Introduction
microRNAs (miRNAs) are a group of small non-coding
RNAs 21–25 nucleotides in length that negatively regulate the
expression of targeted genes in animals, plants and viruses by
interacting with complementary sites in the 3′ untranslated region
(3′UTR) and degrading mRNA or suppressing the translation of target
genes (1). More than 60% of human
protein-encoding genes are under selective pressure to maintain
pairing with miRNAs (2), suggesting
that miRNAs may play important roles in a wide array of
physiological and pathological processes, including human
oncogenesis, cell differentiation, proliferation and apoptosis. The
dysregulation of these processes is also a hallmark of cancer.
Mutations affecting miRNAs or their functional interactions with
oncogenes and tumor-suppressor genes may be involved in
tumorigenesis.
miR-16-1 is located at chromosome 13q14, which is
deleted in 68% of chronic lymphocytic leukemia (CLL) cases
(3), and is downregulated in most
solid tumors (4–7). miR-16 inhibits cell proliferation and
induces cell cycle arrest, and increases cell apoptosis by
negatively regulating Bcl2 in CLL, prostate and hepatocellular
carcinoma cancer cells (6,8,9).
Moreover, several studies have shown that p53, as a nuclear protein
that is essential for cell cycle control, DNA repair and induction
of apoptosis under many stresses, also enhances the maturation of
several miRNAs at the post-transcriptional levels, including miR-16
(10,11). The p53 protein has also been shown
to play important roles in the oncogenesis of colon cancer. These
findings indicate that miR-16, a miRNA related to p53, has a
critical function in human carcinogenesis. However, the possible
roles and mechanisms of miR-16 and its relationship with p53 in CRC
cells are still not well established.
In this study, we sought to investigate the
expression of miR-16 in CRC and normal adjacent tissues and
elucidate the effect of miR-16 on the growth of CRC cells in
vitro and examine its downstream target. We found that miR-16
was downregulated in 67% of CRC tissues, and overexpression of
miR-16 inhibited cell proliferation and induced cell apoptosis in
CRC cells. We further identified the oncogene survivin as a direct
and functional target of miR-16. Furthermore, we found that miR-16
regulated the p53/survivin signaling pathway in CRC cells in
vitro. Our findings provide further evidence for the
implication of dysregulated miRNAs in CRC, and miR-16 may serve as
a molecular target for CRC, which to date has a dismal outcome with
limited therapeutic options.
Materials and methods
Cells and acquisition of tissue
specimens
Human CRC cell lines HCT116 and LoVo were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China) and maintained in RPMI-1640 with 10% fetal bovine serum
(Invitrogen, Carlsbad, CA, USA) and 1% antibiotics (100 U/ml
penicillin and 100 μg/ml streptomycin sulfate) in a humidified
atmosphere containing 5% CO2 at 37°C.
Surgically excised tumor specimens and normal
adjacent tissues from 52 patients with World Health Organization
(WHO) grade I–IV CRC were collected at the Department of
Gastroenterology, Huizhou Center Hospital, Huizhou, China. Each
tumor specimen was divided into two parts. One part was immediately
snap-frozen in liquid nitrogen and stored at −80°C, the other part
was used for pathological assesment by which the tumor
differentiation was graded as well, moderate or poor (including
undifferentiated) (12). The
protocol for this study and acquisition of tissue specimens were
approved by the local institutional review boards at the affiliated
institutions of the authors. Human tissue acquisition and use in
this study complied with the National Regulations on the Use of
Clinical Samples in China.
Quantitative real-time PCR
Total RNA was isolated using TRIzol reagent
(Invitrogen) according to the manufacturer’s protocol. First-strand
cDNA synthesis was carried out using TaqMan MicroRNA Reverse
Transcription kit and miR-16 RT primers (Applied Biosystems, Foster
City, CA, USA). Real-time PCR was carried out with TaqMan Universal
Master Mix II and miR-16 TaqMan MicroRNA Assay Mix on an ABI PRISM
7500 (Applied Biosystems). miR-16 expression was normalized against
U6. In addition, cDNA was synthesized from 1.5 μg total RNA with
M-MLV reverse transcriptase (Invitrogen) in a 20-μl volume, and
mRNA levels of survivin were detected using a SYBR-Green I Mix
(Roche, Switzerland) in a 20-μl reaction volume on Light Cycler
480II/96 (Roche) as instructed by the manufacturer. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was used as an endogenous
control. Data analysis was carried out using the
2−ΔΔCt method. The primer sequences for
real-time PCR were: survivin, 5′-CCACCGCATCTCTACATTCA-3′ (sense)
and 5′-TATGTTCCTCTATGGGGTCG-3′ (antisense); GAPDH,
5′-GTCAACGGATTTGGTCGTATTG-3′ (sense) and
5′-CTCCTGGAAGATGGTGATGGG-3′ (antisense).
miRNA mimics, inhibitors, small
interfering RNA (siRNA), plasmids and transfections
miR-16 mimics and inhibitors and scrambled controls
were purchased from GenePharma (Shanghai, China). The sequences of
these miRNAs were: miR-16 mimics, 5′-UAGCAGCACGUAAAUAUUGGCG-3′;
miR-16 inhibitors, 5′-CGCCAAUAUUUACGUGCUGCUA-3′; scrambled control,
5′-UUCUCCGAACGUGUCACGUTT-3′. The p53 gene was amplified from
pGEMT-p53. The amplified p53 gene products were digested by
NotI and BamHI and inserted into pcDNA3.1 to obtain
pcDNA3.1-p53. Cells were transfected with appropriate miRNA, siRNA
oligonucleotides and plasmids using Lipofectamine 2000 reagent
(Invitrogen) following the manufacturer’s instructions. The medium
was replenished 6 h after transfection.
CCK-8 cell proliferation assays
Cell proliferation was detected by Cell Counting
Kit-8 (CCK-8) (Beyotime, China) according to the manufacturer’s
protocol. Briefly, cells were plated in 96-well plates at
0.5×104 cells/well. CCK-8 reagent (10 μl) was added at
0, 24, 48 and 72 h after transfection, and after a 1-h incubation
at 37°C, the optical density (OD) was read at 450 nm by a
microplate luminometer reader.
Apoptosis assays
Apoptosis assays were performed using a commercial
Annexin V apoptosis detection kit according to the manufacturer’s
protocol (BD Biosciences). Briefly, transfected cells were
collected, washed with PBS and resuspended in binding buffer
containing 10 mM HEPES (pH 7.4); 2.5 mM CaCl2, and 140
mM NaCl. Annexin V-PE and 7-AAD were added and FACS was performed
after 15 min of incubation at room temperature. Experiments were
performed in triplicates and at least three times
independently.
Luciferase assays
The 3′-UTRs of the target gene survivin were
amplified by PCR using the following primers:
5′-atctGGTACCGGAGAAAGTGCGCCGTGCCA-3′ and
5′-tcctAAGCTTGRGGAAGGCTCTGCCCACGC-3′. The PCR products were then
gel purified, digested and inserted into pGL3-basic vector
(Promega, Madison, WI, USA). Target site mutations were generated
using the PCR products with the appropriate primers containing
point substitutions. The sequences were confirmed by
sequencing.
We performed the luciferase assays using HCT116
cells transiently transfected with 0.1 μg reporter plasmid and 0.65
pmol miRNA mimics or control miRNA in 96-well plates or with
Renilla constructs (as an internal control). Luciferase
assays were performed 48 h later using the Dual-Luciferase Reporter
Assay system according to the manufacturer’s instructions
(Promega). All luciferase activity readings were normalized
relative to the activity of the Renilla luciferase control.
All experiments were performed in triplicate.
Western blot assays
Total protein was extracted using a commercially
available whole cell lysis buffer (Beyotime). Immunoblotting was
performed as previously described (13). The following antibodies were used:
anti-survivin and anti-caspase-9 antibodies (Cell Signaling
Technology, Beverly, MA, USA), anti-p53 antibodies (Santa Cruz
Biotechnology, Santa Cruz, CA, USA), anti-GAPDH antibodies
(Zhongshan Goldenbridge Biotechnology, Guangzhou, China),
anti-cyclin D1 and anti-CDK6 antibodies (Santa Cruz Biotechnology),
anti-capase-3, and anti-caspase-8 antibodies (BD Biosciences).
Protein bands were visualized using ECL substrates (Millipore,
USA).
Statistical analysis
Data are expressed as means ± SD and analyzed using
the SPSS 13.0 software (SPSS Inc., Chicago, IL, USA). The Wilcoxon
or t-test was used for comparing two groups, and one-way ANOVA was
used for comparing multiple groups where the Bonferroni or
Tamhane’s T2 test was used to compare differences between groups. A
P-value <0.05 was considered to indicate a statistically
significant result.
Results
miR-16 is downregulated and is correlated
with histological differentiation in primary colorectal cancer
tissues
miR-16 has been shown to be downregulated in CLL,
lung, prostate and ovarian cancer (4–7), but
whether miR-16 expression in CRC is dysregulated remains to be
elucidated. In this study, we sought to examine miR-16 expression
in an independent primary CRC cohort. We examined the expression of
miR-16 in 52 patients with WHO grade I–IV CRC and normal adjacent
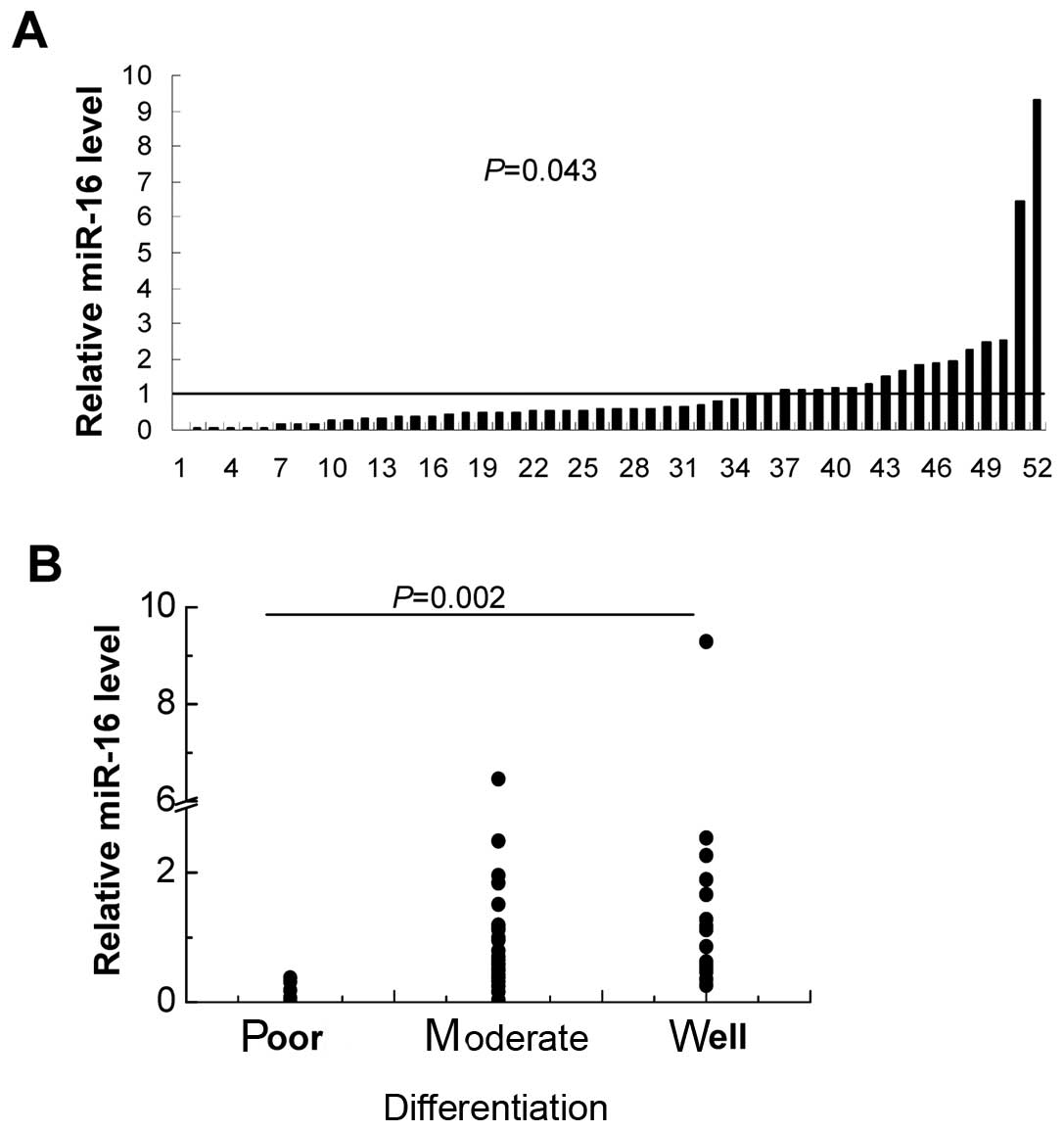
tissues by real-time RT-PCR. We found that miR-16 was downregulated
in 67% (35/52) of the CRC tissues (Fig.
1), and the relative expression level of miRNA transcripts
(median 0.59, 25% quartile 0.33, 75% quartile 1.17) was lower than
that of adjacent normal tissues (P=0.043). In addition, miR-16
expression was inversely correlated to histological differentiation
(P=0.002). miR-16 levels expressed as a median (25–75% quartile) in
tumor tissues of poor, moderate and well differentiation were: 0.13
(0.05–0.33), 0.59 (0.36–1.12) and 0.99 (0.52–1.83),
respectively.
miR-16 overexpression inhibits the
proliferation of colorectal cancer cells in vitro
To further elucidate the effect of miR-16 on CRC, we
first examined its effect on the proliferation of CRC cells. We
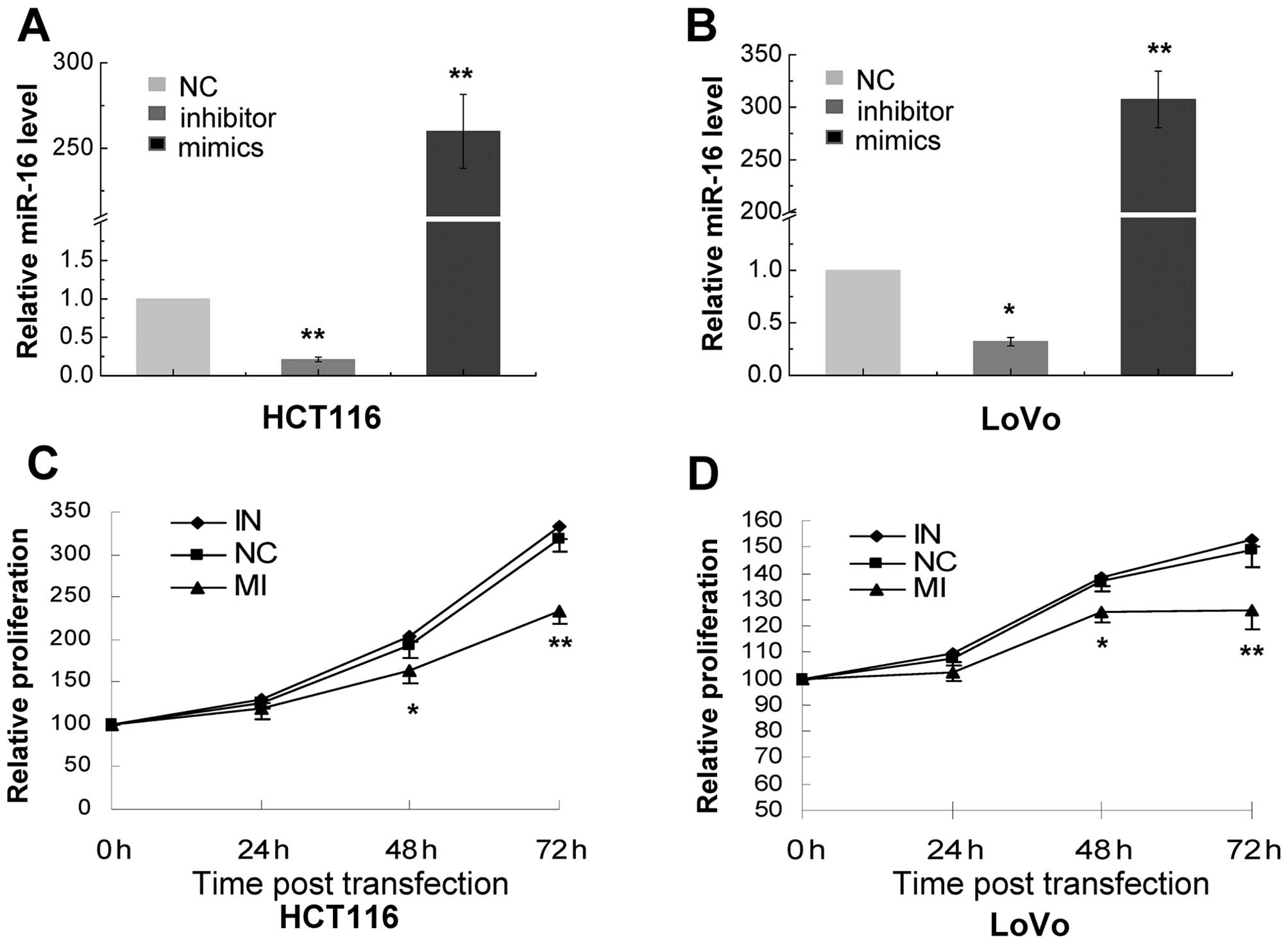
transiently transfected HCT116 and LoVo cells with miR-16
inhibitors and mimics. Our RT-PCR assays showed that cells
transfected with miR-16 mimics exhibited a markedly higher level of
miR-16 than cells transfected with scrambled RNA (259.83±21.63 and
307.46±26.93, respectively) (Fig. 2A
and B). On the other hand, miR-16 inhibitors caused a marked
reduction in the levels of miR-16 in the transfected cells
(0.21±0.03 and 0.32±0.04, respectively) (Fig. 2A and B). We further found that
miR-16 inhibitors exerted no apparent effect on the proliferation
of HCT116 and LoVo cells while miR-16 mimics caused a significant
decrease in the proliferation of the transfected cells at 48- and
72-h post-transfection (Fig. 2C and
D).
miR-16 overexpression enhances apoptosis
of colorectal cancer cells in vitro
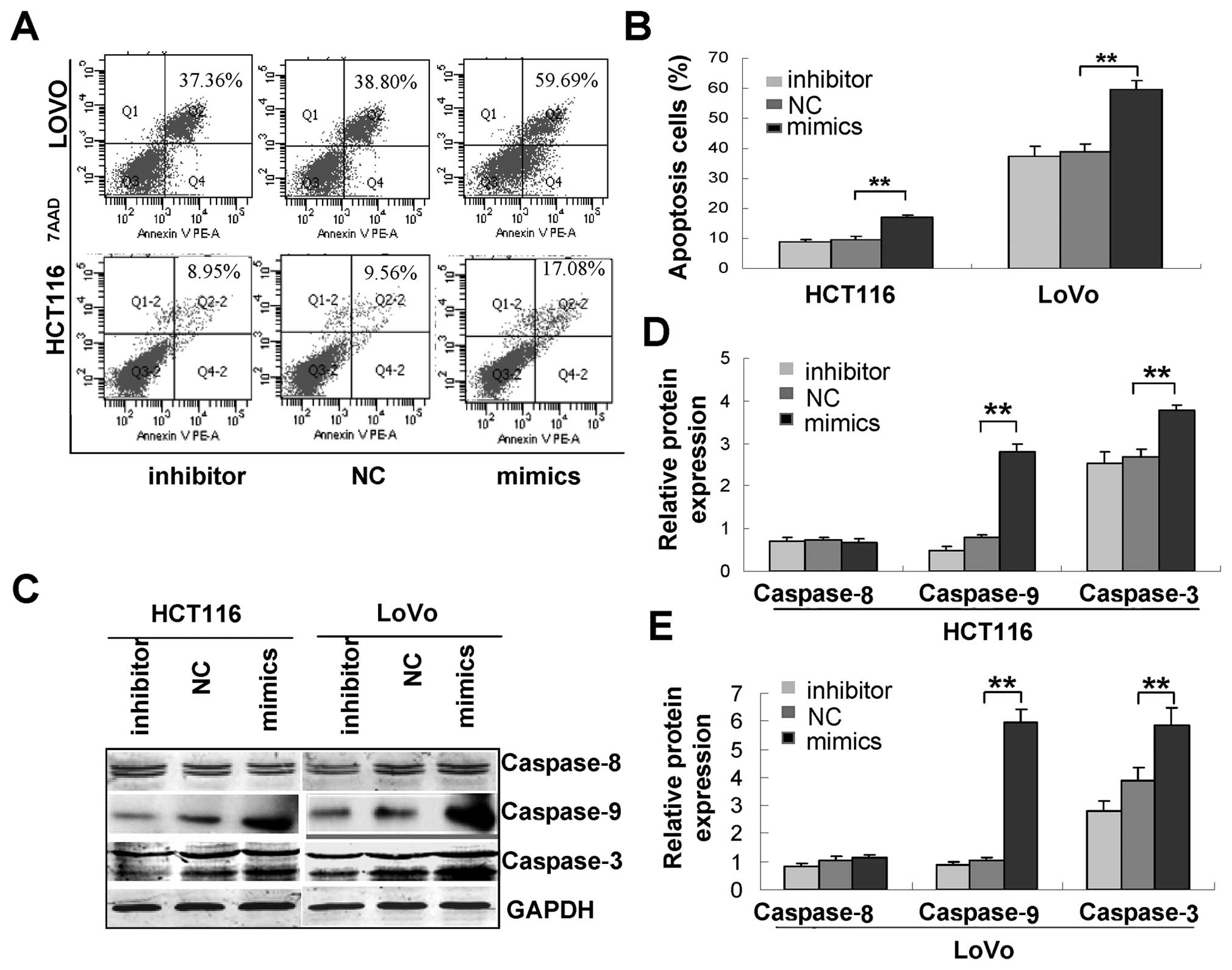
Next, we examined whether miR-16 induces the
apoptosis of CRC cells. Flow cytometric analysis of Annexin
V-stained cells transfected with miR-16 inhibitors or mimics or
scrambled control showed that miR-16 caused a significant increase
in the percentage of apoptotic cells from 9.56±1.13 to 17.08±0.64%
in HCT116 cells and from 38.80±2.63 to 59.69±2.99% in LoVo cells
(P<0.05 in both) while miR-16 inhibitors caused no apparent
change in the percentage of apoptotic CRC cells (Fig. 3A and B). Immunoblotting assays
further revealed that miR-16 caused a marked increase in the
expression of cleaved caspase-9 and -3 (Fig. 3C), suggesting that miR-16 may induce
apoptosis through the intrinsic apoptosis pathway.
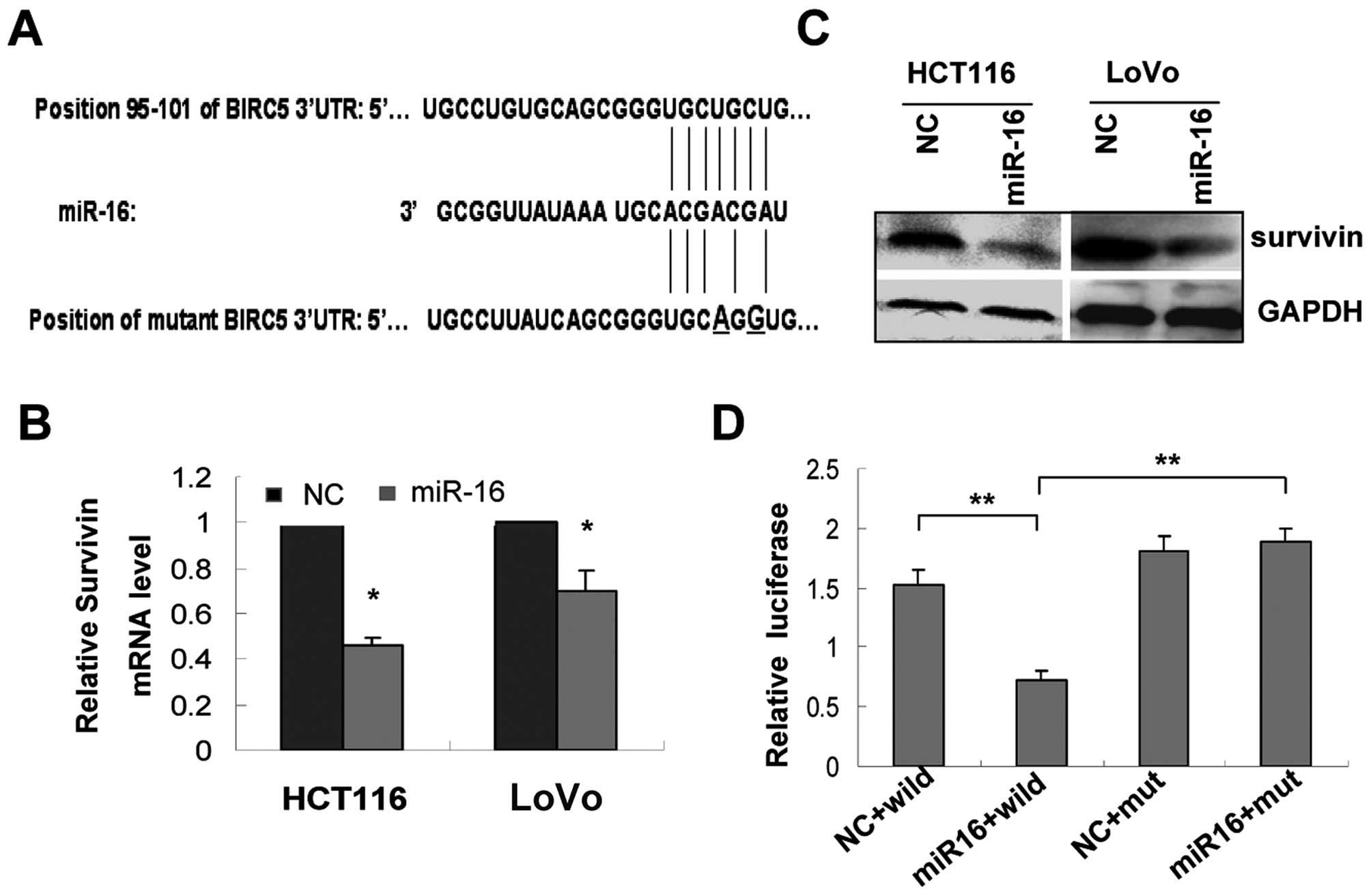
miR-16 directly targets survivin
Survivin is implicated in regulating the intrinsic
apoptotic pathway. We examined whether the survivin (also
named as BIRC5) mRNA sequence (NM-001012270) contains target sites
of miR-16 through RNA22, and the miR-16 binding site was found in
the survivin sequence (Fig.
4A). We then investigated whether miR-16 affects the protein or
mRNA expression of survivin. We found that transfection of CRC
cells with miR-16 mimics caused a significant reduction in the
survivin mRNA transcript levels (Fig.
4B), a finding that was further confirmed by western blot
assays (Fig. 4C), which showed an
apparent decrease in the levels of survivin in HCT116 and LoVo
cells transfected with miR-16 mimics. Transient transfection of
HCT116 cells with the reporter plasmid containing the seed sequence
of the 3′-UTR of the survivin gene and miR-16 mimics
produced a significant reduction in luciferase reporter gene
activities (P<0.001 vs. the scrambled control) (Fig. 4D). By contrast, transient
transfection of HCT116 cells with the reporter plasmid-containing
mutations in the seed sequence of the 3′-UTR of the survivin
gene (Fig. 4A) and miR-16 mimics
did not decrease the activities of the mutant luciferase reporter.
These findings suggest that the 3′-UTR of the survivin gene
was a functional target site of miR-16, which, when overexpressed,
suppressed the expression of survivin at both the mRNA and protein
levels.
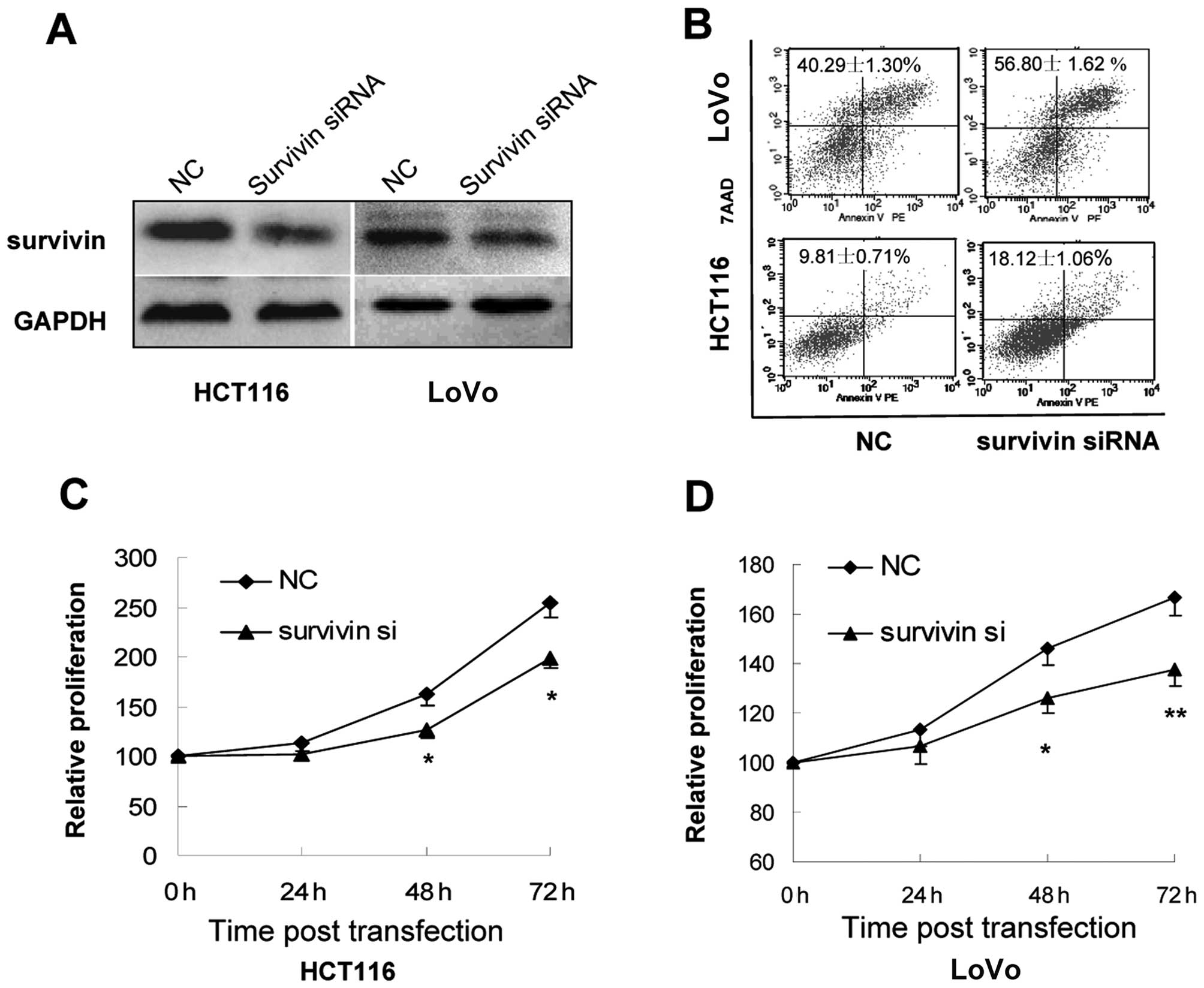
Knockdown of survivin expression by siRNA
represses growth and induces apoptosis of colorectal cancer cells
in vitro
To further confirm the potential relationship
between miR-16 and the downstream target survivin, we knocked down
the expression of survivin in CRC cells by transfection with siRNA
against survivin (Fig. 5A) and then
examined the effect on the proliferation and apoptosis of the
transfected cells. Flow cytometry of Annexin V-stained CRC cells
indicated that survivin downregulation by siRNA caused a marked
increase in the percentage of apoptotic cells (Fig. 5B). CCK-8 assays further showed that
survivin downregulation was associated with markedly impaired
growth of HCT116 and LoVo cells (Fig.
5C and D), which is consistent with the phenotype of CRC cells
overexpressing miR-16.
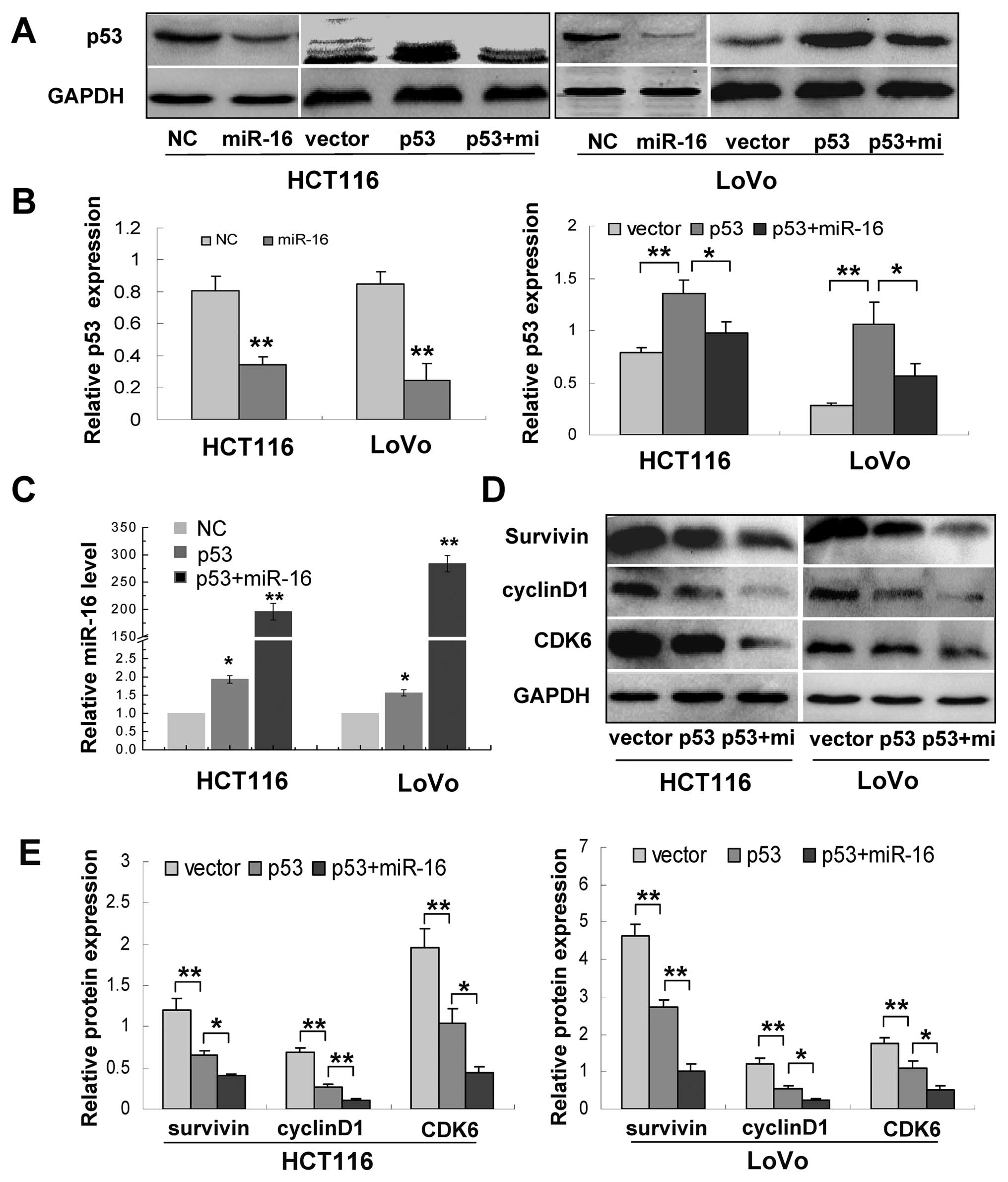
miR-16 regulates the p53/survivin
signaling pathway
It has been reported that p53 is a target of miR-16,
and p53 regulates maturation of miR-16 (11,14).
We also showed here that survivin is a direct target of miR-16. We
subsequently performed assays to determine the association of p53,
miR-16 and survivin. We first examined p53 expression in cells
overexpressing miR-16 and found that miR-16 overexpression
noticeably inhibited p53 levels in both HCT116 and LoVo cells, and
cells cotransfected with the miR-16 mimics and p53 repressed p53
expression compared to cells transfected with p53 alone (Fig. 6A and B). Then, we transfected HCT116
and LoVo cells with plasmids expressing wild-type p53. Our PCR
assays showed that, compared with cells transfected with the
control vector, p53 overexpression caused a 2- and 1.5-fold
increase in the levels of miR-16 in HCT116 and LoVo cells,
respectively (Fig. 6A–C).
Meanwhile, the levels of several miR-16 targets cyclin D1, CDK6 and
survivin were also reduced in cells treated with the wild-type p53
plasmid, whereas miR-16 and p53 cotransfection exerted stronger
inhibition of these targets (Fig. 6D
and E). Taken together, these results indicate that miR-16
regulates the p53/survivin signaling pathway.
Discussion
Deregulation of miR-16 has been demonstrated in
several tumors. miR-16 has been described as a tumor suppressor and
is consistently downregulated in CLL, non-small cell lung cancer
and pituitary adenomas (4,8,15).
However, the association between p53, miR-16 and survivin remains
largely unknown. Our results clearly showed that miR-16 was
downregulated in CRC tissues and was correlated with the degree of
histological differentiation, which was a stage-independent
prognostic factor in CRC patients (12). We further demonstrated that miR-16
inhibited proliferation and induced apoptosis of CRC cells through
the intrinsic apoptosis pathway. Survivin is undetectable in
normal, differentiated adult tissues, but is abundantly expressed
in cancer tissues (16). Shen et
al constructed an oncolytic adenovirus with a survivin-targeted
small hairpin and found that adenovirus mediated survivin knockdown
and suppressed human colorectal carcinoma growth in vitro
and in vivo(17). In the
present study, we found that survivin was a direct target of
miR-16. Moreover, miR-16 downregulated p53 expression while p53
upregulated miR-16 levels, suggesting the presence of a regulatory
loop between the two molecules. p53 also concomitantly inhibited
the expression of cyclin D1, CDK6 and survivin, indicating that p53
regulates cyclin D1, CDK6 and survivin through miR-16.
Survivin is a member of the inhibitor of apoptosis
(IAP) gene family, which was discovered in 1997. It is
implicated in multiple essential functions, including cell
division, checkpoint mechanisms of genomic integrity and apoptosis
(18). Survivin also plays an
important role in transition from adenoma with low dysplasia to
high dysplasia during human colorectal tumorigenesis (19). Our data indicated that miR-16
regulated survivin expression at both the mRNA and protein levels;
and miR-16 expression inhibited survivin expression and induced
apoptosis in CRC cells. Additionally, expression of miR-16 in
HCT116 cells repressed 3′-UTR reporter activity of vectors
expressing the wild-type miR-16 binding site in the 3′-UTR of
survivin, but not that of vectors expressing the mutated seed
sequence. These results confirmed that survivin was one of the
direct targets of miR-16.
Survivin expression was also reported to correlate
with the p53 status (20). Mutant
p53 increases while wild-type p53 represses the
expression of survivin (21).
Wild-type p53 transcriptionally repressed survivin by
binding to its promoter (22),
histone deacetylases and methylation of its promoter (23–25).
However, another study did not find that p53 could physically
associate with the survivin promoter (24). Studies have indicated that survivin
inhibited by p53 is not correlated with methylation of the survivin
promoter (25,26). Therefore, the mechanisms whereby p53
represses survivin are still unclear due to the complexity of its
regulation. In our study, we first showed that p53 repressed
survivin expression by upregulating miR-16 in CRC cells. The
relationship between p53 and miR-16 is similar to that observed in
this study where p53 upregulates miR-145 (27). In other words, p53 possibly
regulates survivin through the miR-16 pathway in CRC cells. Taken
together, miR-16 and their regulators comprise an intricate network
that could be intimately implicated in oncogenesis and tumor
progression. In summary, our results demonstrated that miR-16 was
downregulated and inversely related to histological differentiation
in CRC tissues, and overexpression of miR-16 inhibited
proliferation and induced apoptosis of CRC cells. Furthermore, we
showed that survivin was a direct target of miR-16 and miR-16
regulated the p53/survivin signaling pathway. Our data suggest that
miR-16 may be an important tumor suppressor in CRC cells.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
miR
|
microRNA
|
|
CLL
|
chronic lymphocytic leukemia
|
|
CCK-8
|
Cell Counting Kit-8
|
|
IAP
|
the inhibitor of apoptosis
|
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bandi N, Zbinden S, Gugger M, et al:
miR-15a and miR-16 are implicated in cell cycle regulation in a
Rb-dependent manner and are frequently deleted or down-regulated in
non-small cell lung cancer. Cancer Res. 69:5553–5559. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhattacharya R, Nicoloso M, Arvizo R, et
al: miR-15a and miR-16 control Bmi-1 expression in ovarian cancer.
Cancer Res. 69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bonci D, Coppola V, Musumeci M, et al: The
miR-15a-miR-16-1 cluster controls prostate cancer by targeting
multiple oncogenic activities. Nat Med. 14:1271–1277. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cimmino A, Calin GA, Fabbri M, et al:
miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsang WP and Kwok TT: Epigallocatechin
gallate up-regulation of miR-16 and induction of apoptosis in human
cancer cells. J Nutr Biochem. 21:140–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boominathan L: The tumor suppressors p53,
p63, and p73 are regulators of microRNA processing complex. PLoS
One. 5:e106152010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alexander D, Jhala N, Chatla C, et al:
High-grade tumor differentiation is an indicator of poor prognosis
in African Americans with colonic adenocarcinomas. Cancer.
103:2163–2170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng S, Cong S, Zhang X, et al:
MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation
and induces cell apoptosis in lung cancer cells. Nucleic Acids Res.
39:6669–6678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fabbri M, Bottoni A, Shimizu M, et al:
Association of a microRNA/TP53 feedback circuitry with pathogenesis
and outcome of B-cell chronic lymphocytic leukemia. JAMA.
305:59–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bottoni A, Piccin D, Tagliati F, Luchin A,
Zatelli MC and degli Uberti EC: miR-15a and miR-16-1
down-regulation in pituitary adenomas. J Cell Physiol. 204:280–285.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yip KW, Shi W, Pintilie M, et al:
Prognostic significance of the Epstein-Barr virus, p53, Bcl-2, and
survivin in nasopharyngeal cancer. Clin Cancer Res. 12:5726–5732.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen W, Wang CY, Wang XH and Fu ZX:
Oncolytic adenovirus mediated Survivin knockdown by RNA
interference suppresses human colorectal carcinoma growth in vitro
and in vivo. J Exp Clin Cancer Res. 28:812009. View Article : Google Scholar
|
|
18
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawasaki H, Toyoda M, Shinohara H, et al:
Expression of survivin correlates with apoptosis, proliferation,
and angiogenesis during human colorectal tumorigenesis. Cancer.
91:2026–2032. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lynch CJ and Milner J: Loss of one p53
allele results in four-fold reduction of p53 mRNA and protein: a
basis for p53 haplo-insufficiency. Oncogene. 25:3463–3470. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vegran F, Boidot R, Oudin C, Defrain C,
Rebucci M and Lizard-Nacol S: Association of p53 gene alterations
with the expression of antiapoptotic survivin splice variants in
breast cancer. Oncogene. 26:290–297. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hoffman WH, Biade S, Zilfou JT, Chen J and
Murphy M: Transcriptional repression of the anti-apoptotic survivin
gene by wild type p53. J Biol Chem. 277:3247–3257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murphy M, Ahn J, Walker KK, et al:
Transcriptional repression by wild-type p53 utilizes histone
deacetylases, mediated by interaction with mSin3a. Genes Dev.
13:2490–2501. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mirza A, McGuirk M, Hockenberry TN, et al:
Human survivin is negatively regulated by wild-type p53 and
participates in p53-dependent apoptotic pathway. Oncogene.
21:2613–2622. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raj D, Liu T, Samadashwily G, Li F and
Grossman D: Survivin repression by p53, Rb and E2F2 in normal human
melanocytes. Carcinogenesis. 29:194–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang X, Xiong G, Chen X, et al: Survivin
expression in esophageal cancer: correlation with p53 mutations and
promoter polymorphism. Dis Esophagus. 22:223–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suh SO, Chen Y, Zaman MS, et al:
MicroRNA-145 is regulated by DNA methylation and p53 gene mutation
in prostate cancer. Carcinogenesis. 32:772–778. 2011. View Article : Google Scholar : PubMed/NCBI
|