Introduction
Hepatocellular carcinoma (HCC) is a common
malignancy worldwide. Major risk factors include chronic infection
with hepatitis B virus (HBV) or hepatitis C virus (HCV) and
exposure to dietary aflatoxin B1 (1). It is estimated that HBV is responsible
for 50–80%, whereas HCV is associated with 10–25% of HCC cases
(2). HBV is the predominant cause
of HCC in most Asian, African and Latin American countries. By
contrast, HCV is more common than HBV in Europe, Japan and the USA
(3). HCV-associated HCC typically
develops 20–30 years after infection and is usually, but not
always, preceded by cirrhosis (4).
Previous studies have demonstrated that multiple
genetic and epigenetic changes are involved in the molecular
pathogenesis of HCC, including somatic mutations in the p53 tumor
suppressor gene (TP53), which has been reported with a rate
of 14–35% worldwide, depending on the level of aflatoxin exposure
(5,6). TP53 mutations are frequently
observed in HCC cases with HBV or HCV infection (4,7,8). The
identification of the interactions between p53 and virus proteins
is highly significant for therapeutic strategies aimed at reducing
the chronicity and/or carcinogenicity of the virus. However, the
association between TP53 mutations and virus carried state
in the pathogenesis of HCC has yet to be fully elucidated.
The most common missense mutations in human cancer
are known as hotspot mutations. More complex mutations such as
insertion/deletion/nonsense are less frequently described.
Microindels are unique, very rare mutations that result in inserted
and deleted sequences of different sizes (between one and 50
nucleotides) at the same nucleotide position, with relevance to
evolution and the onset of cancer (9). Little is known about the mechanisms
responsible for these mutations (10). As a tumor suppressor gene frequently
mutated in almost any tumor type, TP53 microindels have not
been reported as much in tumor.
In the present study, we identified and analyzed a
novel somatic TP53 microindel in a case of early stage HCC
with HCV but not HBV infection; we also examined the association
between TP53 mutations and HCV or HBV positivity in the
development of HCC with a panel of HCC cases from North China.
Materials and methods
Tissue samples
One hundred and sixteen cases (102 males, 14
females) of HCC excised surgically at Beijing Friendship Hospital,
Capital Medical University and Beijing Youan Hospital, Capital
Medical University between January 2005 and December 2010 were
examined in this study, including 13 HCV-positive cases, 93
HBV-positive cases, and 10 cases that were both HBV and HCV
negative. Three cases were both HBV and HCV positive. The mean age
of the patients was 52.8±9.1 years (range, 22–81 years).
The tumors were immediately frozen and stored at
−80°C or fixed in buffered formalin and embedded in paraffin.
Written informed consent was obtained from all patients for use of
their clinical materials in research. The study protocol was
approved by the Clinical Research Ethics Committee of the Beijing
Friendship Hospital, Capital Medical University.
PCR-SSCP analysis followed by direct
sequencing for TP53 mutations
Genomic DNA was extracted using an EZ DNA FFPE kit™
(Omega) or tissue DNA kit™ (DP304-02, Tiangen, Beijing) according
to the manufacturer's instructions. Prescreening for mutations in
exon 5 to 8 of the TP53 gene by single-strand conformational
polymorphism (SSCP) analysis followed by direct sequencing was
conducted as previously described (11). Samples exhibiting mobility shifts on
SSCP analysis were subsequently re-amplified using the same primers
as for SSCP and sequenced using a BigDye Terminator Cycle
Sequencing kit (ABI PRISM; Applied Biosystems) in an ABI PRISM 3100
DNA sequencer (Applied Biosystems). The identified mutations were
verified by sequencing a second product of amplification on both
strands.
Immunohistochemical assay for expression
of p53, p21WAF/CIP, Bax and Mdm2
Sections (4 μm) were cut for immunohistochemistry
(IHC). Immunohistochemical staining was performed using antibodies
against p53 (monoclonal, sc-126; Santa Cruz Biotechnology, Santa
Cruz, CA, USA; 1:200), p21WAF/CIP (monoclonal, BD556431;
BD Biosciences; 1:200), Mdm2 (monoclonal, Batch 3012; Novocastra;
1:100) and Bax (polyclonal, 06-499; Upstate Biotechnology, Inc.;
1:200) at 4°C overnight. Secondary antibody (anti-mouse IgG, cat.
no. MP-7402; Vector Laboratories) was used at 37°C for 1 h and
antigen-antibody reactions were visualized with
3,3′-diaminobenzidine (SK-4100; Vector Laboratories). Tissue
structures were visualized by counterstaining with hematoxylin.
Statistical analysis
Fisher's exact test (for expected value, ≤5) was
used to identify molecular associations using SAS v9.2 software
(SAS Institute, Inc., Cary, NC, USA). A P-value of <0.05 was
considered to indicate a statistically significant difference in
all tests.
Results
Identification and analysis of TP53
mutations in HCC
One hundred and sixteen cases of HCC were examined
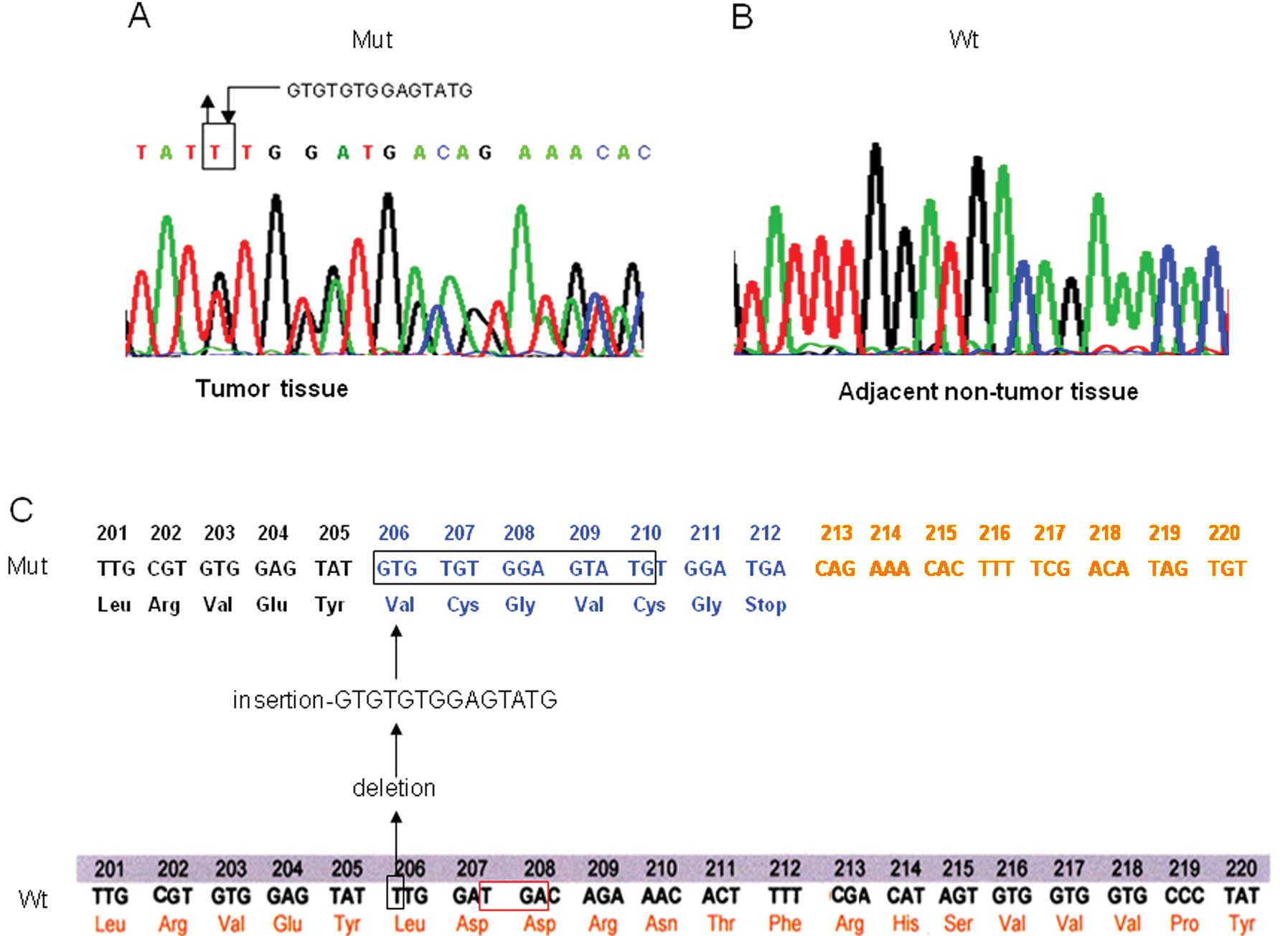
for TP53 mutations. In a case with HCV infection (case 360),
a novel heterozygous mutation, 616ins14del1 (14-1 microindel), was
identified in exon 6 of the TP53 gene; in this mutation, one
base (T) was deleted followed by insertion of a 14 base nucleotide,
GTGTGTGGAGTATG, at locus 616 (Fig.
1A). This TP53 microindel was not observed in DNA from
adjacent non-tumor tissue, indicating that it was a tumor-specific
somatic mutation (Fig. 1B). The
mutation led to a frameshift of TP53 mRNA starting from
locus 616 (codon 206) and generated a stop codon (TGA) at locus 621
(codons 207–208, corresponding to codon 212 in the mutant
sequence), resulting in the generation of a truncated p53 protein
with 211 amino acids (Fig. 1C).
Of the 116 HCC cases analyzed, 19 (16.4%) contained
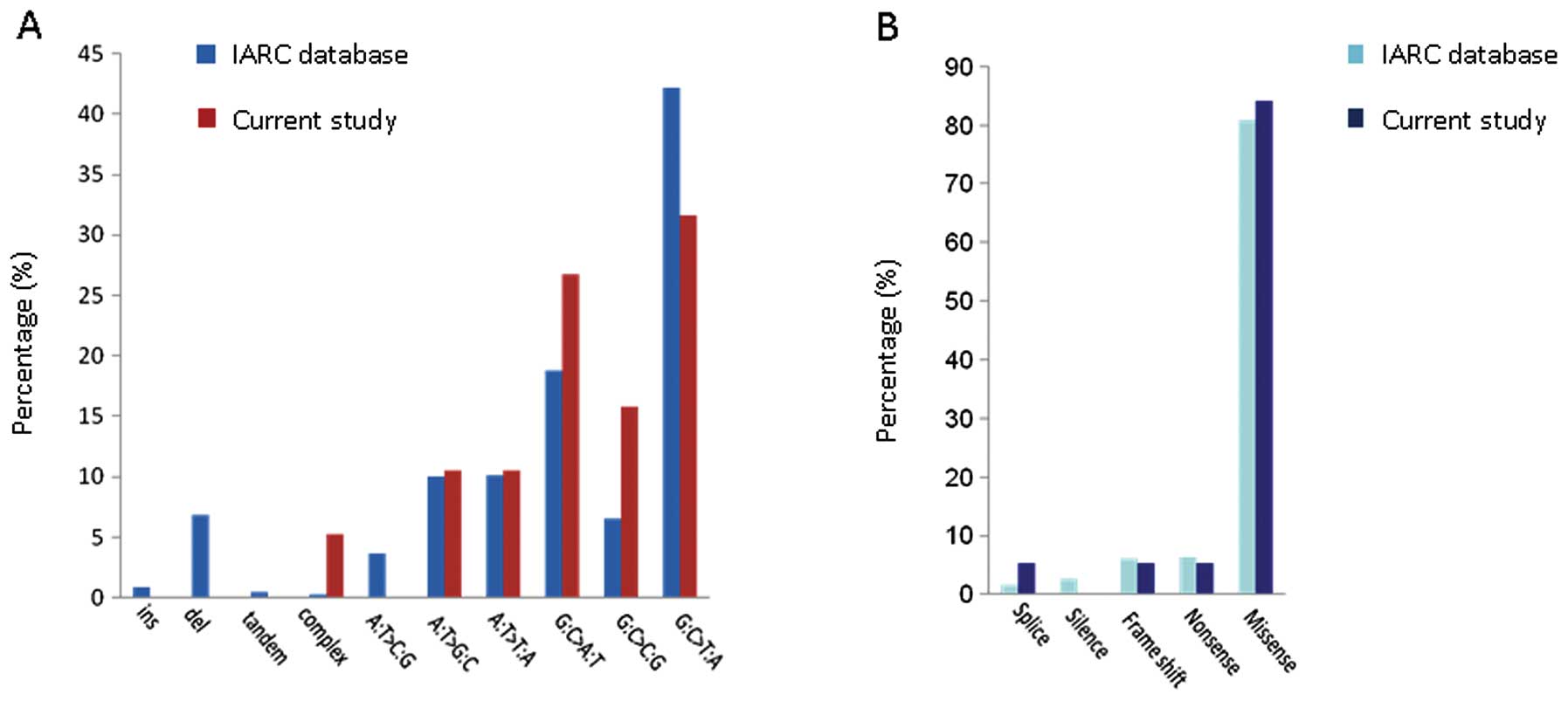
a TP53 mutation (Table I).
Aside from the TP53 14-1 microindel, 18 transition mutations
in the TP53 gene which have been reported in the
International Agency for Research on Cancer (IARC) database (R15
release, http://www-p53.iarc.fr) were also
identified, including 16 missense point mutations, one nonsense
mutation and one splicing mutation. The pattern and effect of
TP53 mutations in HCC identified in the present study are
similar as those reported in the IARC database (Fig. 2).
 | Table IMutations in the TP53 gene
identified in HCC cases. |
Table I
Mutations in the TP53 gene
identified in HCC cases.
| Case | Exon | Codon | Nucleotide
changes | Amino acid
substitution |
|---|
| 1 | 5 | 157 | GTC→TTC | Val→Phe |
| 2 | 5 | 157 | GTC→TTC | Val→Phe |
| 3 | 5 | 175 | CGC→CAC | Arg→His |
| 4 | 6 | 220 | TAT→TGT | Tyr→Cys |
| 5 | 6 | 192 | CAG→CAT | Gln→His |
| 6 | 6 | 206 |
616ins14del1 |
Frameshift |
| 7 | | | IVS6+19G→C | Splicing
mutation |
| 8 | 7 | 245 | GGC→GAC | Gly→Asp |
| 9 | 7 | 248 | CGG→GGG | Arg→Gly |
| 10 | 7 | 248 | CGG→GGG | Arg→Gly |
| 11 | 7 | 248 | CGG→GGG | Arg→Gly |
| 12 | 7 | 249 | AGG→AGT | Arg→Ser |
| 13 | 8 | 301 | CCA→CTA | Pro→Leu |
| 14 | 8 | 298 | CAG→TAG | Gln→Stop |
| 15 | 8 | 285 | GAG→AAG | Glu→Lys |
| 16 | 8 | 285 | GAG→AAG | Glu→Lys |
| 17 | 8 | 273 | CGT→TGT | Arg→Cys |
| 18 | 8 | 273 | CGT→TGT | Arg→Cys |
| 19 | 8 | 286 | GAA→GTA | Arg→Val |
All TP53 mutations were identified in the HCC
cases with HBV or HCV infection. Of the 13 HCV-positive HCC cases,
5 (38.5%) contained a TP53 mutation, i.e. the 14-1
microindel in case 360, R249S in case 212, E285K in case 421, R248G
in case 522 and P301L in case 568, and there was a significant
association between TP53 mutations and HCV positivity
(Table II). By contrast, 15 of the
93 HBV-positive HCC cases contained a TP53 mutation (16.1%),
and there was no significant association between TP53
mutations and HBV positivity (Table
II). In one case with both HCV and HBV positivity, a
TP53 mutation E285K was identified.
 | Table IICorrelation of TP53 mutations
with HBV and/or HCV positivity in hepatocellular carcinoma.a |
Table II
Correlation of TP53 mutations
with HBV and/or HCV positivity in hepatocellular carcinoma.a
| TP53
mutation | |
|---|
|
| |
|---|
| Virus carried
state | Yes | No | Significance
(P-value) |
|---|
| HBV |
| Positive | 15 | 78 | 1.0000 |
| Negative | 4 | 19 | |
| HCV |
| Positive | 5 | 8 | 0.0379 |
| Negative | 14 | 89 | |
| HBV or HCV |
| Positive | 19 | 87 | 0.2116 |
| Negative | 0 | 10 | |
| HBV and HCV |
| Positive | 1 | 2 | 0.4183 |
| Negative | 18 | 95 | |
Functional consequences of the novel TP53
microindel in HCC
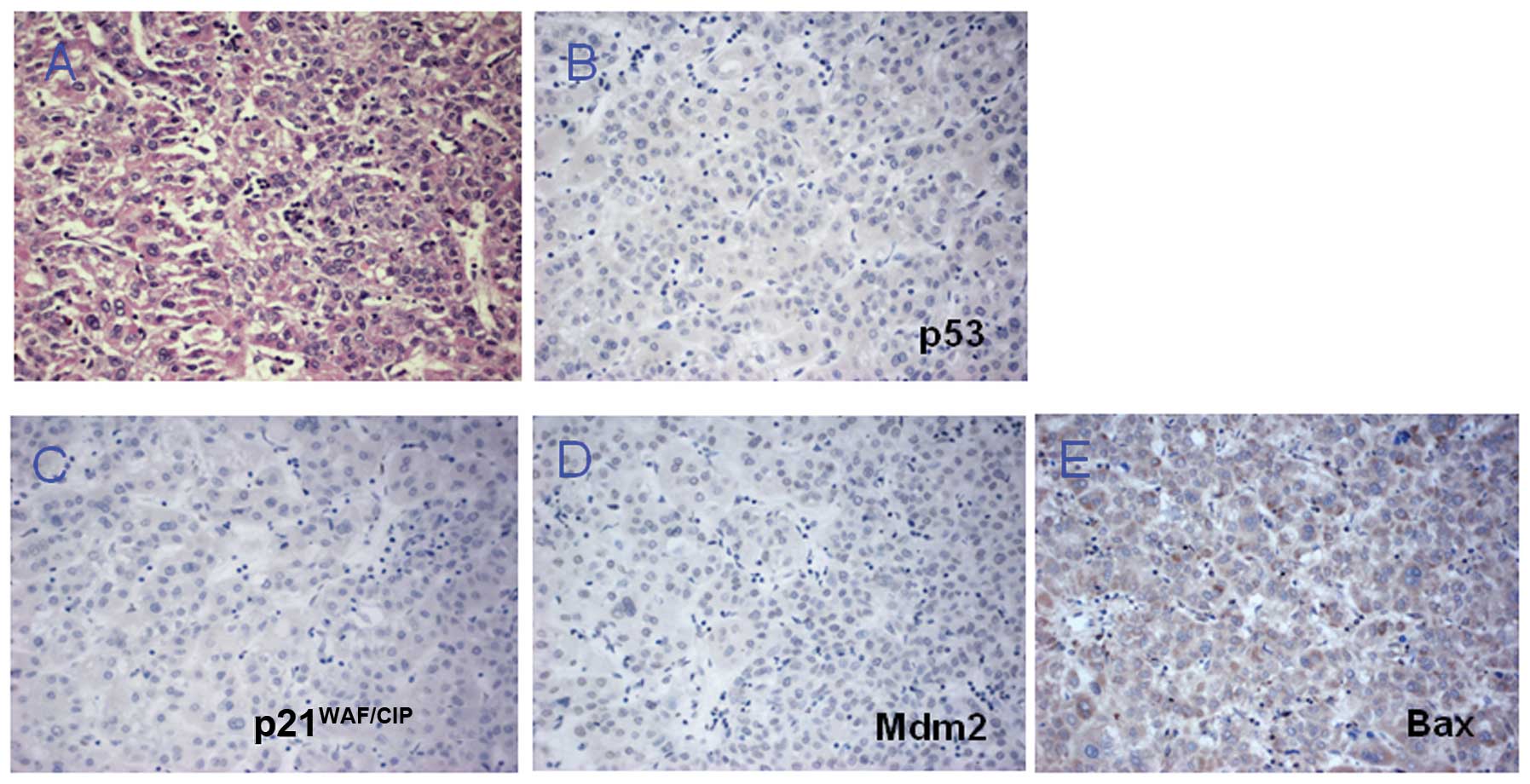
To evaluate the functional consequences of the
TP53 microindel, IHC was conducted to determine the
expression of the p53 protein, as well as its downstream targets
p21WAF/CIP, Bax and Mdm2 in the HCC case. The results
showed no positive staining for p53 (Fig. 3A and B) and negative staining for
p21WAF/CIP, Mdm2 and Bax in tumor cells (Fig. 3C-E). These IHC results suggest that
loss of p53 activity may result in the downregulation of
p21WAF/CIP, Mdm2 and Bax, which are crucial for
inhibition of the cell cycle and for inducing apoptosis. Thus, the
truncated p53 protein caused by the TP53 microindel may
result in loss function of normal p53, leading to loss of control
of the cell cycle and apoptosis.
Discussion
Germline mutations in the TP53 gene have been
identified in patients with Li-Fraumeni syndrome (LFS), which is an
inherited cancer predisposition syndrome characterized by a wide
spectrum of neoplasms (12).
Somatic TP53 mutations were identified in almost all tumor
types including HCC, particularly following exposure to aflatoxin
(7,13). According to the IARC database, 1020
TP53 mutations have been identified in 31.3% of HCC cases.
These mutations comprise 933 base transitions (91.5%), nine
insertions (0.88%), 70 deletions (6.86%), five tandems (0.49%), and
three complex mutations [0.29%; an 8.5-kb DNA rearrangement in a
human hepatoma cell line (Hep3B) identified by restriction fragment
analysis (14), a 248–289dup
(A83-S96dup) in an HCC case from France (15), and a TGAAAAC-AG replacement (exon 3)
in a case from Hong Kong (8); no
detailed clinical data or information on the consequences of the
mutations is provided by any of these studies]. In the present
study, all patients came from North China (regions with low
aflatoxin exposure), and TP53 mutation was identified in
16.4% of total HCC cases, lower than the average rate (31.3%)
reported in the IARC TP53 database. Notably, a special type
of complex TP53 mutation, 616ins14del1 (14-1 microindel),
was identified in HCC with HCV infection in the population of North
China.
Microindels in coding regions often lead to a
frameshift, with devastating consequences for protein function.
Meanwhile, microindels can also be in-frame and thereby alter the
properties of a protein by adding or subtracting a small number of
amino acids (protein tinkering). Protein tinkering can sometimes be
a critical step in carcinogenesis (such as EGFR microindels in lung
cancer, KIT mutation in gastrointestinal stromal tumors) (9,16). To
date, a total of 66 TP53 somatic microindels have been
reported in the context of other mutations in the IARC database.
The majority (79%) of these microindels result in a reading frame
shift; the others are in-frame, resulting in protein tinkering
(17). The novel TP53
microindel identified in the present study brings into frame a
termination codon at the equivalent of codon 207–208, with an
altered C-terminus from codon 206-ter. A truncated and presumably
inactivated product is predicted, lacking a number of key domains
including part of the DNA-binding domain and the tetramerization
domain. We did not examine the loss of heterozygosity (LOH) of
TP53 in the specific HCC case. However, IHC confirmed that
the truncated p53 protein caused by the TP53 microindel may
result in loss function of normal p53, leading to loss of control
of the cell cycle and apoptosis. The loss of p53 protein expression
could be explained by the truncated protein or by damage to the
TP53 mRNA through a nonsense-mediated digestion pathway due
to premature termination of transcription (18).
The patient with the TP53 microindel was a
67-year-old male diagnosed with HCC (T1N0M0, Union for
International Cancer Control standard) following physical
examination. The patient underwent surgical resection two weeks
after diagnosis. The characteristics of the HCC were as follows: i)
location, left lobe of liver, baseline computed tomography scan
revealed no distant dissemination of cancer; ii) size, 4×4×3.8 cm,
with an intact capsule; iii) serology, HCV(+), hepatitis B surface
antigen (−), HBV DNA (−); and iv) histology, moderate
differentiation. The patient had no specific exposure to dietary
aflatoxin and did not consume alcohol excessively. There was no
family history of tumor, inherited disease, or infectious diseases.
According to the above clinical data, the HCC case with the
TP53 microindel was at an early stage, and HCV but not HBV
infection was present; there were no other specific risk factors
for HCC. Since the novel TP53 microindel resulted in the
disruption of the TP53 signaling pathway, it may act as a
driver mutation and contribute to the development of HCC.
Although it is evident that HCV infection may
contribute to tumor initiation and development, the direct
molecular role for HCV in the pathogenesis of HCC and the molecular
processes causing HCC remain unclear (19). To gain insight into HCV-related
hepatocarcinogenesis, microarray analysis has been applied in
several studies (7,20,21).
Analysis of HCV-associated cirrhosis revealed an upregulation of
pro-inflammatory, pro-apoptotic and pro-proliferative genes, which
might reflect groups of genes involved in HCV-related cirrhosis
progressing to HCC (20). Notably,
microarray analysis of numerous TP53 mutant and TP53
wild-type HCC cases showed significant differences in their gene
expression patterns. Cell cycle-related genes (CCNG2 and
BZAP45) and cell proliferation-related genes (SSR1,
ANXA2, S100A10 and PTMA) were overexpressed in
mutant TP53 tumors compared with wild-type TP53
tumors (21). This observation
indicates a higher potential for malignancy in HCV-related HCC with
a TP53 mutation. In addition, it has been postulated that
the HCV protein might cause mutation of the TP53 gene, but
this is controversial (22,23). The above observation suggested a
significant role of co-presence of TP53 mutation and HCV
infection in the pathogenesis of HCC, consistent with the findings
in the present study.
As previously described, HBV is the predominant
cause of HCC in most Asian countries (3), and the majority of the cases analyzed
in the present study were HBV positive (80.2%). Some evidence
supports a direct oncogenic role for HBV in the development of HCC,
i.e. the integration of HBV DNA into the chromosomal DNA of HCC;
the role of the HBV X gene in the pathogenesis of HBV-associated
HCC, in particular, binding to and inactivation of p53 (24), and a recent whole-genome study
showed the role of interferon regulatory factor 2 (IRF2) as a tumor
suppressor in HBV-associated HCC and its function as a regulator of
the p53 pathway (25). However, the
present study did not reveal any relationship between TP53
mutations and HBV infection in HCC in the Chinese population,
consistent with a recent study which showed no correlation between
mutational change in TP53 and the number of HBV integration
events in a Chinese population (26).
Collectively, the present study shows the
co-presence of TP53 mutations and HCV infection but not HBV
infection in HCC in the small subset of HCCs from North China,
suggesting different carcinogenetic pathways between HBV- and
HCV-related hepatocarcinogenesis in relation to disruption of p53.
A large scale deep study is required to fully understand the
molecular role of p53 in virus infection-related
hepatocarcinogenesis.
Acknowledgements
This study was partly supported by a grant from the
National Natural Science Foundation of China (no. 81071973) and a
grant from the Scientific Research Foundation for Returned Overseas
Chinese Scholars, Bureau of Human Resources and Social Security of
Beijing, China (Key project, 2010). The authors thank Dr Magali
Olivier (IARC TP53 Database Manager, Group of Molecular
Mechanisms and Biomarkers, Lyon, France) for the helpful
information on the study.
References
|
1
|
Thorgeirsson SS and Grisham TW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatology. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Raza SA, Clifford GM and Franceschi S:
Worldwide variation in the relative importance of hepatitis B and
hepatitis C viruses in hepatocellular carcinoma: a systematic
review. Br J Cancer. 96:1127–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang TJ and Heller T: Pathogenesis of
hepatitis C-associated hepatocellular carcinoma. Gastroenterology.
127:S62–S71. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mínguez B, Tovar V, Chiang D, Villanueva A
and Llovet J: Pathogenesis of hepatocellular carcinoma and
molecular therapies. Curr Opin Gastroenterol. 25:186–194.
2009.PubMed/NCBI
|
|
6
|
Ozturk M: p53 mutation in hepatocellular
carcinoma after aflatoxin exposure. Lancet. 338:1356–1359. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hussain SP, Schwank J, Staib F, Wang XW
and Harris CC: TP53 mutations and hepatocellular carcinoma:
insights into the etiology and pathogenesis of liver cancer.
Oncogene. 26:2166–2176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen GG, Merchant JL, Lai PBS, et al:
Mutation of p53 in recurrent hepatocellular carcinoma and its
association with the expression of ZBP-89. Am J Pathol.
162:1823–1829. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gonzalez KD, Hill KA, Li K, et al: Somatic
microindels: analysis in mouse soma and comparison with the human
germline. Hum Mutat. 28:69–80. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hill KA, Gonzalez KD, Scaringe WA, Wang JC
and Sommer SS: Preferential occurrence of 1–2 microindels. Hum
Mutat. 27:55–61. 2006.
|
|
11
|
Huang J, Grotzer MA, Watanabe T, Watanabe
T, Hewer E, Pietsch T, Rutkowski S and Ohgaki H: Mutations in the
Nijmegen breakage syndrome gene in medulloblastomas. Clin Cancer
Res. 14:4053–4058. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malkin D, Li FP, Strong LC, et al: Germ
line p53 mutations in a familial syndrome of breast cancer,
sarcomas, and other neoplasms. Science. 250:1233–1238. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bressac B, Katherine M, Galvin T, Liang J,
Isselbacher KJ, Wands JR and Ozturk M: Abnormal structure and
expression of p53 gene in human hepatocellular carcinoma. Proc Natl
Acad Sci USA. 87:1973–1977. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boyault S, Rickman DS, Reynies AD, et al:
Transcriptome classification of HCC is related to gene alterations
and to new therapeutic targets. Hepatology. 45:42–52. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wardelmann E, Merkelbach-Bruse S, Pauls K,
et al: Polyclonal evolution of multiple secondary KIT mutations in
gastrointestinal stromal tumors under treatment with imatinib
mesylate. Clin Cancer Res. 12:1743–1749. 2006. View Article : Google Scholar
|
|
17
|
Scaringe WA, Li K, Gu DQ, Gonzalez KD,
Chen ZB, Hill KA and Sommer SS: Somatic microindels in human
cancer: the insertions are highly error-prone and derive from
nearby but not adjacent sense and antisense templates. Hum Mol
Genet. 17:2910–2918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noensie EN and Dietz HC: A strategy for
disease gene identification through nonsense mediated mRNA decay
inhibition. Nat Biotechnol. 19:434–439. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bouchard MJ and Navas-Martin S: Hepatitis
B and C virus hepatocarcinogenesis: lessons learned and future
challenge. Cancer Lett. 305:123–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bassiouny AE, Nosseir MM, Zoheiry MK, et
al: Differential expression of cell cycle regulators in
HCV-infection and related hepatocellular carcinoma. World J
Hepatol. 2:32–41. 2010.PubMed/NCBI
|
|
21
|
Okada T, Iizuka N, Yamada-Okabe H, et al:
Gene expression profile linked to p53 status in hepatitis C
virus-related hepatocellular carcinoma. FEBS Lett. 555:583–590.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Machida K, Cheng KT, Sung VM, et al:
Hepatitis C virus induces a mutator phenotype: enhanced mutations
of immunoglobulin and proto-oncogenes. Proc Natl Acad Sci USA.
101:4262–4267. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Anzola M and Burgos JJ: Hepatocellular
carcinoma: molecular interactions between hepatitis C virus and p53
in hepatocarcinogenesis. Expert Rev Mol Med. 5:1–16. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang MH, Chen CJ, Lai MS, et al:
Universal hepatitis B vaccination in Taiwan and the incidence of
hepatocellular carcinoma in children. Taiwan Childhood Hepatoma
Study Group. N Engl J Med. 336:1855–1859. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guichard C, Amaddeo G, Imbeaud S, et al:
Integrated analysis of somatic mutations and focal copy-number
changes identifies key genes and pathways in hepatocellular
carcinoma. Nat Genet. 44:694–698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang S, Yang Z, Li W, et al:
Re-evaluation of the carcinogenic significance of hepatitis B virus
integration in hepatocarcinogenesis. Plos One. 7:e403632012.
View Article : Google Scholar : PubMed/NCBI
|