or promoting tumour formation by regulating cell
proliferation, cell differentiation, apoptosis and the expression
of target genes. The transcription factor Snail, a member of the
SNAIL superfamily (#18 and #19), is a zinc finger protein found in
the Drosophila embryo in 1984 (25). Subsequently, Snail and its
homologues have been found in other vertebrates, including humans.
An increasing number of studies have shown that tumour invasion and
metastasis are closely related to the overexpression of Snail.
Thus, Snail can be used as a molecular marker for early diagnosis
and prognosis in cancer. The investigation results by Tetsu and
McCormick (26) (whose citation
frequency ranked second in this field) showed that β-catenin
activates transcription from the cyclin D1 promoter, and the
promoter sequences related to consensus TCF/LEF-binding sites are
necessary for activation.
Our map analysis showed that another small cluster
was closely related to Field A, which involves several
tumour-suppressor genes: p53, p63 and p14. The p53
tumour-suppressor protein plays a vital role in regulating cell
growth following exposure to various stress stimuli. In normal
cells, p53 induces either growth arrest, preventing the replication
of damaged DNA, or apoptosis, which is important for eliminating
defective cells (27). As a tumour
suppressor, p53 is activated by a number of stressors to induce
apoptosis and cell cycle arrest (28). In at least half of all cancers, the
p53 gene is mutated (29). Many
oncogenes belong to the same gene family in the human genome. The
p53 gene family has become one of the most comprehensive and
thorough focuses of gene family research. p63, a homologue of p53,
is more complex than p53. In recent years, the search for a
relationship between the abnormal expression of p63 and
tumorigenesis has been a hotspot of tumour research. Another newly
discovered tumour-suppressor gene, p14ARF, is an important member
of the cell cycle network. The cell cycle is monitored by p14ARF
via the p53-MDM2 and non-p53 pathway to inhibit tumour
development.
The other small clusters that co-cited closely with
Field A include the chromatin-remodelling protein Brg1 (#4), von
(#11), p21 (#13) and human telomerase (#15 and #16). As a ribosomal
nuclear protease, new tumour marker and anticancer target,
telomerase has become a hot topic in breast cancer research.
Telomerase has the ability to maintain the normal karyotype after a
cell exceeds its normal lifespan in vitro, revealing the
mechanism of abnormal division in cancer cells (30).
Research front 2
Tumour metastasis and angiogenesis (
Fig. 3, Field B)
Many studies have indicated that angiogenesis is the
basis for tumour metastasis, and malignant tumours often grow
slowly without angiogenesis.
Our map shows that many scholars are interested in
metastatic renal cell carcinoma and metastatic colorectal cancer.
With the continuous exploration of the mechanism of renal cell
carcinoma development, many of the hypoxia-inducible factor
(HIF)-associated proteins, including VEGF (vascular endothelial
growth factor), PDGF (platelet-derived growth factor) and TGF-α
(transforming growth factor-α), are involved in renal cell
carcinoma. Endothelial cell receptors, periderm cell receptors, and
tumour cell receptors have also been shown to play roles in renal
cell carcinoma.
In recent years, the molecular-targeted therapy of
metastatic renal cell cancer and metastatic colorectal cancer has
become a hotspot in oncology research. For example, bevacizumab, a
main molecular-targeted drug, is a monoclonal antibody against
vascular endothelial growth factor (31). In addition, as an inhibitor of
angiogenesis, cilengitide has a function in preventing the
production of blood vessel production (32).
Presently, molecular-targeted drug treatment is a
new approach to inhibit tumour development, invasion and
metastasis. There are many outstanding research results in this
research area. For instance, the investigations of Paez et
al(33) and Lynch et
al(34) indicate that EGFR
mutations can predict sensitivity to gefitinib. These studies
provide the foundation for the application of this
molecular-targeted drug.
In Fig. 3, two
clusters can be seen, endostatin (#44) and VEGF (#45), each of
which have an intense co-citation relationship with metastatic
colorectal cancer and metastatic renal cell carcinoma, indicating
that these clusters are closely related. In 1971, Folkman (35) first proposed that tumour growth and
metastasis rely on angiogenesis. According to recent relevant
publications, the inhibition of tumour angiogenesis and induction
of tumour hibernation or apoptosis are effective methods for
treating tumours. For example, endostatin plays an anti-angiogenic
role and exhibits broad-spectrum, low toxicity and non-resistant
characteristics. The application of endostatin is a new strategy
for treating cancer, and anti-VEGF therapy is predicted to become a
mainstream therapy for cancer treatment.
Fig. 3 also shows
that tumour metastasis co-cites with another cluster (protease
ADAM9), indicating that these clusters are closely related. The
expression of ADAM9 in colon cancer tissue was found to be
significantly higher than that in adjacent normal tissue and has a
correlation with the disease progression of metastatic colon cancer
and tumour angiogenesis (36).
Research front 3
The relationship between cancer cells and
apoptosis (
Fig. 3, Field C)
Apoptosis, also known as programmed cell death, was
first described in 1972 by Kerr (37), an American pathologist. In recent
years, researchers have been paying close attention to apoptotic
mechanisms. Commonly, the occurrence of tumours results from the
unlimited growth of tumour tissue, which is caused by unregulated
apoptosis. Mitochondria, the only organelles containing their own
genetic material in mammalian cells (38), play a very important role in the
process of tumour cell apoptosis. Caspase activation is closely
linked with mitochondrial outer membrane permeabilisation in the
mitochondrial pathway of apoptosis (39).
Based on in-depth studies of apoptosis and tumour
pathogenesis, a number of ligands and receptors that induce tumour
cell apoptosis have been identified, all of which belong to the
tumour necrosis factor (TNF) supergene family. Regarding the tumour
necrosis factor family, the TRAIL (TNF-related apoptosis inducing
ligand) system is one of the research hotspots of recent years. The
overexpression of Trail (40) can
selectively kill a variety of tumour cells yet shows no significant
toxicity to most normal cells, suggesting its possible use as a new
anticancer drug. A study published by Walczak et al(41) demonstrated that the growth of a
human mammary adenocarcinoma cell line could be inhibited by
LZ-TRAIL without significant toxicity to normal tissues. Another
outstanding experiment performed by Ashkenazi et al(42) showed that Apo2L (or TRAIL) has
potent anticancer activity but leaves normal cells unharmed.
Research front 4
Tumour vaccines (
Fig.
3, Field D)
With the development of tumour immunology and
molecular biology, the research areas involving the interaction
between the tumour and the body, tumour immune tolerance and tumour
antigen identification have made great progress and have
contributed to the development of cancer vaccines. To date, many
tumour vaccines have been developed via animal experiments and
clinical trials. Various effective vaccines induce cytotoxic
T-lymphocytes with adverse reactions. Therefore, as an efficient
tumour immunotherapy with low toxicity, tumour vaccines are
potentially effective drugs that must be developed.
A study by Cao et al(43) suggeted that it is possible to fight
solid tumours by modulating antitumour immunity. Their research
provided a solid foundation for theories on tumour vaccines. An
example of a cancer vaccine is the immunodominant peptide of the
gp100 melanoma-associated antigen constructed by Rosenberg et
al(44). His experiment
initiated the development of novel cancer immunotherapies. In
another example of an effective tumour vaccine, BERH-2 tumours can
be eradicated by hybrid cells made from the fusion of BERH-2 rat
hepatocellular carcinoma cells with activated B cells (45).
As mentioned above, the difference between tumour
vaccines and conventional vaccines is that tumour vaccines are
active immunotherapies for treatment and do not prevent infection
with pathogens. With the development of tumour immunology, the
tumour mechanism of immune tolerance has been gradually recognised,
and an increasing number of tumour antigens have been identified.
These antigens are helpful in the research and development of
tumour vaccines. At present, the majority of tumour vaccines, such
as the gastric tumour cell vaccine, lung tumour cell vaccine, and
melanoma cell vaccine, have been tested in animal models, and only
a few have been studied in clinical trials, such as the cervical
cancer vaccine and breast cancer vaccine. Preparation of tumour
vaccines has gradually improved and developed to include genetic
modifications and cell fusion methods. In the near future, tumour
vaccines may become a new means of cancer treatment after surgery,
radiotherapy and chemotherapy.
In conclusion, having analysed the results, we
understand that 30 major international oncology journals do not
represent the overall productivity in the field of oncology; thus,
there may be a few disadvantages regarding the research front based
on the authors’ outputs. Otherwise, compared to the disadvantages,
this network offers many advantages as it is more practical, easy
to handle, and effective, and will be more instructive for
scientific research evaluation work.
However, by mapping the knowledge domains of
oncology research, we found that the citation frequency for the US
accounted for 53.8% of the total citation frequency of the top 20
core countries performing oncology research. China is the sole
developing country to enter the top 10. Within the core
institutions of oncology research, the US accounted for 18 of the
top 20 institutions, and its total citation frequency accounted for
93.5% of the top 20. The research strength of the University of
Texas, the NCI and Harvard University has steadily grown for nearly
10 years, and most countries and institutions have separate and
distinct research directions. It is clear that the United States is
the leading country in oncology research.
We found that the current oncology research fronts
are focused in four fields: i) the mechanism of abnormal oncogene
expression; ii) tumour metastasis and angiogenesis; iii) the
relationship between cancer cells and apoptosis; and iv) tumour
vaccines. The four research frontiers included 55 branch fields. We
also identified the 36 most collaborative academic communities and
determined that their oncology research strength is basically in
balance. Multiple myeloma, angiogenesis, and acute lymphocytic
leukaemia were the primary focuses of research collaborations in
oncology from 2001 to 2010.
After our analysis of the main research centres with
respect to institutions and countries, we traced oncology articles
published in the studied time frame. By document co-citation
analysis, we accurately identified the research fronts, hotspots,
and classic milestone publications that provide the foundation in
the field. From our co-authorship analysis, we determined the most
collaborative academic communities. Through the interpretation of
our knowledge maps, we depicted the overall image and trends of
oncology research progress in an objective, scientific and
systematic manner. We also demonstrated the inherent mainstream
areas and knowledge structure in the oncology discipline so that
countries with developing research programs can track the
international research forefronts, grasp the correct direction for
their research, and identify entry points for their own studies.
These maps also provide scientific evidence and suggestions for
policymakers to select the most prolific academic groups and
individuals in the oncology research community and establish a more
efficient system for managing and financing oncology research in
the future. The study not only provides support for important
decisions in the science and technology field but also supplies a
basis for technology planning and evaluation and a quantitative
basis for the selection of research projects. An improvement in
international oncology research has important theoretical
significance and academic value.
There are many issues worthy of further analysis in
oncology research. We are planning to supplement our current data
with that from 1981 to 2000 to further study the changing trends in
the oncology research fronts and hotspots and the changing
scientific laws in the field of oncology. Because the present
scientometric methods have not identified relevant disease-specific
or clinical activities, we will combine the meta-analysis with the
present methods to evaluate disease identification in our next
study.
Acknowledgements
This study was supported by the National Nature
Science Foundation of China under grant numbers, No. 71240006 and
No. 71103114; the Taiyuan City Science and Technology Innovation
Project Program under grant number, No. 110271. In addition, the
authors would like to thank other member of the authors’ team for
helpful discussions, particularly Professor Dawei Guo.
References
|
1
|
Borner K, Chen CM and Boyack KW:
Visualizing knowledge domains. Annu Rev Inf Sci Technol.
37:179–255. 2003. View Article : Google Scholar
|
|
2
|
Chen Y and Liu ZY: The rise of mapping
knowledge domain. Studies in Science of Science. 23:149–154.
2005.
|
|
3
|
Jankovic MP, Kaufmann M and Kindler CH:
Active research fields in anesthesia: a document co-citation
analysis of the anesthetic literature. Anesth Analg. 5:1524–1533.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jarneving B: A comparison of two
bibliometric methods for mapping of the research front.
Scientometrics. 2:245–263. 2005. View Article : Google Scholar
|
|
5
|
Lee MR and Chen TT: Revealing research
themes and trends in knowledge management: from 1995 to 2010.
Knowledge-Based System. 28:47–58. 2012. View Article : Google Scholar
|
|
6
|
Zhao R and Wang J: Visualizing the
research on pervasive and ubiquitous computing. Scientometrics.
86:593–612. 2011. View Article : Google Scholar
|
|
7
|
Chen CM, McCain K, White H and Lin X:
Mapping Scientometrics (1981–2001). In: Proc 65th Asist Annual
Meeting; 39. pp. 25–34. 2002
|
|
8
|
An XY and Wu QQ: Co-word analysis of the
trends in stem cell field based on subject heading weighting.
Scientometrics. 88:133–144. 2011. View Article : Google Scholar
|
|
9
|
Chen CM: CiteSpace II: detecting and
visualizing emerging trends and transient patterns in scientific
literature. J Am Soc Inf Sci Technol. 57:359–377. 2006. View Article : Google Scholar
|
|
10
|
Chen C, Ibekwe-San Juan F and Hou J: The
structure and dynamics of cocitation clusters: a
multiple-perspective cocitation analysis. J Am Soc Inf Sci Technol.
61:1386–1409. 2010. View Article : Google Scholar
|
|
11
|
Chen CM: Searching for intellectual
turning points: progressive knowledge domain visualization. Proc
Natl Acad Sci USA. 101:5303–5310. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CM, Cribbin T, Macredie R and Morar
S: Visualizing and tracking the growth of competing paradigms: two
case studies. J Am Soc Inf Sci Technol. 53:678–689. 2002.
View Article : Google Scholar
|
|
13
|
Chen C, Hu Z, Liu S and Tseng H: Emerging
trends in regenerative medicine: a scientometric analysis in
CiteSpace. Expert Opin Biol Ther. 12:593–608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Small H: Co-citation in the scientific
literature: a new measure of the relationship between two
documents. J Am Soc Info Sci. 24:265–269. 1973. View Article : Google Scholar
|
|
15
|
Liu Z, Chen Y and Chen H: Mapping
Knowledge Domains Methods and Application. 1st edition. People
Publishing House; Peking: pp. 34–81. 2008
|
|
16
|
Liu J: An introduction to Social Network
Analysis. 1st edition. Social Science Academic Press; (China),
Beijing: pp. 4–25. 2004
|
|
17
|
Katz JS and Martin BR: What is research
collaboration? Res Policy. 26:1–18. 1997. View Article : Google Scholar
|
|
18
|
Gossart C and Ozman M: Co-authorship
networks in social sciences: the case of Turkey. Scientometrics.
78:323–345. 2009. View Article : Google Scholar
|
|
19
|
Lu HY and Feng YQ: A measure of authors’
centrality in co-authorship networks based on the distribution of
collaborative relationships. Scientometrics. 81:499–511. 2009.
|
|
20
|
Yu Q, Shao H and Duan Z: Research groups
of oncology co-authorship network in China. Scientometrics.
89:553–567. 2011. View Article : Google Scholar
|
|
21
|
Morel CM, Serruya SJ, Penna GO and
Guimarães R: Co-authorship network analysis: a powerful tool for
strategic planning of research, development and capacity building
programs on neglected diseases. PLoS Negl Trop Dis. 3:82009.
View Article : Google Scholar
|
|
22
|
Liu B, Nicolaides NC, Markowitz S, Willson
JKV, Parsons RE, Jen J, Papadopolous N, Peltomäki P, de la Chapelle
A, Hamilton SR, Kinzler KW and Vogelstein B: Mismatch repair gene
defects in sporadic colorectal cancers with microsatellite
instability. Nat Genet. 9:48–55. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Issa JP, Baylin SB and Herman JG: DNA
methylation changes in hematologic malignancies: biologic and
clinical implications. Leukemia. 11:S7–S11. 1997.PubMed/NCBI
|
|
24
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
25
|
Sun C and Wang H: The role of SNAIL gene
in gynecologic cacinoma metastasis mechanism. J Int Obstet Gynecol.
4:260–263. 2010.
|
|
26
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sionov RV and Haupt Y: The cellular
response to p53: the decision between life and death. Oncogene.
45:6145–6157. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tyner SD, Venkatachalam S, Choi J, Jones
S, Ghebranious N, Igelmann H, Lu XB, Soron G, Cooper B, Brayton C,
Park SH, Thompson T, et al: p53 mutant mice that display early
ageing-associated phenotypes. Nature. 415:45–53. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bond GL, Hu WW, Bond EE, Robins H, Lutzker
SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K,
Yip L, Hwang SJ, Strong LC, Lozano G and Levine AJ: A single
nucleotide polymorphism in the MDM2 promoter attenuates the p53
tumor suppressor pathway and accelerates tumor formation in humans.
Cell. 119:591–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bodnar AG, Ouellette M, Frolkis M, Holt
SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S and
Wright WE: Extension of life-span by introduction of telomerase
into normal human cells. Science. 279:349–352. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hurwitz H, Fehrenbacher L, Novotny W,
Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S,
Holmgren E, Ferrara N, Fyfe G, et al: Bevacizumab plus irinotecan,
fluorouracil, and leucovorin for metastatic colorectal cancer. N
Engl J Med. 350:2335–2342. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo QS and Liu YX: Lung cancer target to
treatment medicine progress. World Clin Drugs. 5:282–285. 2005.
|
|
33
|
Paez JG, Janne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon T,
Naoki K, Sasaki H, et al: EGFR mutations in lung cancer:
correlation with clinical response to gefitinib therapy. Science.
304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, Louis DN, Christiani DC, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Folkman J: Tumor angiogenesis: therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi W and Li JS: Clinical significance of
the expression of ADAM9 in colon carcer. J Southeast University
(Medical Science Edition). 4:274–278. 2009.
|
|
37
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis-basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu LJ, Peng JX, Hong HZ, Ye W and Qiao
YY: Mitochondrial changes and role in apoptosis. Chin J Cell Biol.
27:117–120. 2005.
|
|
39
|
Green DR and Kroemer G: The
pathophysiology of mitochondrial cell death. Science. 305:626–629.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deng C and Shao ZW: Apoptosis of
osteosarcoma cells induced by a combination of TRAIL, adriamycin
and IFN-γ. Cancer Res Prev Treat. 1:1–4. 2010.PubMed/NCBI
|
|
41
|
Walczak H, Miller RE, Ariail K, Gliniak B,
Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C,
Smolak P, et al: Tumoricidal activity of tumor necrosis factor
related apoptosis-inducing ligand in vivo. Nat Med. 5:157–163.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Masters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, DeForge L, Koumenis IL, et al: Safety and antitumor activity of
recombinant soluble Apo2 ligand. J Clin Invest. 104:155–162. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao ZA, Daniel D and Hanahan D: Sub-lethal
radiation enhances anti-tumor immunotherapy in a transgenic mouse
model of pancreatic cancer. BMC Cancer. 2:112002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rosenberg SA, Yang JC, Schwartzentruber
DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME,
Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, et al:
Immunologic and therapeutic evaluation of a synthetic peptide
vaccine for the treatment of patients with metastatic melanoma. Nat
Med. 4:321–327. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo Y, Wu M, Chen H, Wang X, Liu G, Li G,
Ma J and Sy MS: Effective tumor vaccine generated by fusion of
hepatoma cells with activated B cells. Science. 263:518–520. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He TC, Chan TA, Vogelstein B and Kinzler
KW: PPAR delta is an APC-regulated target of non-steroidal
anti-inflammatory drugs. Cell. 99:335–345. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Collado M, Blasco MA and Serrano M:
Cellular senescence in cancer and aging. Cell. 130:223–233. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Therasse P, Arbuck SG, Eisenhauer EA,
Wanders J, Kaplan RS, Rubinstein L, et al: New guidelines to
evaluate the response to treatment in solid tumors. J Natl Cancer
Inst. 92:205–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, et al: EGF receptor gene mutations are common
in lung cancers from ‘never smokers’ and are associated with
sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad
Sci USA. 101:13306–13311. 2004.
|
|
50
|
Blann AD, Lip GY, Islim IF and Beevers DG:
Evidence of platelet activation in hypertension. J Hum Hypertens.
11:607–609. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chou JJ, Li HL, Salvesen GS, Yuan JY and
Wagner G: Solution structure of BID, an intracellular amplifier of
apoptotic signaling. Cell. 96:615–624. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 144:646–674. 2000. View Article : Google Scholar
|
|
53
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, et al: Mutations of the BRAF gene in human
cancer. Nature. 417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jemal A, Siegel R, Ward E, Hao YP, Xu JQ,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
55
|
Chan TA, Hermeking H, Lengauer C, Kinzler
KW and Vogelstein B: 14-3-3 Sigma is required to prevent mitotic
catastrophe after DNA damage. Nature. 401:616–620. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chenevix-Trench G, Spurdle AB, Gatei M,
Kelly H, Sambrook J, et al: Dominant negative ATM mutations in
breast cancer families. J Natl Cancer Inst. 94:205–215. 2002.
View Article : Google Scholar : PubMed/NCBI
|