Introduction
Hypermethylation of the CpG islands in
tumor-suppressor gene (TSG) promoters is one hallmark of the cancer
epigenome. This aberrant hypermethylation silences many important
TSGs that regulate cell proliferation, differentiation, death,
invasion and other cellular processes, promoting cancer initiation,
progression and metastasis (1).
Therefore, reactivation of TSG expression by reversing promoter CpG
island hypermethylation has attracted considerable attention in
cancer research.
At present, nucleoside analogs such as 5-azacytidine
and 5-aza-2′-deoxycytidine are widely used to reduce DNA
methylation. These analogs become incorporated into the DNA of
rapidly growing cells during replication, where the modified
cytosine rings form covalent complexes with DNA methyltransferases,
which traps and leads to depletion of these enzymes, thus
inhibiting DNA methylation (1).
However, targeting DNA methyltransferases leads to the problem of
loss of specificity. Apart from demethylating the CpG islands in
TSG promoters, nucleoside analogs also induce repetitive sequence
hypomethylation, an event that promotes the malignant phenotype
(1–3). For example, treatment of rat
chondrosarcoma cells with 5-aza-2-deoxycytidine led to
hypomethylation of LINEs and satellite DNA sequences, accompanied
by enhanced tumorigenesis and invasiveness of the cells (2). Treatment of human acute lymphoblastic
leukemia cells with 5-aza-2-deoxycytidine induced hypomethylation
of LINE-1 repeat elements and elicited their retrotransposition,
which subsequently activated the proto-oncogene
c-MET(3). Therefore,
specific demethylation of the hypermethylated CpG islands in TSG
promoters would provide a promising safe method for cancer therapy
based on epigenetic mechanisms.
Active DNA demethylation is a direct enzymatic
process that converts 5-methylcytosine to cytosine or
5-hydroxymethylcytosine (4). A
number of active demethylation mechanisms exist in animal oocytes.
In mouse, pig, bovine and human zygotes, oocyte components catalyze
rapid demethylation of the paternal genome within a few hours of
fertilization (5). When a somatic
nucleus is transplanted into an enucleated oocyte, the ooplasm also
drives demethylation of the somatic nuclear DNA before replication
begins (6). High levels of this
demethylation activity can be retained in oocyte extract and
exhibit genomic element specificity. Bian et al(7) induced specific demethylation of the
OCT4 promoter without affecting the methylation levels of
major satellite repeats and imprinted gene H19, by incubating
reversibly permeabilized mouse fibroblasts in axolotl oocyte
extract. Treatment of reversibly permeabilized fibroblasts with
porcine oocyte extract by Miyamoto et al(8) led to demethylation of the NANOG
promoter, but did not change the methylation status of centromeric
satellite regions. However, no study has investigated whether the
active demethylation activity of oocyte extract can be used to
specifically reverse the CpG island hypermethylation in the
promoters of TSGs in cancer cells.
In this study, for the first time we incubated
reversibly permeabilized human lung cancer cells with bovine oocyte
extract. The treatment led to significant demethylation of the
hypermethylated CpG islands in TSG promoters, while the methylation
levels of repetitive sequences were not affected. The demethylation
upregulated the transcription of the repressed TSGs and inhibited
the malignant phenotype of the cancer cells.
Materials and methods
Bovine oocyte collection and
maturation
Ovaries were collected from freshly slaughtered
healthy cows, and transported to the laboratory within 3 h. Cumulus
oocyte complexes (COCs) were aspired from follicles 2–8 mm in
diameter. COCs with compact cumulus cells were collected and
incubated in TCM-199 supplemented with 10% fetal bovine serum
(FBS), 0.38 mmol/l sodium pyruvate, 10 μg/ml follicle-stimulating
hormone, 5 μg/ml luteinizing hormone and 1 μg/ml 17-β-estradiol for
22 h at 38.5°C in 5% CO2 in humidified air.
Bovine oocyte extract preparation
Bovine oocyte extract was prepared as described
previously with minor changes (8).
Briefly, the matured COCs were digested with 0.1% hyaluronidase to
remove cumulus cells. Oocytes with a first polar body and even
cytoplasm were digested with 0.5% pronase to remove the zona
pellucida. The zona-free oocytes were transferred into 1.5-ml tubes
and washed three times with extraction buffer (5 mM
MgCl2, 60 mM NaCl, 2 mM β-mercaptoethanol, protease
inhibitor cocktail, 5 mM EGTA and 50 mM HEPES, pH 7.4) containing
an energy regenerating system (2 mM ATP, 20 mM phosphocreatine, 20
U/ml creatine kinase and 2 mM GTP). The washed oocytes were
resuspended (100 oocytes/μl) in extraction buffer containing the
energy-regenerating system, and disrupted by centrifugation twice
at 20,800 × g for 30 min at 4°C. The lysate was mixed by pipetting,
and subsequently centrifuged at 5,000 × g for 10 min at 4°C. The
supernatant was used as the oocyte extract.
Oocyte extract treatment
The human non-small cell lung cancer (NSCLC) cell
line NCI-H460 was cultured in RPMI-1640 medium supplemented with
10% FBS at 37°C with 5% CO2. For permeabilization,
2×106 H460 cells were collected and suspended in 20
μg/ml digitonin solution for 2 min. Permeabilization was assessed
by monitoring the uptake of 70-kDa FITC-dextran (40 μg/ml) in a
separate sample after resealing the plasma membranes (9). The permeabilized cells were suspended
in bovine oocyte extract (5,000 cells/10 μl extract;
extract-treated cells) or an equal volume of extraction buffer
(buffer-treated cells) at 38.5°C for 3.5 h with occasional tapping.
For membrane resealing, the cell suspension was diluted with 1 ml
RPMI-1640 containing 2 mM CaCl2, and incubated for 2 h
at 37°C. After pelleting by centrifugation at 400 × g for 5 min,
the cells were subjected to normal culture or used directly for
assays.
Bisulfite sequencing
DNA was isolated from cells using the
Wizard® SV genomic DNA purification system (Promega
Corporation, Madison, WI, USA). Bisulfite conversion was performed
using the EZ DNA methylation kit (Zymo Research, Irvine, CA, USA )
according to the vendor’s recommendations. Bisulfite converted DNA
was amplified with the primers listed in Table I. The PCR products were then cloned
into the vector pEASY-T1 (Transgene, China), with at least 10
clones from each sample subjected to sequencing.
 | Table IBisulfite sequencing primers. |
Table I
Bisulfite sequencing primers.
| Gene name | Forward primer
(F)
Reverse primer (R) |
|---|
| RUNX3 | F:
TAGTTTTGTAGAGGGTTTTTTAGTG
R: AATTAAAACCAACATTAACCTAAAC |
| CDH1 | F:
AATAAAAGAATTTAGTTAAGTGT
R: GTTGTTGTTGTTGTAGGTATT |
| RASSF1A | F:
TTATTTAGTGGGTAGGTTAAGTGTGTT
R: AAACCTAAATACAAAAACTATAAAACCC |
| WIF1 | F:
GAGTGATGTCCCAGGGGTCTCT
R: CCTAAATACCAAAAAACCTAC |
| Alpha
satellites | F:
TAATTAATTAAACCCCTTT
R: TTTTTATGTTTAAGATTGG |
| Retroviral LTR of
minisatellite MS32 | F:
CCACTCAAACATAAATTTAA
R: GTTTAGATTGTGTAGTTTAA |
Quantitative real-time PCR
Total RNA was isolated using RNAiso reagent, and
reverse transcribed using the PrimeScript RT reagent kit (both from
Takara Bio, Inc., Shiga, Japan). Real-time PCR was performed using
the primers described in Table II
and SYBR Premix Ex Taq (Takara Bio, Inc.) in triplicate on a 7300
Real-Time PCR system (Applied Biosystems, Foster City, CA, USA).
The relative expression levels of each gene were calculated using
the ΔΔCt method using GAPDH as an endogenous control.
 | Table IIReal-time PCR primers. |
Table II
Real-time PCR primers.
| Gene name | Forward primer
(F)
Reverse primer (R) |
|---|
| GAPDH | F:
TGTCCCCACTGCCAACGTGTCA
R: GCGTCAAAGGTGGAGGAGTGGGT |
| RUNX3 | F:
CAGCACCACAAGCCACTTCA
R: GGTCGGAGAATGGGTTCAGTT |
| CDH1 | F:
CGGGAATGCAGTTGAGGATC
R: AGGATGGTGTAAGCGATGGC |
| RASSF1A | F:
ACCTCTGTGGCGACTTCATCT
R: AGGTGAACTTGCAATGCG |
| WIF1 | F:
TGAATTTTACCTGGCAAGCTG
R: GGACATTGACGGTTGGATCT |
Soft agar assay
A 0.6% (wt/vol) solution of low melting point agar
was prepared in normal medium, placed into 6-well culture plates,
and allowed to solidify. A layer of 0.3% low melting point agar
containing 1×104 cells was gently placed on top. The
cells were incubated in a humidified atmosphere (5% CO2)
at 37°C, and the number of colonies larger than 100 μm was counted
after 3 weeks.
Cell migration and invasion assay
Migration and invasion assays were performed using
24-well Transwell chambers (polycarbonate membrane, 8-μm pore size;
Costar). For the migration assay, 4×104 cells suspended
in 200 μl serum-free RPMI-1640 were placed into each upper chamber,
and 600 μl of RPMI-1640 containing 20% FCS was placed into each
lower chamber as a chemoattractant. The plates were incubated for
24 h at 37°C, then the medium was removed from the Transwell
chambers, and the cells on the upper surface of the Transwell
membrane were wiped off. Cells that had migrated to the lower
surface of the Transwell membrane were fixed, stained with
hematoxylin, and the number of cells in five randomly selected
fields at ×200 magnification were counted. Three independent
experiments were performed.
For the invasion assay, the upper surfaces of the
Transwell membranes were pre-coated with Matrigel (diluted 1:7; BD
Biosciences, Franklin Lakes, NJ, USA) which was allowed to gel at
37°C for 4 h, then the cells were added to the Transwell membranes
and the assay was conducted using the same method as that used for
the migration assay.
Statistical analysis
Bisulfite sequencing data were analyzed using the
χ2 test; real-time PCR data were analyzed using one-way
ANOVA; soft agar assay, cell migration assay and cell invasion
assay data were analyzed using the unpaired Student’s t-test.
Significance was accepted at p<0.05.
Results
Permeabilization assessment
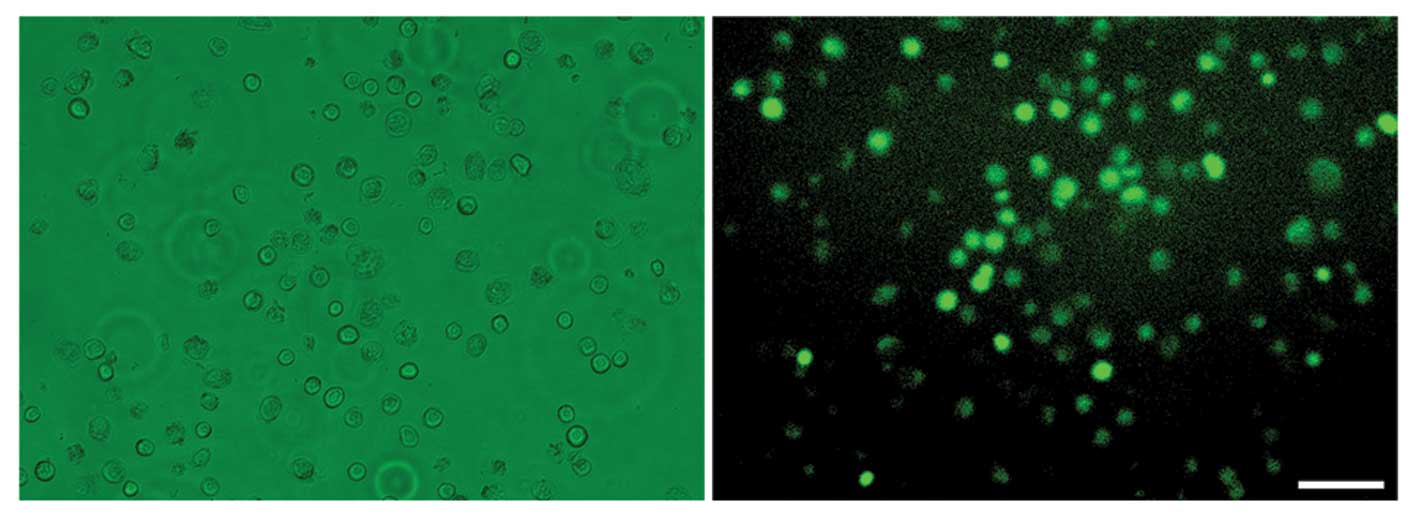
In order to introduce the demethylation factors from
bovine oocyte extract into H460 cells, we first permeabilized the
membranes of the H460 cells using digitonin, an efficient and mild
permeabilization agent. Over 80% of the permeabilized cells showed
uptake of 70-kDa FITC-dextran (Fig.
1), indicating that the large molecules in bovine oocyte
extract could readily enter the permeabilized H460 cells.
Bovine oocyte extract reverses
hypermethylation of TSG promoter CpG islands
The TSGs RUNX3, CDH1, RASSF1A and WIF1
are frequently silenced in lung cancer cells due to
hypermethylation of the CpG islands in their promoters. We analyzed
the effects of bovine oocyte extract treatment on the methylation
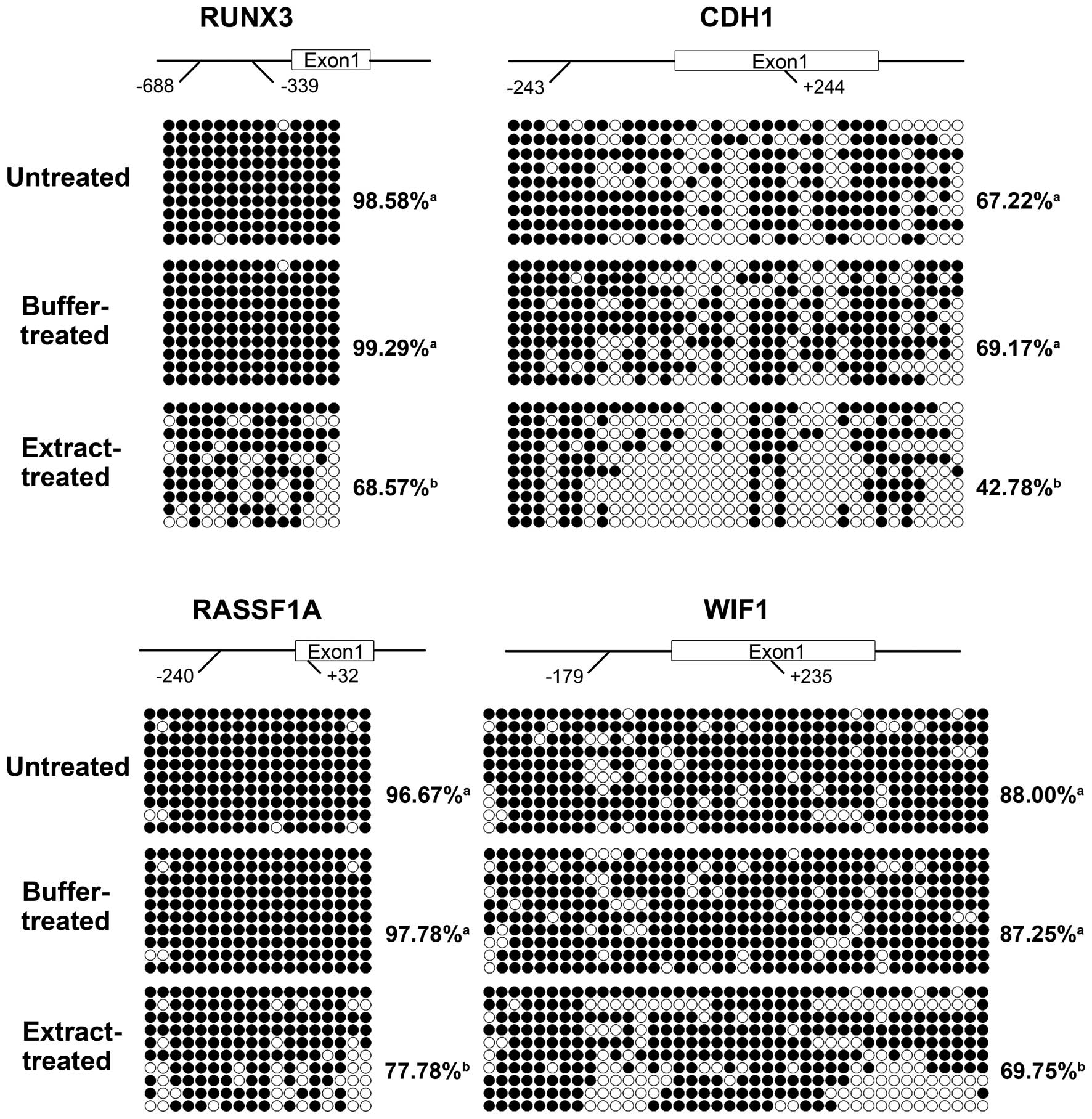
of the promoter CpG islands of these four TSGs. As shown in
Fig. 2, the promoter CpG islands of
the four TSGs were hypermethylated in the untreated H460 cells. The
methylation levels of the CpG islands of the four TSGs were similar
in the buffer-treated cells and untreated cells (Fig. 2). In the extract-treated cells,
incubation in the bovine oocyte extract induced significant
demethylation of the CpG islands in the four TSGs (Fig. 2). The methylation levels were
decreased by 30.94% for RUNX3, by 38.15% for CDH1, by
20.45% for RASSF1A and by 20.06% for WIF1, compared
to the buffer-treated cells.
The extract-induced demethylation at the CpG islands
was not randomly distributed among the CpG sites. The demethylation
focused on CpG sites 7, 13 and 14 for RUNX3; sites 4, 7, 10,
11, 13, 17 and 25 for CDH1; sites 2, 11, 17 and 18 for
RASSF1A; and sites 10–12 and 30–35 for WIF1. Many of
these CpG sites reside in transcription factor binding regions,
particularly the binding regions for the transcription factor Sp1
(Table III).
 | Table IIIDemethylated CpG sites residing in the
transcription factor binding regions. |
Table III
Demethylated CpG sites residing in the
transcription factor binding regions.
| RUNX3 | CDH1 | RASSF1A | WIF1 |
|---|
| Sp1 | 7, 14 | 17 | 11, 17 | 12 |
| CP2 | 14 | - | - | - |
| GATA-2 | - | - | 2 | 32 |
| SRY | - | 12 | - | - |
| Tst-1 | - | 4 | - | - |
| p300 | - | - | - | 30 |
These results indicated that the bovine oocyte
extract induced efficient and site-specific demethylation at the
CpG islands in the TSG promoters.
Bovine oocyte extract does not
demethylate repetitive sequences
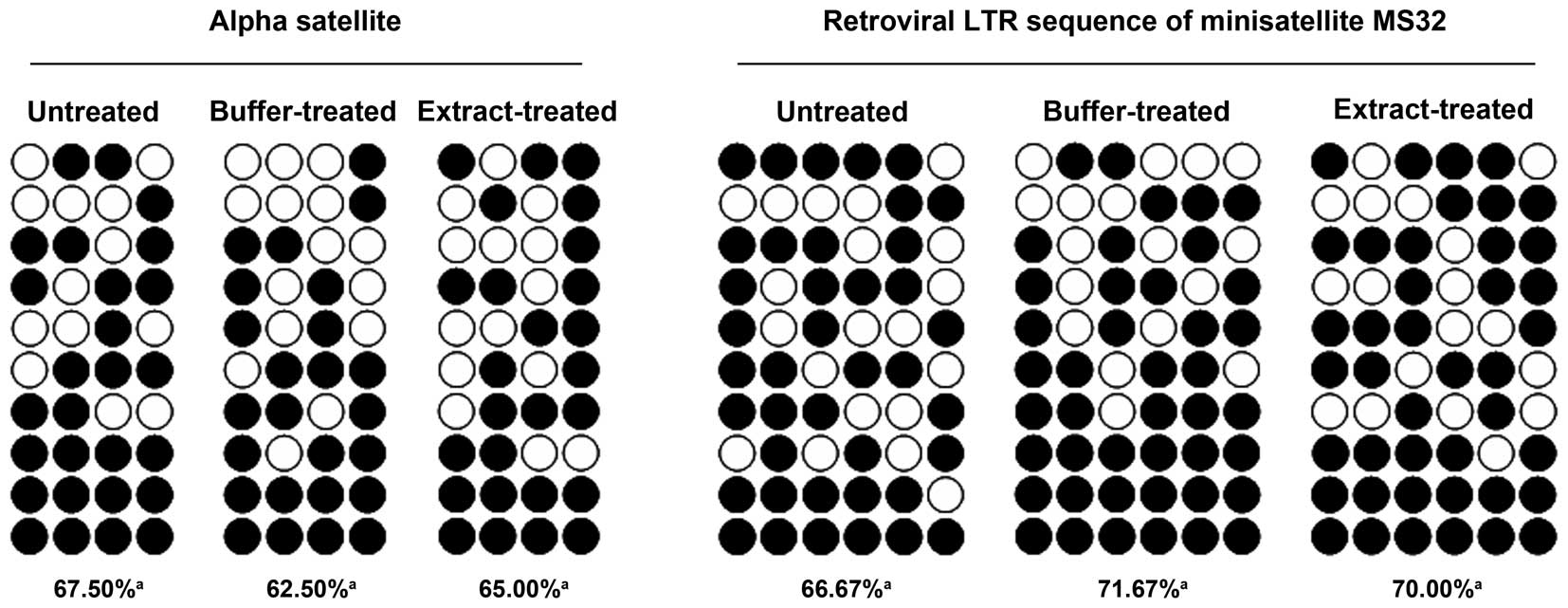
We next examined whether the bovine oocyte extract
treatment led to demethylation of repetitive sequences. The alpha
satellite sequence and retroviral long terminal repeat (LTR)
sequence of minisatellite MS32 were selected for analysis.
Bisulfite sequencing demonstrated that the methylation levels of
these sequences were similar in untreated, buffer-treated and
extract-treated cells (Fig. 3),
suggesting that the bovine oocyte extract did not induce
hypomethylation of repetitive sequences.
Bovine oocyte extract upregulates
transcription of the four TSGs
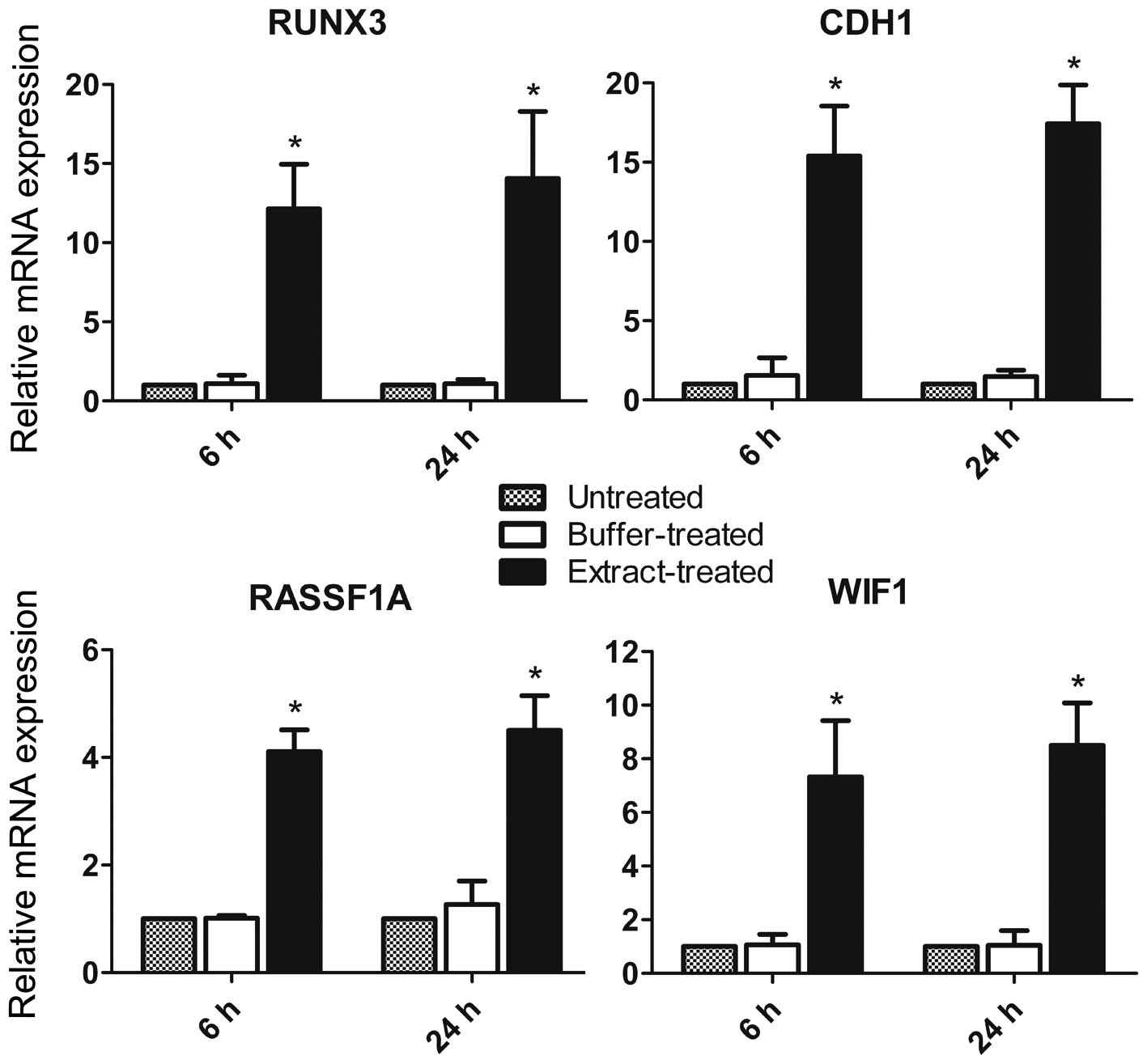
Since DNA hypermethylation of promoter CpG islands
silences gene transcription, we investigated whether the bovine
oocyte extract-induced demethylation activates transcription of the
four tested TSGs. After returning to culture for 6 h, the
extract-treated cells displayed a marked increase in transcription
of the four TSGs, compared to the untreated and buffer-treated
cells (Fig. 4). The increase was
more obvious for RUNX3 and CDH1, consistent with the
greater degree of demethylation observed in their promoter CpG
islands in response to the bovine oocyte extract. After return to
culture for 24 h, transcription of the four TSGs was further
upregulated in extract-treated cells (Fig. 4). The expression levels of the four
TSGs were not significantly different in the buffer-treated and
untreated cells, indicating that the permeabilization process did
not upregulated the expression of the four TSGs.
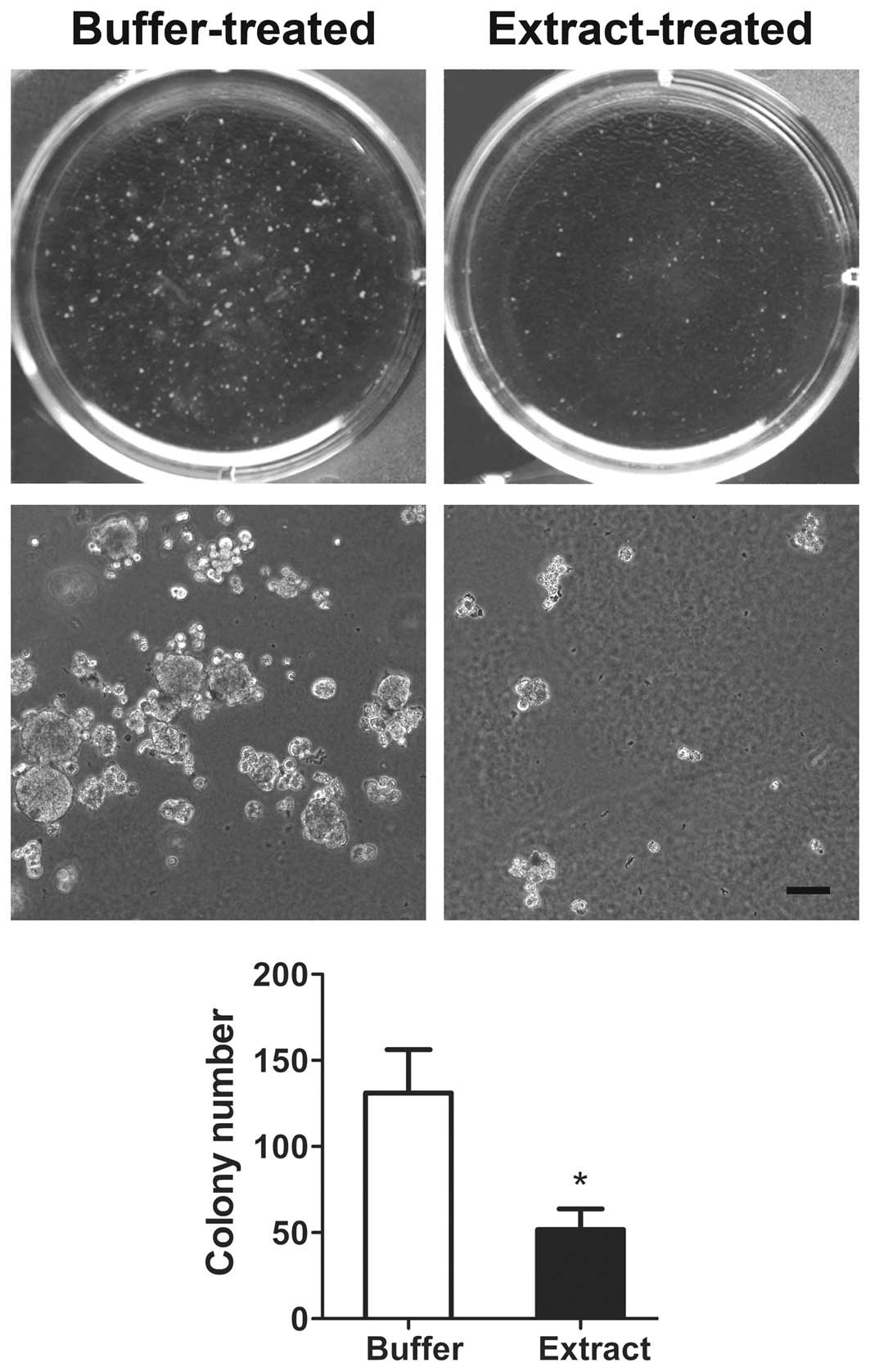
Bovine oocyte extract inhibits the
malignant phenotype of H460 cells
The TSGs RUNX3, CDH1, RASSF1A and WIF1
play important roles in the regulation of cell proliferation,
migration and invasion. Thus, we next investigated the effect of
bovine oocyte extract treatment on the malignant phenotype of H460
cells. Buffer-treated cells were used as a control. In the soft
agar assay, the extract-treated cells formed significantly fewer
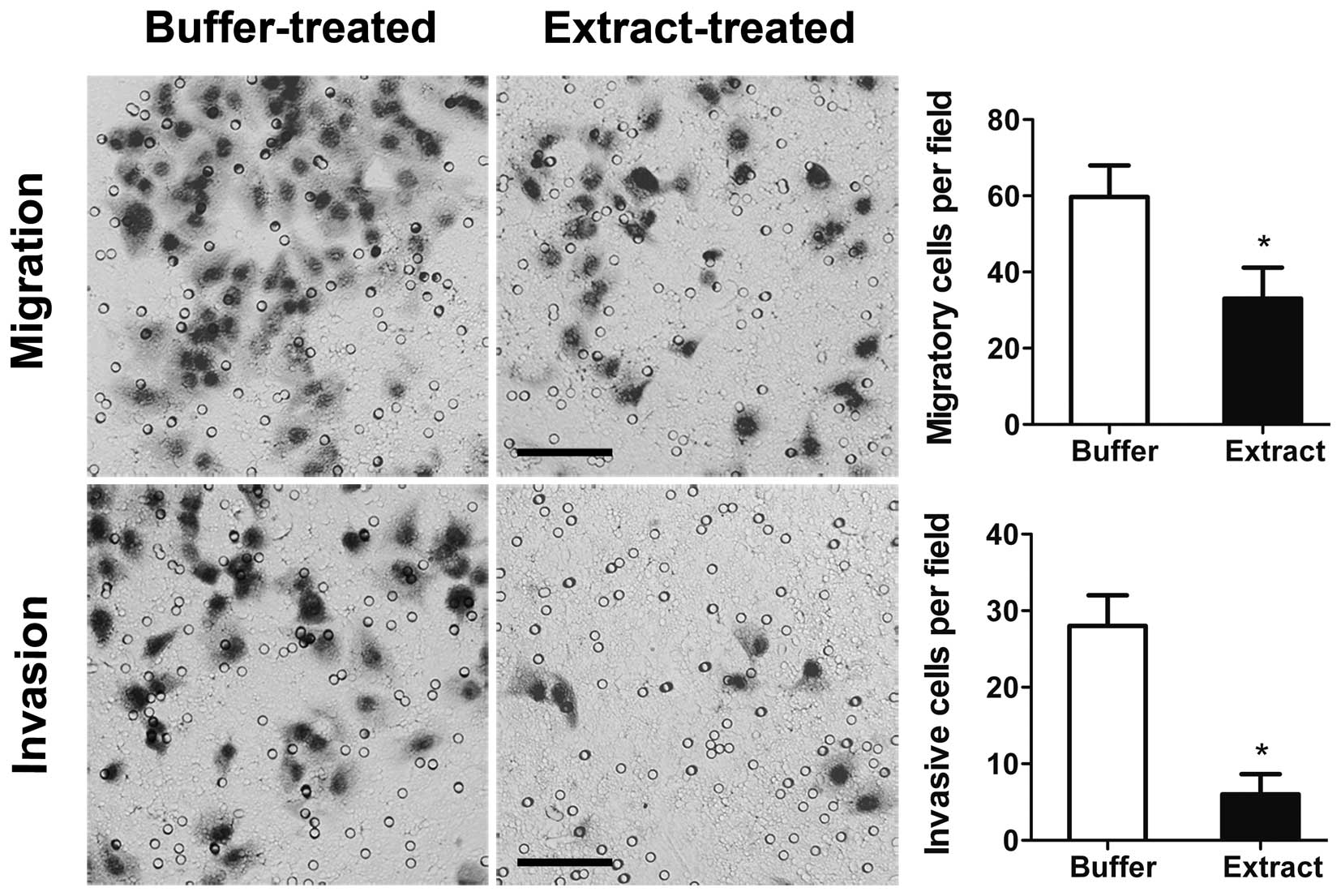
and smaller colonies than in the buffer-treated cells (Fig. 5). In the migration assay, the number
of extract-treated cells that migrated through the Transwell
membrane was 40.72% lower than the number of migrating
buffer-treated cells (Fig. 6). In
the invasion assay, the number of extract-treated cells that passed
through the Matrigel barrier was 78.05% lower than the number of
invasive buffer-treated cells (Fig.
6). Thus, bovine oocyte extract treatment strongly reduced the
anchorage-independent proliferation, migration and invasive
capacity of H460 cells.
Discussion
A number of innate active DNA demethylation
mechanisms exist in mammals to drive rapid DNA demethylation in
specific sequences at various developmental stages. Exposure of
cancer cells to these active demethylation mechanisms may help to
understand the epigenetic mechanisms leading to repression of TSG
expression, and help explore new epigenetic-based anti-cancer
approaches. In this study, we investigated the specific reversal of
CpG island hypermethylation in TSG promoters using the active DNA
demethylation activity of bovine oocytes.
The extract-treated cells displayed significant
demethylation of the TSG promoter CpG islands after only a 3.5-h
incubation with the bovine oocyte extract. As this incubation time
was short and the cells were suspended in a serum-free environment
during incubation, the extract-induced demethylation was an active
process independent of DNA replication. Of the molecules which have
been identified to possess active demethylation activity, at least
methyl-CpG-binding domain protein 4 (MBD4) (10), activation-induced cytidine deaminase
(AID) (11), teneleven
translocation 3 (TET3) (12),
elongator complex protein 3 (ELP3) (4), DNA methyltransferase 3A (DNMT3A)
(10) and DNA methyltransferase 3B
(DNMT3B) (10) are expressed in
oocytes. These proteins, except for DNMT3A and DNMT3B, have a
molecular weight lower than 70 kDa, particularly AID, whose
molecular weight is only 24 kDa. Therefore, these proteins readily
enter permeabilized cells to catalyze active demethylation,
converting 5-methylcytosine to cytosine or
5-hydroxymethylcytosine.
The bovine oocyte extract upregulated expression of
the repressed tumor-suppressor genes by demethylating several
specific CpG sites rather than all of the CpG sites within each CpG
island. This avoids aggravating the genome hypomethylation in
cancer cells, and favors revealing key regulatory sequences in the
CpG islands. Many of the demethylated CpG sites reside in
transcription factor binding regions, suggesting that demethylation
of the CpG sites within transcription factor binding regions plays
an important role in transcriptional activation. It may be that the
methylation modification of these CpG sites hinders transcription
factors from binding to the promoters or provides proper binding
sites for chromatin condensation factors.
Studies have demonstrated that apart from reversing
the hypermethylation of TSG promoters, nucleoside analogs also
induce hypomethylation of repetitive sequences such as satellite
DNA sequences (2,13). Hypomethylation of repetitive
sequences increases genome instability by promoting chromatin
rearrangement and activating transposable elements, thereby
inducing or aggravating the malignant phenotype. Notably, bovine
oocyte extract treatment did not affect the DNA methylation levels
of repetitive sequences; rather it specifically demethylated the
hypermethylated TSG promoters. This suggests that bovine oocyte
extract treatment may provide a safe demethylation approach.
Paternally methylated imprinted genes, intracisternal A particle
retrotransposons, and heterochromatin in and around the centromeres
remain unaffected during the demethylation of the paternal genome
by ooplasm after fertilization. Therefore, the process of
ooplasm-driven demethylation is intrinsically genome
element-selective, and this genome element-selective attribute was
well preserved during the treatment of cancer cells with bovine
oocyte extract.
Bisulfite sequencing showed that the promoter CpG
islands of RUNX3, CDH1, RASSF1A and WIF1 were all
hypermethylated in untreated H460 cells. Hypermethylation of
promoter CpG islands induces closed chromatin conformations, which
prevent transcription factors from binding to the gene promoters,
resulting in transcriptional repression. Previous studies have
demonstrated that the expression of the four TSGs is silenced in
H460 cells (14–17). However, the extract-treated cells
displayed significant transcriptional upregulation of the four TSGs
after being returned to culture for 6 h. This observation suggests
that bovine oocyte extract treatment can rapidly alter the aberrant
chromatin structure and quickly reestablish a relatively open
chromatin conformation around these four TSG promoters in H460
cells. In contrast, nucleoside analog-induced TSG activation
usually requires a treatment time of 3 days or longer, with
treatment for shorter times such as 24 h often generating no effect
(15).
Anchorage-independent colony formation in soft agar
is a highly clinically relevant model for evaluating NSCLC cell
proliferation (18). Compared to
the buffer-treated cells, the extract-treated cells only formed a
few small clones after a 3-week culture in soft agar. Thus, the
upregulation of TSG expression by bovine oocyte extract treatment
led to long-term inhibition of cancer cell proliferation.
The vaccinia vaccine has been reliably administered
to human beings for centuries. Bovine insulin and thymic peptides
have also been widely used in the clinical therapy of human
diseases, and these treatments are safe and have a high efficacy.
In this study, we demonstrated that bovine oocyte extract can
specifically reverse promoter CpG island hypermethylation of TSGs
without inducing hypomethylation of repetitive sequences.
The promoter demethylation upregulates the
expression of epigenetically repressed TSGs and inhibits the
malignant phenotype of H460 cells. Thus, bovine oocyte extract may
provide a useful tool for investigating the epigenetic alterations
in cancer cells, and may represent a promising source of drugs for
epigenetic-based therapy of human diseases.
Acknowledgements
This study was supported by a grant from the Major
Program of the Inner Mongolia Natural Science Foundation (No.
2012ZD04), and a grant from NSFC (No. 30860185).
References
|
1
|
Huang YW, Kuo CT, Stoner K, Huang TH and
Wang LS: An overview of epigenetics and chemoprevention. FEBS Lett.
585:2129–2136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamm CA, Xie H, Costa FF, Vanin EF, Seftor
EA, Sredni ST, Bischof J, Wang D, Bonaldo MF, Hendrix MJ and Soares
MB: Global demethylation of rat chondrosarcoma cells after
treatment with 5-aza-2′-deoxycytidine results in increased
tumorigenicity. PLoS One. 4:e83402009.PubMed/NCBI
|
|
3
|
Roman-Gomez J, Jimenez-Velasco A, Agirre
X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA,
Navarro G, Colomer D, Prosper F, Heiniger A and Torres A: Promoter
hypomethylation of the LINE-1 retrotransposable elements activates
sense/antisense transcription and marks the progression of chronic
myeloid leukemia. Oncogene. 24:7213–7223. 2005. View Article : Google Scholar
|
|
4
|
Wu SC and Zhang Y: Active DNA
demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol.
11:607–620. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morgan HD, Santos F, Green K, Dean W and
Reik W: Epigenetic reprogramming in mammals. Hum Mol Genet.
14:R47–R58. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dean W, Santos F, Stojkovic M,
Zakhartchenko V, Walter J, Wolf E and Reik W: Conservation of
methylation reprogramming in mammalian development: aberrant
reprogramming in cloned embryos. Proc Natl Acad Sci USA.
98:13734–13738. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bian Y, Alberio R, Allegrucci C, Campbell
KH and Johnson AD: Epigenetic marks in somatic chromatin are
remodelled to resemble pluripotent nuclei by amphibian oocyte
extracts. Epigenetics. 4:194–202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miyamoto K, Tsukiyama T, Yang Y, Li N,
Minami N, Yamada M and Imai H: Cell-free extracts from mammalian
oocytes partially induce nuclear reprogramming in somatic cells.
Biol Reprod. 80:935–943. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ostrup O, Hyttel P, Klaerke DA and Collas
P: Remodeling of ribosomal genes in somatic cells by Xenopus
egg extract. Biochem Biophys Res Commun. 412:487–493. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oliveri RS, Kalisz M, Schjerling CK,
Andersen CY, Borup R and Byskov AG: Evaluation in mammalian oocytes
of gene transcripts linked to epigenetic reprogramming.
Reproduction. 134:549–558. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morgan HD, Dean W, Coker HA, Reik W and
Petersen-Mahrt SK: Activation-induced cytidine deaminase deaminates
5-methylcytosine in DNA and is expressed in pluripotent tissues:
implications for epigenetic reprogramming. J Biol Chem.
279:52353–52360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iqbal K, Jin SG, Pfeifer GP and Szabó PE:
Reprogramming of the paternal genome upon fertilization involves
genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA.
108:3642–3647. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daskalos A, Nikolaidis G, Xinarianos G,
Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field
JK and Liloglou T: Hypomethylation of retrotransposable elements
correlates with genomic instability in non-small cell lung cancer.
Int J Cancer. 124:81–87. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li QL, Kim HR, Kim WJ, Choi JK, Lee YH,
Kim HM, Li LS, Kim H, Chang J, Ito Y, Youl Lee K and Bae SC:
Transcriptional silencing of the RUNX3 gene by CpG hypermethylation
is associated with lung cancer. Biochem Biophys Res Commun.
314:223–228. 2004. View Article : Google Scholar
|
|
15
|
Reinhold WC, Reimers MA, Lorenzi P, Ho J,
Shankavaram UT, Ziegler MS, Bussey KJ, Nishizuka S, Ikediobi O,
Pommier YG and Weinstein JN: Multifactorial regulation of
E-cadherin expression: an integrative study. Mol Cancer Ther.
9:1–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Endoh H, Yatabe Y, Shimizu S, Tajima K,
Kuwano H, Takahashi T and Mitsudomi T: RASSF1A gene inactivation in
non-small cell lung cancer and its clinical implication. Int J
Cancer. 106:45–51. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mazieres J, He B, You L, Xu Z, Lee AY,
Mikami I, Reguart N, Rosell R, McCormick F and Jablons DM: Wnt
inhibitory factor-1 is silenced by promoter hypermethylation in
human lung cancer. Cancer Res. 64:4717–4720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takezawa K, Okamoto I, Yonesaka K,
Hatashita E, Yamada Y, Fukuoka M and Nakagawa K: Sorafenib inhibits
non-small cell lung cancer cell growth by targeting B-RAF in KRAS
wild-type cells and C-RAF in KRAS mutant cells. Cancer Res.
69:6515–6521. 2009. View Article : Google Scholar : PubMed/NCBI
|