Introduction
Mucin 1 (MUC1) is a transmembrane glycoprotein that
is expressed in most epithelial cells and is aberrantly
overexpressed in many types of human adenocarcinomas and
hematologic malignancies (1). MUC1
consists of a large extracellular N-terminal subunit and a
C-terminal subunit that resides on the cell surface as a
heterodimeric complex via strong noncovalent interactions. The
N-terminal subunit consists of a variable number of 20-amino acid
tandem repeats (VNTR) that comprise the majority of the
extracellular domain. The C-terminal subunit is composed of a
58-amino acid extracellular domain, a 28-amino acid transmembrane
domain (TM) and a 72-amino acid cytoplasmic tail (CT) (2–4). The
MUC1-CT is highly conserved among different species and possesses 7
tyrosine residues that can be potentially phosphorylated by
multiple kinases (5–8); this region also associates with
certain transcription factors (9–11).
Available evidence indicates that the MUC1-CT is involved in many
signaling pathways, including the Wnt/β-catenin (5,12,13),
p53 (9,11) and NF-κB (14) pathways. β-catenin, a major effector
of the Wnt signaling pathway, interacts with the MUC1-CT at an
SXXXXXSSLS site. This interaction blocks GSK3β-induced degradation
of β-catenin and promotes the translocation of β-catenin to the
nucleus, where it forms complexes with the LEF/TCF (lymphoid
enhancer factor/T cell factor) transcription factors and activates
transcription of Wnt-responsive genes such as cyclin D1 and c-Myc
to regulate cell proliferation (15,16).
Previous studies have shown that MUC1 plays a role
in a diverse array of cellular processes including differentiation
(17) motility or inhibition of
cell-cell and cell-matrix adhesion (18–21)
and immune regulation (22).
Recently, the majority of studies have shown that the MUC1-CT
contributes to malignant transformation as an oncoprotein.
Overexpression of the MUC1-CT in 3Y1 fibroblasts induced cellular
transformation and promoted tumor formation in nude mice (23). Mutation of the MUC1-CT (Y46F, Y60F)
abrogated MUC1-induced anchorage-independent growth and
tumorigenicity in human colon carcinoma cells (24,25).
Inhibition of the MUC1-CT induced cancer cell death and tumor
regression (26,27). The deletion of MUC1 expression from
MMTV-Wnt-1 transgenic mice resulted in a significant increase in
the time to mammary gland tumor onset (28). However, several studies have shown
an inverse association between MUC1 and cell proliferation and
adhesion. For example, Hattrup and Gendler (29) and Costa et al(30) demonstrated that MUC1 downregulation
in human BT20 breast carcinoma cells or human gastric carcinoma
MKN45 cells increased proliferation. Considering these
contradictory findings, the role of MUC1 in cancer progression has
yet to be clarified.
In this study, we investigated the effects and
related mechanisms of MUC1 on cancer-related characteristics of B16
cells by stable expression of the human full-length MUC1 in B16
cells. We found that MUC1 expression in B16 cells inhibited cell
proliferation, decreased cell migration and invasion, and
suppressed tumor growth in BALB/c nude mice. These results suggest
that the modulatory effects of MUC1 in tumor cells may be more
complex than previously appreciated, which reinforces the
importance of understanding alternative regulatory mechanisms of
MUC1.
Materials and methods
Cell line, plasmids and animals
The B16 cell line was purchased from ATCC (Manassas,
VA, USA) and cultured in Iscove’s modified Dulbecco’s medium (IMDM)
supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and
10% fetal bovine serum (FBS) (Gibco-BRL, Carlsbad, CA, USA) in a
humidified atmosphere of 5% CO2 at 37°C. The pcDNA3-MUC1
plasmid, which contains the full-length human MUC1 consisting of 22
TR, was a gift from Dr O.J. Finn of the University of Pittsburgh
(Pittsburgh, PA, USA). BALB/c nude mice (4–6 weeks old) were
purchased from Vital River Laboratories (Vital River, China).
Animals were maintained in specific pathogen-free conditions and
were fed sterile water and food ad libitum. All animals were
treated in accordance with the National Institutes of Health Guide
for the Care and Use of Laboratory Animals and with the approval of
the Scientific Investigation Board of Science and Technology of
Jilin Province.
Cell transfection
B16 cells growing in an exponential phase were
seeded in a 6-well plate. When cells reached 80–90% confluence, 1.0
μg of pcDNA3-MUC1 plasmid was transfected with Lipofectamine™ 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s protocol. Two stable MUC1-positive clones (B16-MUC1
9–12, B16-MUC1 9–23) were selected in 1,000 μg/ml G418 (Gibco-BRL),
and the concentration was decreased to 600 μg/ml to maintain
filtrate efficacy. Meanwhile, a negative control B16-neo cell line
was prepared by transfecting the pcDNA3 empty vector into the B16
cells.
Reverse transcription-polymerase chain
reaction (RT-PCR)
MUC1, cyclin D1 and c-Myc mRNA levels were analyzed
by RT-PCR. Total RNA was extracted from cells using Trizol reagent
(Invitrogen Life Technologies). Total RNA was converted to cDNA
using M-MLV reverse transcriptase and Oligo(dT) primers (Promega
Corporation, Madison, WI, USA) according to the manufacturer’s
protocol. The reverse transcribed products were used to amplify
MUC1, cyclin D1 and c-Myc by PCR using Ex-Taq DNA polymerase
(Takara Bio, Inc., Shiga, Japan), and β-actin was used as an
internal control gene. The primer sequences and reaction parameters
are shown in Table I. Amplified
products were analyzed on a 1.5% agarose gel, and DNA was
visualized by a Gel Image System (Tanon). The final value was
expressed as a ratio of the relative density of the target gene to
β-actin from three independent experiments (means ± SD).
 | Table IPrimer sequences and reaction
parameters used for PCR analysis. |
Table I
Primer sequences and reaction
parameters used for PCR analysis.
| Gene name | Primers (5′-3′,
forward and reverse) | Annealing
temperature (°C) | Cycles | Product size
(bp) |
|---|
| Human MUC1 | Forward:
5′-TGAGTGATGTGCCATTTCC-3′ | | | |
| Reverse:
5′-CTGCCCGTAGTTCTTTCG-3′ | 56 | 30 | 158 |
| Mouse β-actin | Forward:
5′-TGTCCACCTTCCAGCAGATGT-3′ | | | |
| Reverse:
5′-AGCTCAGTAACAGTCCGCCTAG-3′ | 54 | 25 | 101 |
| Mouse cyclin
D1 | Forward:
5′-AGCAGAAGTGCGAAGAGG-3′ | | | |
| Reverse:
5′-GCAGTCAAGGGAATGGTC-3′ | 52 | 30 | 154 |
| Mouse c-Myc | Forward:
5′-AAGGGAAGACGATGACGG-3′ | | | |
| Reverse:
5′-TGAGAAACCGCTCCACATA-3′ | 52 | 40 | 172 |
Flow cytometry
To analyze MUC1 expression of stable transfectants,
cells (1×106) were fixed with paraformaldehyde for 1 h
and washed twice with fluorescence-activated cell sorter (FACS)
solution (PBS containing 2% FCS and 0.1% NaN3).
Subsequently, the cells were incubated with a mouse monoclonal
antibody against MUC1 tandem repeats (HMPV; BD Biosciences,
Franklin Lakes, NJ, USA) on ice for 30 min, washed twice with FACS
solution and stained with fluorescein isothiocyanate
(FITC)-conjugated goat anti-mouse secondary antibody (Proteintech
Group, Chicago, IL, USA) at a dilution of 1:100 for 30 min on ice
in the dark. After washing twice with FACS solution, the expression
of MUC1 was analyzed by flow cytometry (FACSCalibur; BD
Biosciences).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and
permeabilized with 0.2% Triton X-100. After blocking with 2% bovine
serum albumin (BSA), the cells were incubated with a mouse
anti-MUC1 monoclonal antibody (GP1.4, NeoMarkers) overnight at 4°C.
After washing, PE-conjugated goat anti-mouse IgG (Proteintech
Group) was added for 1 h at 37°C in the dark. The nuclei were
stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI)
(Sigma-Aldrich, St. Louis, MO, USA) for 10 min, and the cells were
visualized using an inverted fluorescence microscope (Olympus,
IX71). Tumors were fixed in 10% neutral-buffered formalin and
embedded in paraffin. Serial paraffin sections (5-μm) were cut and
mounted on slides for immunofluorescence staining. Sections were
treated with 1.5% rabbit serum at 37°C for 30 min. Following that,
the sections were incubated with primary antibody GP1.4 and
PE-conjugated goat anti-mouse IgG as described above. The nuclei
were stained with DAPI and the sections were visualized by inverted
fluorescence microscopy.
Cell proliferation assay
Cell viability was determined using a WST-1 cell
proliferation assay according to the manufacturer’s protocol (Roche
Diagnostics, Mannheim, Germany). Briefly, cells
(5×103/well) were seeded in triplicate in 96-well plates
and cultured at 37°C with 5% CO2 in a humidified
atmosphere for 96 h. WST-1 reagent was added at 24, 48, 72 or 96 h,
and incubation was continued for an additional 1–2 h. Then, the
absorbance was measured using a microplate reader at a wavelength
of 450 nm (BioTek Instruments, Inc., Winooski, VT, USA). The
resulting values were calculated as a ratio of B16-MUC1 to B16-neo
and were the average from three independent experiments (means ±
SD).
Cell cycle analysis
Cells (1×106) were harvested and then
permeabilized with 70% ice-cold ethanol on ice for 30 min. Cells
were then washed and incubated in staining buffer with 50 μg/ml
propidium iodide (PI), 10 μg/ml RNase A and 0.1% Triton X-100 for
30 min in the dark. Subsequently, the cell cycle was analyzed by
flow cytometry (FACSCalibur; BD Biosciences).
Cell migration and invasion assay
Cell migration and Matrigel invasion assays were
performed using Transwell chambers with 8-μm pore size filters
(Corning Incorporated, Corning, NY, USA) coated with or without
Matrigel matrix (BD Biosciences) in a 24-well plate. In each well,
6×104 cells were seeded to the upper chamber in 200 μl
IMDM containing 1% FBS, and 600 μl IMDM containing 10% FBS was
added to the bottom chamber as a chemoattractant. The cells were
incubated at 37°C in a 5% CO2 atmosphere and allowed to
migrate or invade for 36 h. Following the incubation period, the
remaining cells in the upper chamber were removed gently with a
cotton swab. The cells on the lower surface of the chamber were
fixed with methanol for 20 min, and then stained with 1% crystal
violet in 20% methanol for 30 min. The cells that had migrated or
invaded through the filters were counted in five random fields
under a microscope.
Coimmunoprecipitation analysis
B16-MUC1 cells were lysed with RIPA lysis buffer
containing 50 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% sodium
deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 0.1 mM
ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethyl sulfonyl
fluoride (PMSF), 0.23 U/ml aprotinin, and 10 μM leupeptin
(Sigma-Aldrich). Protein concentrations were measured using a BCA
protein assay kit (Beyotime Biotechnology, Jiangsu, China). Equal
protein aliquots were subjected to immunoprecipitation with 1.0 μg
of mouse IgG or anti-MUC1-CT antibody (Ab-5; Neomarker) for 16 h at
4°C followed by precipitation with Protein G agarose beads (Promega
Corporation). Immunoprecipitated proteins and total cell lysates
were resolved by 10% sodium dodecyl sulphate-polyacrylamide gel
electrophoresis (SDS-PAGE) and subjected to immunoblot analysis
with anti-β-catenin (1:1000; BD Biosciences) for 16 h at 4°C.
Following incubation, the reactivity was detected with horseradish
peroxidase-conjugated secondary antibodies (1:2000; Sigma-Aldrich)
and ECL reagents (GE Healthcare).
Luciferase reporter assay
Cells were seeded in 6-well plates. When cells
reached 90% confluence, 1.0 μg of TOPflash and FOPflash plasmids
(Upstate Biotechnology, Inc., Lake Placid, NY, USA) were
transiently transfected with Lipofectamine™ according to the
manufacturer’s instructions. To normalize the transfection
efficiency, the cells were cotransfected with 0.05 μg of pRL-TK
(Promega Corporation). Forty eight hours post-transfection, the
luciferase assay was performed with the Dual Luciferase Assay
System kit (Promega Corporation). Relative luciferase activity was
calculated as the fold induction after normalization for
transfection efficiency.
Western blot analysis
Cells were lysed with RIPA lysis buffer as described
above. Nuclear and cytoplasmic protein extracts were isolated using
a cytoplasmic and nuclear protein extraction kit (Thermo
Scientific) according to the manufacturer’s protocol. Protein
concentrations were measured using a BCA protein assay kit
(Beyotime Biotechnology). Equal amounts of protein were separated
by 10% SDS-PAGE and transferred to PVDF membranes (Millipore,
Billerica, MA, USA), and the membranes were blocked in 3% BSA
overnight at 4°C. The membranes were then incubated with primary
antibodies against MUC1 (GP1.4) (1:2000), c-Myc (1:1000), cyclin D1
(1:1000; both from Epitomics, Burlingame, CA, USA), β-catenin
(1:1000; BD Transdution Labs) or E-cadherin (1:800, Proteintech)
for 2 h at room temperature, with the antibodies against β-actin
(1:2000), IκBα (1:2000) and Lamin B1 (1:2000, all from Epitomics)
as loading controls. Then, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies (1:2000;
Sigma-Aldrich) for another 2 h at room temperature. The membranes
were developed using ECL reagents (GE Healthcare). Each experiment
was repeated at least 3 times.
In vivo tumor growth assays
To determine the effects of MUC1 expression on
tumorigenesis in vivo, BALB/c female nude mice (4–6 weeks
old) were used to establish a subcutaneous transplant tumor model.
Mice were randomly divided into 2 groups (5 animals/group) that
were designated as the B16-neo group and the B16-MUC1 group.
B16-MUC1 cells or B16-neo cells (2×106) were
subcutaneously injected into the right flank of each mouse. Tumor
size was measured by calipers every 2 days. On day 12 post
injection, the tumors were removed and weighed.
Statistical analysis
All statistical analyses were performed using
unpaired Student’s t-tests, and P<0.05 was considered to
indicate a statistically significant result.
Results
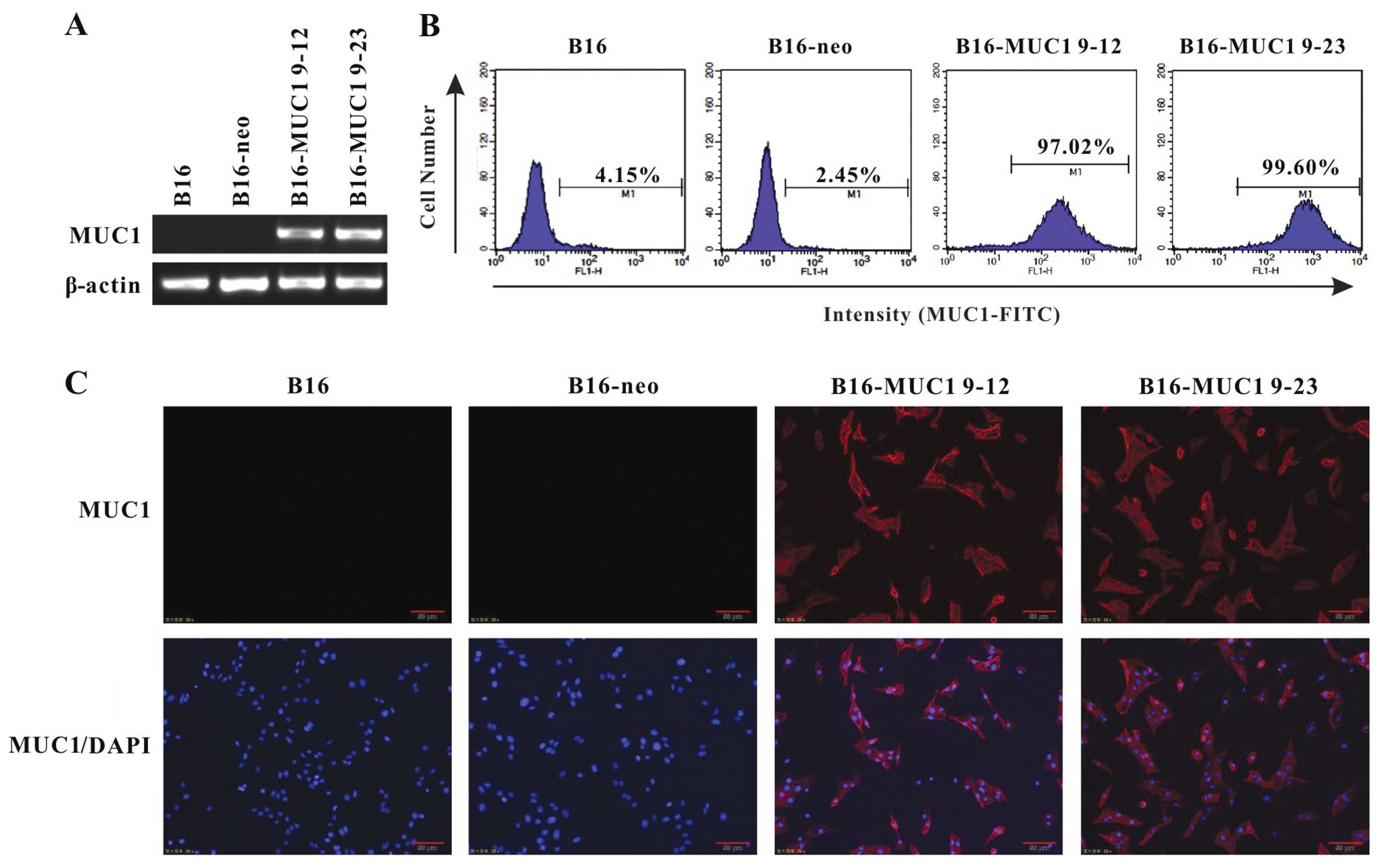
Stable MUC1 expression in mouse melanoma
B16 cells
To assess the effects of MUC1 expression, mouse B16
cells were transfected with a vector encoding human full-length
MUC1 containing 22 TR (pcDNA3-MUC1) or an empty pcDNA3 vector.
Stable transfectants were selected with G418 (1,000 μg/ml) and
analyzed for MUC1 expression by RT-PCR, flow cytometry and
immunofluorescence. Two MUC1-positive clones (B16-MUC1 9–12 and
B16-MUC1 9–23) expressed higher MUC1 mRNA levels and were selected
for further study. By contrast, there was no detectable MUC1
expression in the B16-neo cells transfected with the empty vector
(Fig. 1A). Flow cytometry (Fig. 1B) and immunofluorescence staining
(Fig. 1C) with anti-MUC1 tandem
repeat peptide antibodies (HMPV and GP1.4) verified that MUC1 was
expressed on the cell surface of 97.0 and 99.0% of B16-MUC1 9–12
and B16-MUC1 9–23 cells, respectively.
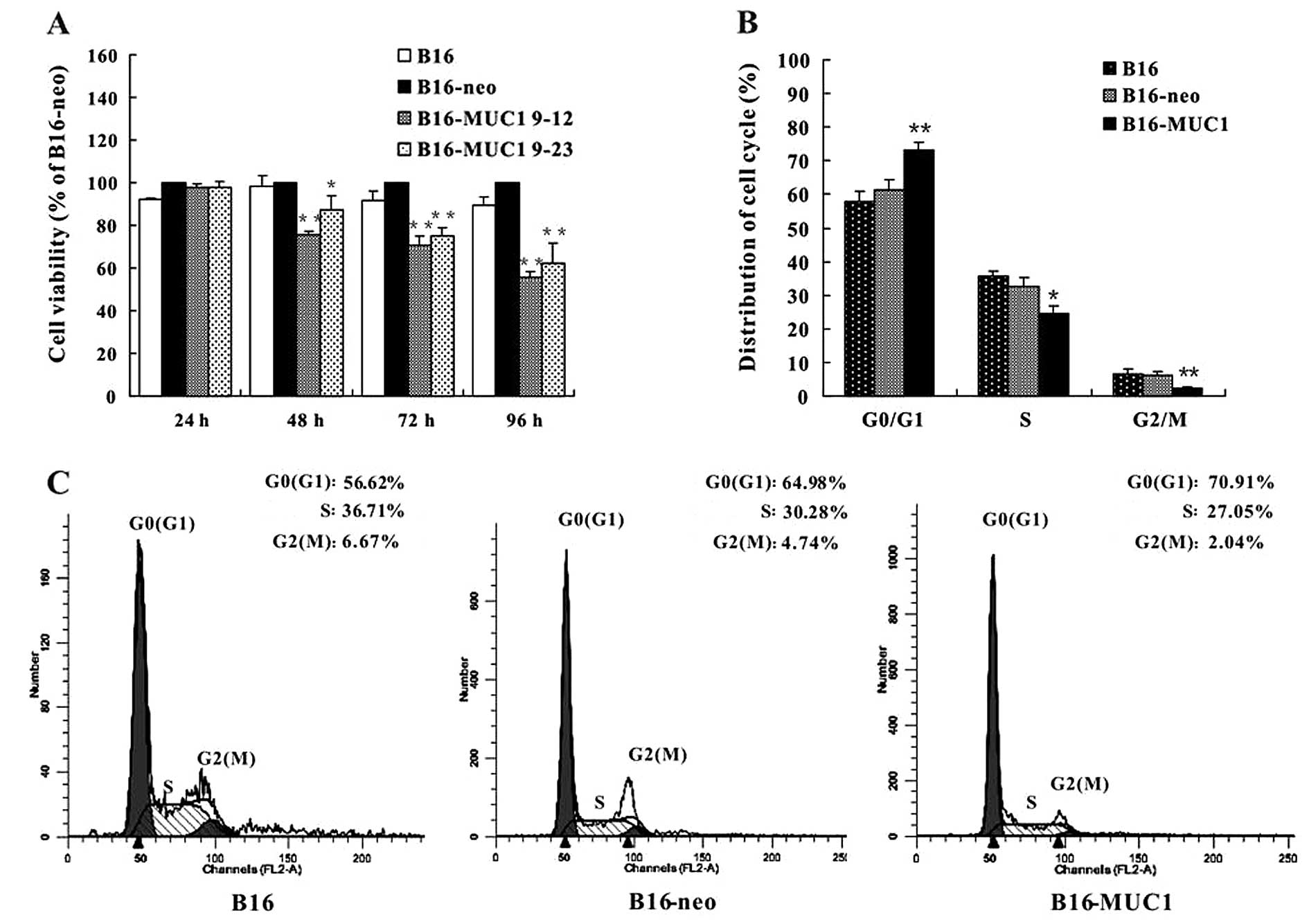
MUC1 expression inhibits cell
proliferation and induces G1-phase arrest in vitro
To determine the effect of MUC1 expression on cell
growth in vitro, equal numbers of B16-MUC1, B16-neo and B16
cells were seeded in 96-well plates and cultured for 96 h in a
humidified atmosphere of 5% CO2 at 37°C. Cell viability
was evaluated by the WST-1 assay. The viability of B16-MUC1 cells
was significantly reduced in a time-dependent manner when compared
with the viability of the B16-neo or B16 cells (P<0.01)
(Fig. 2A). We further analyzed the
cell cycle of B16-MUC1, B16-neo and B16 cells by flow cytometry.
The results showed that B16-MUC1 cells had a higher percentage of
cells in the G0/G1 phase (73.22±2.13%) and fewer in the G2/M phase
(2.32±0.39%) when compared with B16-neo cells (61.2±3.34% in G0/G1)
(6.07±1.26% in G2/M) or B16 cells (57.68±3.41% in G0/G1) (6.39±1.7%
in G2/M) (Fig. 2B and C). These
results indicate that MUC1 expression in B16 cells inhibited cell
proliferation and induced cell cycle arrest at the G1 phase.
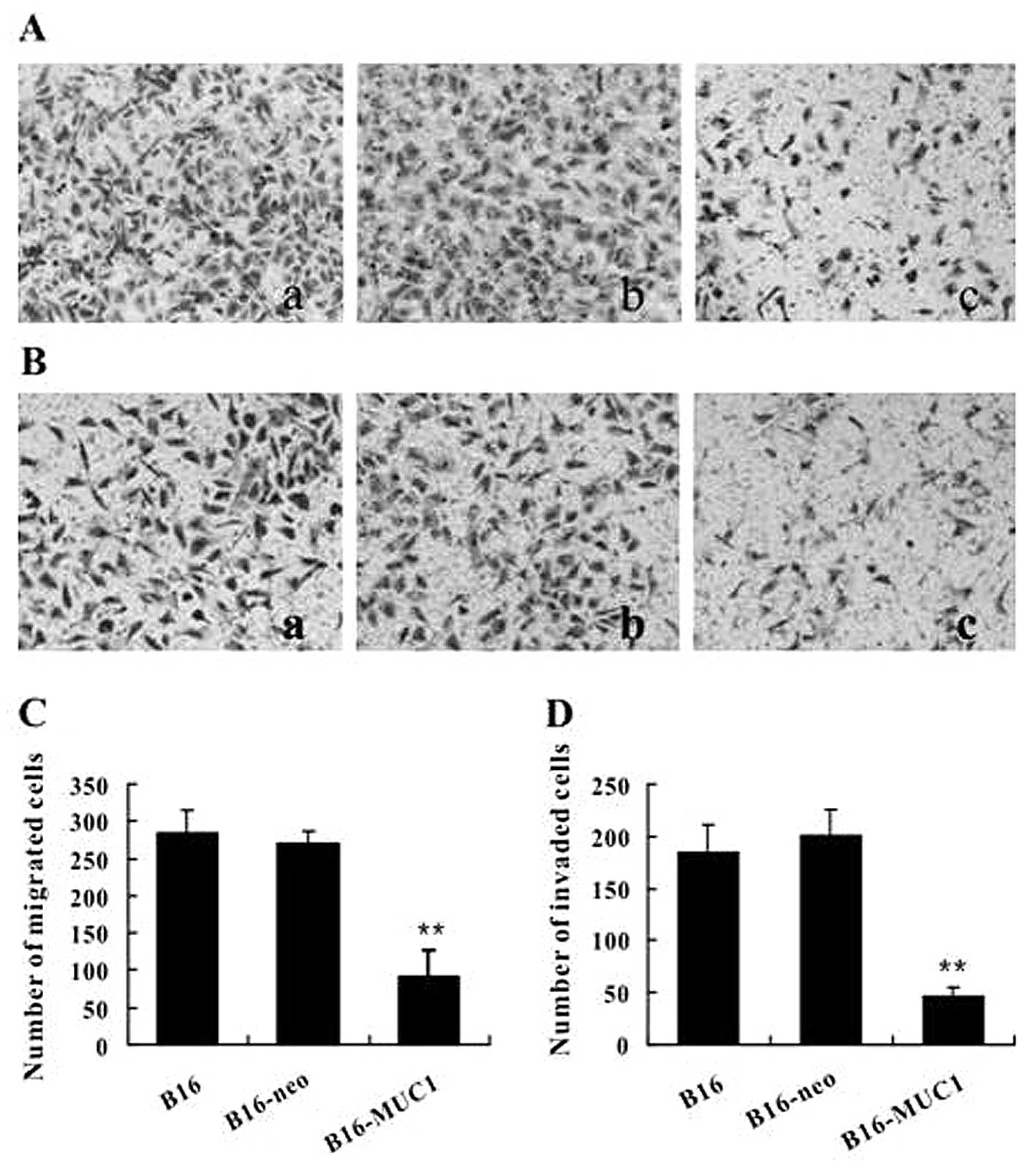
MUC1 expression inhibits cell migration
and invasion in vitro
To investigate whether MUC1 expression affects the
motility of B16 cells, the migratory capacity of cells was
evaluated using the Transwell migration assay. The results showed
that the number of B16-MUC1 cells that migrated into the lower
chamber was significantly decreased when compared to the number of
migrating B16-neo or B16 cells (Fig. 3A
and B) (P<0.01). We further performed a Matrigel invasion
assay to qualitatively observe the effect of MUC1 expression on the
invasive potential of cells. The results showed that the number of
B16-MUC1 cells that invaded through the Matrigel-coated membrane
was also significantly less than the number of invading B16-neo or
B16 cells (Fig. 3C and D)
(P<0.01). These results show that MUC1 expression inhibited cell
migration and invasion in vitro.
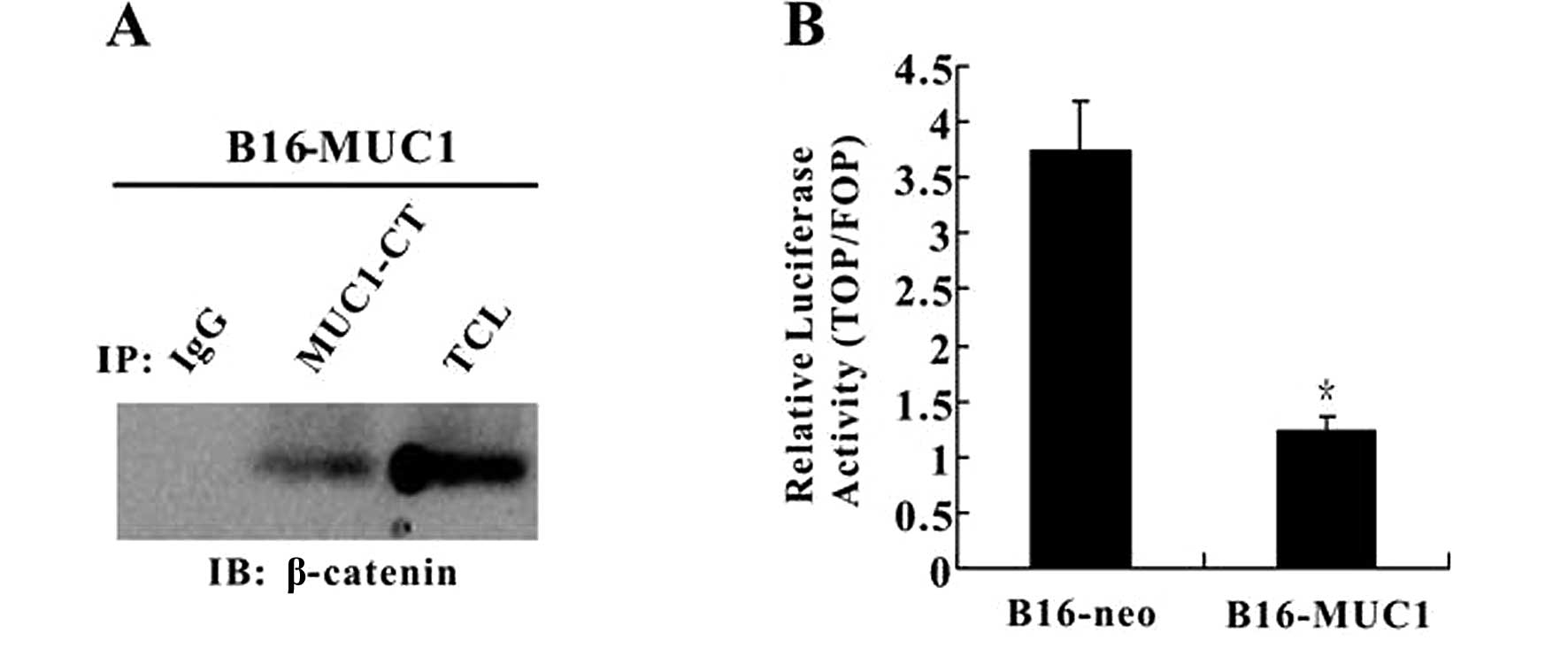
MUC1-CT interacts with β-catenin and
reduces the activity of T cell factor (TCF)
Numerous reports have confirmed that MUC1 binds to
β-catenin and is involved in the β-catenin signaling pathway.
Therefore, we performed coimmunoprecipitation to investigate
whether or not the interaction between MUC1 and β-catenin was also
observed in B16-MUC1 cells. Immunoprecipitation of MUC1 from
B16-MUC1 cell lysates using anti-MUC1-CT antibody (Ab-5) followed
by immunoblot analysis using anti-β-catenin antibody revealed a
protein band that co-migrated with β-catenin in total cell lysates;
no β-catenin band was detected in the control immunoprecipitates
with IgG (Fig. 4A). The results
showed that MUC1-CT binds directly to β-catenin in B16-MUC1 cells.
Since the Wnt pathway is known to be involved in tumor cell
proliferation, to determine the effect of the interaction between
the MUC1-CT and β-catenin on the activation of Wnt signaling, a
luciferase reporter assay was performed. The results showed that
Topflash/Fopflash reporter activity in B16-MUC1 cells was lower
than that in the B16-neo cells (P<0.05) (Fig. 4B). The result indicates that the
interaction between MUC1-CT and β-catenin reduces the activity of
TCF in B16-MUC1 cells when compared with that in the B16-neo
cells.
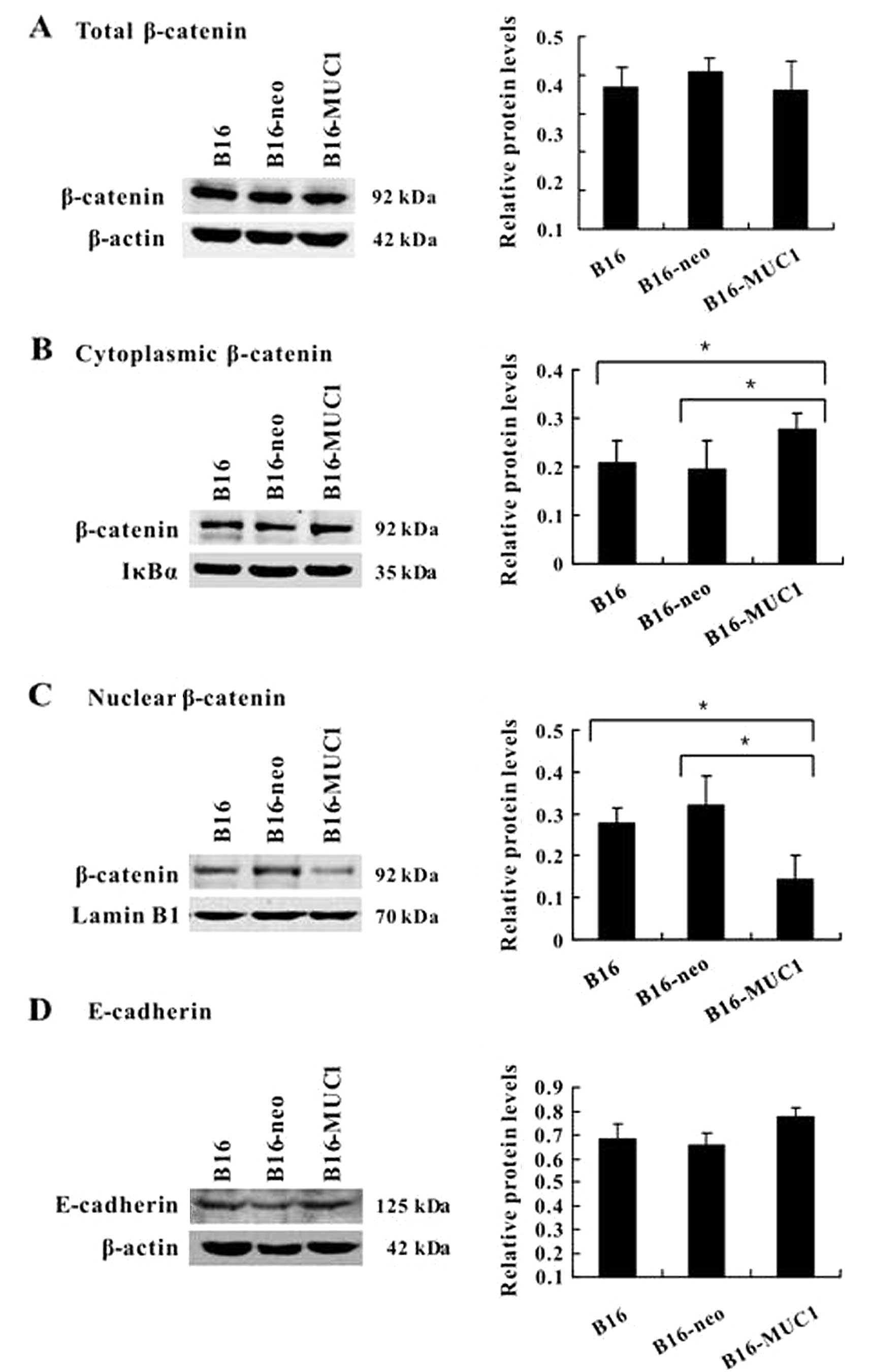
MUC1 expression blocks β-catenin
translocation to the nucleus
Since β-catenin is involved in MUC1 signal
transduction, to evaluate the effect of MUC1 expression on
β-catenin subcellular localization, equivalent protein aliquots of
total cell lysates or purified nuclear or cytosolic fractions from
cells were immunoblotted with the β-catenin antibody. Immunoblot
analysis demonstrated that the total β-catenin levels were
unchanged (Fig. 5A), the
cytoplasmic β-catenin levels were increased (Fig. 5B) (P<0.05) and the nuclear
β-catenin levels were reduced (Fig.
5C) (P<0.05) in B16-MUC1 cells compared to B16-neo or B16
cells. The results indicate that MUC1 expression blocks the
translocation of β-catenin to the nucleus. We also analyzed the
level of E-cadherin, a molecular chaperone of β-catenin that plays
an important role in cell adhesion. The immunoblot results show
that E-cadherin expression was slightly upregulated in the
MUC1-transfected B16 cells when compared with that of the negative
control cells, although there was no statistical significance
(Fig. 5D).
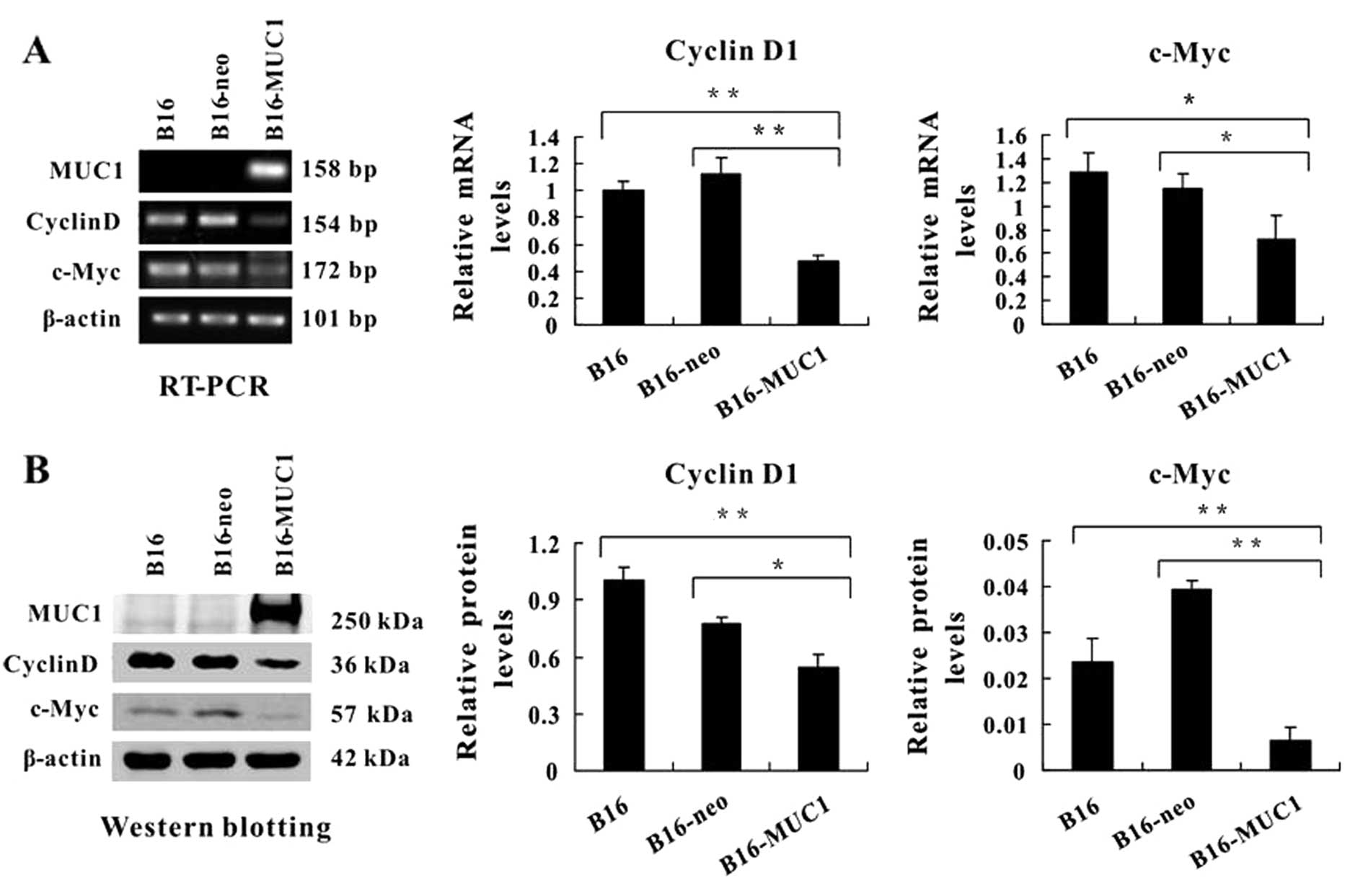
MUC1 expression downregulates both cyclin
D1 and c-Myc
Nuclear translocation of β-catenin can activate
cyclin D1 and c-Myc expression and stimulate cell proliferation.
Our results demonstrated that MUC1 expression reduced levels of
nuclear β-catenin and inhibited cell proliferation. Therefore, we
carried out RT-PCR and western blotting to detect the expression of
cyclin D1 and c-Myc. The PCR results showed that mRNA levels of
cyclin D1 and c-Myc were significantly decreased in the B16-MUC1
cells when compared with levels in the B16 or B16-neo cells
(P<0.01 and P<0.05, respectively; Fig. 6A). Immunoblot analysis showed
similar results (Fig. 6B). These
findings indicate that MUC1 expression downregulated the levels of
cyclin D1 and c-Myc.
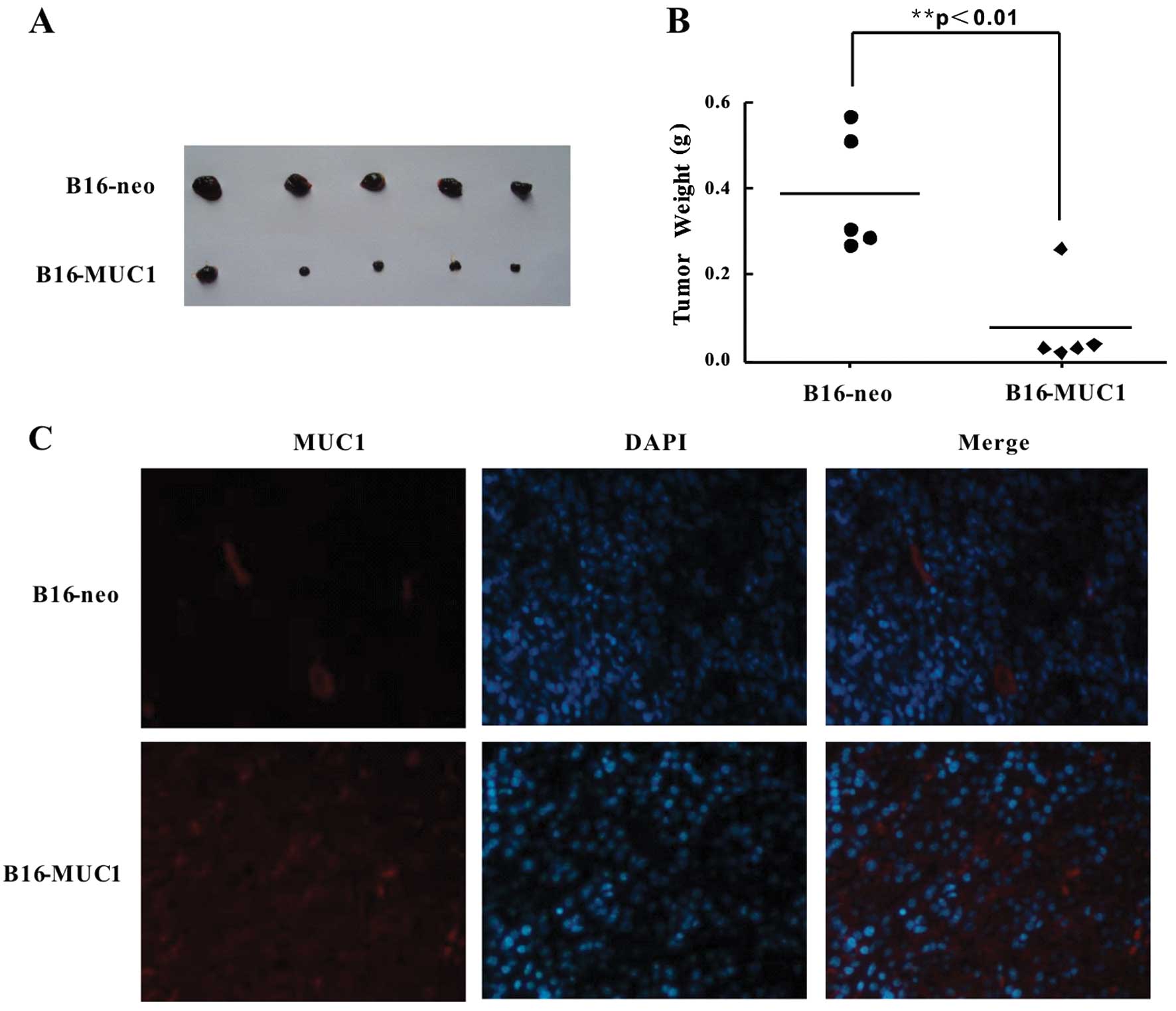
MUC1 expression suppresses tumor growth
in vivo
To evaluate the effects of MUC1 expression on
tumorigenesis in vivo, B16-MUC1 and B16-neo cells
(2×106) were inoculated subcutaneously into BALB/c nude
mice to establish a subcutaneous transplant tumor model. Tumor
growth was monitored for 12 days, and the B16-MUC1 tumors grew more
slowly than the B16-neo tumors. The B16-MUC1 tumors were
significantly smaller than the B16-neo tumors, and the average
weight of B16-MUC1 tumors (0.08±0.05 g) was significantly lower
than that of the B16-neo tumors (0.39±0.03 g) (Fig. 7A and B) (P<0.01). To determine
whether MUC1 was expressed in the tumors, immunofluorescence
staining was performed. The results showed strong positive staining
for MUC1 in the B16-MUC1 tumors, while no MUC1 expression was
detected in the B16-neo tumors (Fig.
7C). These results indicate that MUC1 expression in B16 cells
significantly suppressed tumor growth in a BALB/c nude mouse
transplant tumor model.
Discussion
In the present study, we investigated the effects of
MUC1 on malignancy behavior both in vitro and in vivo
by stable expression of human full-length MUC1 in the B16 mouse
melanoma cell line. We established two MUC1-positive clones,
B16-MUC1 9–12 and B16-MUC1 9–23, and one empty vector control
clone, B16-neo. These cells were characterized in vitro for
MUC1 expression, cell proliferation, cell cycle distribution,
migration and invasion and evaluated in vivo for the effects
of MUC1 expression on tumor growth in a mouse transplant tumor
model.
We found that MUC1 expression in B16 cells
significantly inhibited cell proliferation and induced cell cycle
arrest. These results conflict with most previous reports showing
that MUC1 is an oncogene (31–33).
However, this is not the only report demonstrating that MUC1
expression is associated with inhibited cell proliferation; several
published studies have shown similar results (29,30).
We also found that migration and invasion of B16-MUC1 cells were
significantly decreased compared to B16 and B16-neo cells, opposing
previous findings that MUC1 overexpression is associated with
increased cell migration and invasion in breast, lung and
pancreatic carcinoma cell lines (34,35).
Several published studies have shown that the
MUC1-CT can interact with β-catenin to form a complex that
contributes to tumorigenesis and tumor progression (36,37).
Our present study showed that expression of the human full-length
MUC1 in B16 cells increased the cytoplasmic levels of β-catenin,
but reduced nuclear translocation of β-catenin and decreased cell
proliferation. These results are similar to those described by
Lillehoj et al(38), who
showed that overexpression of MUC1 in HEK293T cells decreased the
nuclear levels of β-catenin and inhibited cell proliferation.
Cyclin D1 and c-Myc are two important transcriptional targets of
the Wnt/β-catenin pathway (39–41),
both of which are involved in regulating cell cycle progression and
promoting cellular proliferation and transformation. In our
studies, MUC1 expression in B16 cells decreased the levels of
nuclear β-catenin, reduced the activity of TCF, downregulated the
expression of cyclin D1 and c-Myc and arrested the cell cycle at G1
phase. These results may provide a possible mechanistic explanation
for how MUC1 expression decreased the proliferation of B16 cells
in vitro. E-cadherin is a cell adhesion molecule that forms
a complex with β-catenin and contributes to cell-cell adhesion
(42), thereby preventing cell
migration and invasion. Therefore, we examined the expression of
E-cadherin in B16-MUC1, B16-neo and B16 cells. The results showed
that E-cadherin expression was slightly increased in cells
expressing MUC1 compared with the control cells, although the data
did not reach statistical significance. These results suggest that
the inhibition of cell migration and invasion may be associated
with the upregulation of E-cadherin.
To investigate the effects of expressing human
full-length MUC1 in B16 cells on tumorigenesis and tumor
progression in vivo, a tumor growth assay was performed
using BALB/c nude mice. We observed a strong reduction in the
growth of B16-MUC1 tumors when compared with B16-neo tumors
(Fig. 7A and B). These results
agreed with the in vitro cell proliferation assays,
suggesting that the decreased growth of MUC1-expressing primary
tumors in nude mice is primarily due to decreased proliferative
activity of the cells themselves. Premaratne et al(43) demonstrated that MUC1 expression in
the prostate cancer cell line C4-2B4 had similar results. These
findings suggest that MUC1 expression in the two types of cell
lines displayed a negative effect on tumor growth, opposing most
previous reports that MUC1 acts as an oncoprotein. Currently, there
is no exact regulatory molecular mechanism to explain the
conflicting data generated in different laboratories. Hattrup and
Gendler (29) cautioned against
overgeneralization of the results from individual cell lines on
MUC1-mediated cancer progression since the functional regulation of
MUC1 in different cell lines may vary depending on diverse factors
such as cell type and signaling context.
In addition, it is frequently assumed that the
MUC1-CT functions as an oncogene, but the effect of the MUC1-N in
cell transformation and tumorigenesis is not yet clear. Several
reports show that the variable number tandem repeat
(VNTR)-containing extracellular domain of MUC1 regulates the
transcription of several genes (44), providing a new insight for
understanding the function of MUC1-N. In a study conducted by
Lillehoj et al(38), an
engineered variant of the MUC1-CT, CD8/MUC1, that lacks the
VNTR-containing extracellular domain was transfected into HEK293
cells resulting in decreased cell proliferation. In our study, we
obtained similar results. Although we transfected full-length human
MUC1 into B16 cells, it may merely be equivalent to transfection of
the mouse MUC1-CT, since the homology with the human protein is
only 34% in the extracellular tandem repeat domain, whereas it is
87% in the transmembrane and cytoplasmic domains (45). Moreover, the MUC1-CT is identical in
normal and tumor cells. Based on these findings, we propose that
the VNTR-containing extracellular domain of MUC1 may play an
important role in regulating the tumor-promoting effects in various
types of cancers, but further studies are needed.
In summary, we demonstrated that MUC1 expression in
B16 cells inhibited cell proliferation, migration and invasion and
suppressed tumor growth in a mouse transplant tumor model. These
results may be associated with several MUC1-related molecules of
the β-catenin signaling pathway. It is suggested that the
regulatory mechanisms of MUC1 as a oncoprotein are more complex
than previously appreciated, which reinforces the importance of
understanding alternative mechanisms that may regulate MUC1.
Acknowledgements
We would like to thank Dr O.J. Finn for the
pcDNA3-MUC1 plasmid, which was used to transfect the B16 cell line.
This study was supported by grants from the China National Natural
Science Foundation (no. 30972782) and the Major Development
Programs for New Drugs of the Chinese Academy of Sciences during
the 12th Five-Year Plan Period (no. 2011ZX09102-001-36).
References
|
1
|
Kufe DW: Mucins in cancer: function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gendler S, Taylor-Papadimitriou J, Duhig
T, Rothbard J and Burchel J: A highly immunogenic region of a human
polymorphic epithelial mucin expressed by carcinomas is made up of
tandem repeats. J Biol Chem. 263:12820–12823. 1988.PubMed/NCBI
|
|
3
|
Ligtenberg MJ, Kruijshaar L, Buijs F, van
Meijer M, Litvinov SV and Hilkens J: Cell-associated episialin is a
complex containing two proteins derived from a common precursor. J
Biol Chem. 267:6171–6177. 1992.PubMed/NCBI
|
|
4
|
Kufe DW: Targeting the human MUC1
oncoprotein: a tale of two proteins. Cancer Biol Ther. 7:81–84.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Bharti A, Chen D, Gong J and Kufe D:
Interaction of glycogen synthase kinase 3beta with the DF3/MUC1
carcinoma-associated antigen and beta-catenin. Mol Cell Biol.
18:7216–7224. 1998.PubMed/NCBI
|
|
6
|
Li Y, Kuwahara H, Ren J, Wen G and Kufe D:
The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1
carcinoma-associated antigen with GSK3 beta and beta-catenin. J
Biol Chem. 276:6061–6064. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren J, Li Y and Kufe D: Protein kinase C
delta regulates function of the DF3/MUC1 carcinoma antigen in
beta-catenin signaling. J Biol Chem. 277:17616–17622. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Raina D, Ahmad R, Kumar S, et al: MUC1
oncoprotein blocks nuclear targeting of c-Abl in the apoptotic
response to DNA damage. EMBO J. 25:3774–3783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei X, Xu H and Kufe D: Human MUC1
oncoprotein regulates p53-responsive gene transcription in the
genotoxic stress response. Cancer Cell. 7:167–178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei X, Xu H and Kufe D: MUC1 oncoprotein
stabilizes and activates estrogen receptor alpha. Mol Cell.
21:295–305. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei X, Xu H and Kufe D: Human mucin 1
oncoprotein represses transcription of the p53 tumor suppressor
gene. Cancer Res. 67:1853–1858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamamoto M, Bharti A, Li Y and Kufe D:
Interaction of the DF3/MUC1 breast carcinoma-associated antigen and
beta-catenin in cell adhesion. J Biol Chem. 272:12492–12494. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang L, Chen D, Liu D, Yin L, Kharbanda S
and Kufe D: MUC1 oncoprotein blocks glycogen synthase kinase
3beta-mediated phosphorylation and degradation of beta-catenin.
Cancer Res. 65:10413–10422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahmad R, Raina D, Trivedi V, et al: MUC1
oncoprotein activates the IkappaB kinase beta complex and
constitutive NF-kappaB signaling. Nat Cell Biol. 9:1419–1427. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu XF, Yang E, Li J and Xing PX: MUC1
cytoplasmic tail: a potential therapeutic target for ovarian
carcinoma. Expert Rev Anticancer Ther. 6:1261–1271. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kufe DW: Functional targeting of the MUC1
oncogene in human cancers. Cancer Biol Ther. 8:1197–1203. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shin CY, Park KH, Ryu BK, Choi EY, Kim KC
and Ko KH: Squamous differentiation downregulates Muc1 mucin in
hamster tracheal surface epithelial cell. Biochem Biophys Res
Commun. 271:641–646. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ligtenberg MJ, Buijs F, Vos HL and Hilkens
J: Suppression of cellular aggregation by high levels of episialin.
Cancer Res. 52:2318–2324. 1992.PubMed/NCBI
|
|
19
|
Wesseling J, van der Valk SW, Vos HL,
Sonnenberg A and Hilkens J: Episialin (MUC1) overexpression
inhibits integrin-mediated cell adhesion to extracellular matrix
components. J Cell Biol. 129:255–265. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Satoh S, Hinoda Y, Hayashi T, Burdick MD,
Imai K and Hollingsworth MA: Enhancement of metastatic properties
of pancreatic cancer cells by MUC1 gene encoding an anti-adhesion
molecule. Int J Cancer. 88:507–518. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Regimbald LH, Pilarski LM, Longenecker BM,
Reddish MA, Zimmermann G and Hugh JC: The breast mucin MUC1 as a
novel adhesion ligand for endothelial intracellular adhesion
molecule 1 in breast cancer. Cancer Res. 56:4244–4249.
1996.PubMed/NCBI
|
|
22
|
Agrawal B and Longenecker BM: MUC1
mucin-mediated regulation of human T cells. Int Immunol.
17:391–399. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Liu D, Chen D, Kharbanda S and Kufe
D: Human DF3/MUC1 carcinoma-associated protein functions as an
oncogene. Oncogene. 22:6107–6110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang L, Ren J, Chen D, Li Y, Kharbanda S
and Kufe D: MUC1 cytoplasmic domain coactivates Wnt target gene
transcription and confers transformation. Cancer Biol Ther.
2:702–706. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raina D, Ahmad R, Chen D, Kumar S,
Kharbanda S and Kufe D: MUC1 oncoprotein suppresses activation of
the ARF-MDM2-p53 pathway. Cancer Biol Ther. 7:1959–1967. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Ahmad R, Kosugi M, et al: Survival
of human multiple myeloma cells is dependent on MUC1 C-terminal
transmembrane subunit oncoprotein function. Mol Pharmacol.
78:166–174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Raina D, Kosugi M, Ahmad R, et al:
Dependence on the MUC1-C oncoprotein in non-small cell lung cancer
cells. Mol Cancer Ther. 10:806–816. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schroeder JA, Adriance MC, Thompson MC,
Camenisch TD and Gendler SJ: MUC1 alters beta-catenin-dependent
tumor formation and promotes cellular invasion. Oncogene.
22:1324–1332. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hattrup CL and Gendler SJ: MUC1 alters
oncogenic events and transcription in human breast cancer cells.
Breast Cancer Res. 8:R372006. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Costa NR, Paulo P, Caffrey T,
Hollingsworth MA and Santos-Silva F: Impact of MUC1 mucin
downregulation in the phenotypic characteristics of MKN45 gastric
carcinoma cell line. PLoS One. 6:e269702011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan Z, Liu X, Wong S, Machan JT and Chung
MA: MUC1 knockdown with RNA interference inhibits pancreatic cancer
growth. J Surg Res. 157:e39–e46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schroeder JA, Masri AA, Adriance MC, et
al: MUC1 overexpression results in mammary gland tumorigenesis and
prolonged alveolar differentiation. Oncogene. 23:5739–5747. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brossart P, Schneider A, Dill P, et al:
The epithelial tumor antigen MUC1 is expressed in hematological
malignancies and is recognized by MUC1-specific cytotoxic
T-lymphocytes. Cancer Res. 61:6846–6850. 2001.PubMed/NCBI
|
|
34
|
Yuan Z, Wong S, Borrelli A and Chung MA:
Down-regulation of MUC1 in cancer cells inhibits cell migration by
promoting E-cadherin/catenin complex formation. Biochem Biophys Res
Commun. 362:740–746. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao J, McConnell MJ, Yu B, et al: MUC1 is
a downstream target of STAT3 and regulates lung cancer cell
survival and invasion. Int J Oncol. 35:337–345. 2009.PubMed/NCBI
|
|
36
|
Kufe D: Oncogenic function of the MUC1
receptor subunit in gene regulation. Oncogene. 29:5663–5666. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Retterspitz MF, Mönig SP, Schreckenberg S,
et al: Expression of {beta}-catenin, MUC1 and c-met in diffuse-type
gastric carcinomas: correlations with tumour progression and
prognosis. Anticancer Res. 30:4635–4641. 2010.
|
|
38
|
Lillehoj EP, Lu W, Kiser T, Goldblum SE
and Kim KC: MUC1 inhibits cell proliferation by a
beta-catenin-dependent mechanism. Biochim Biophys Acta.
1773:1028–1038. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sutherland RL and Musgrove EA: Cyclin D1
and mammary carcinoma: new insights from transgenic mouse models.
Breast Cancer Res. 4:14–17. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lavergne E, Hendaoui I, Coulouarn C, et
al: Blocking Wnt signaling by SFRP-like molecules inhibits in vivo
cell proliferation and tumor growth in cells carrying active
β-catenin. Oncogene. 30:423–433. 2011.PubMed/NCBI
|
|
42
|
Wijnhoven BP, Dinjens WN and Pignatelli M:
E-cadherin-catenin cell-cell adhesion complex and human cancer. Br
J Surg. 87:992–1005. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Premaratne P, Welén K, Damber JE, Hansson
GC and Bäckström M: O-glycosylation of MUC1 mucin in
prostate cancer and the effects of its expression on tumor growth
in a prostate cancer xenograft model. Tumour Biol. 32:203–213.
2011. View Article : Google Scholar
|
|
44
|
Cascio S, Zhang L and Finn OJ: MUC1
protein expression in tumor cells regulates transcription of
proinflammatory cytokines by forming a complex with nuclear
factor-κB p65 and binding to cytokine promoters: importance of
extracellular domain. J Biol Chem. 286:42248–42256. 2011.PubMed/NCBI
|
|
45
|
Spicer AP, Parry G, Patton S and Gendler
SJ: Molecular cloning and analysis of the mouse homologue of the
tumor-associated mucin, MUC1, reveals conservation of potential
O-glycosylation sites, transmembrane, and cytoplasmic
domains and a loss of minisatellite-like polymorphism. J Biol Chem.
266:15099–15109. 1991.PubMed/NCBI
|