Introduction
Cell biology is an important field of study and a
valuable tool used to investigate cell proliferation,
differentiation, development, aging, apoptosis or death,
carcinogenesis and metastasis (1).
Researchers have established thousands of cell lines as models to
explore the corresponding tissues. However, with the increasing
number of immortalized cell lines established, cross-contamination
of cell lines is also becoming a more frequently encountered
problem (2,3). Cell line cross-contamination may alter
the characteristics of the cultures, leading to inaccurate results
and affecting the credit score of the research group (3,4).
Therefore, the identification of immortalized cell lines is key for
cell biology, particularly for cancer cell biology, as most
immortalized cell lines are cancer cell lines.
Cell lines derived from different individuals are
generally easier to be identified as they normally have different
short tandem repeat (STR) profiling (5,6).
However, for the cell lines derived from the same individual with
different biological properties, it is unclear whether they have
contaminated each other as they have the same STR loci. Thus, it is
necessary to identify the molecular markers as well as the distinct
biological characteristics in certain aspects to determine the
homologous cancer cell lines.
In the present study, we introduced the comparable
salivary adenoid cystic carcinoma (SACC) cell lines, the parental
cell line SACC-83, with a low lung metastasis rate, and its
daughter cell line SACC-LM, with a high lung metastasis rate
(7,8). SACC-83 was isolated from a patient
pathologically diagnosed in 1983 with SACC in the sublingual gland,
and SACC-LM was generated from lung metastatic lesions through
5-round selections starting from intravenous injection of SACC-83
cells (8).
Cell cross-contamination is becoming an increasingly
serious issue (3,4) and although the homologous SACC cell
lines SACC-83 and SACC-LM have recently been used as a SACC model
to investigate the underlying mechanism of metastasis (8–12), the
molecular features of these SACC cell lines remain unclear. In this
study, we screened the expression pattern of 136 genes in these two
cell lines in order to find the molecular properties of the two
SACC cells and to avoid the misuse of the two cell lines in the
future.
Materials and methods
Cell culture
Human SACC cell lines, SACC-83 and SACC-LM,
established in our laboratory, were cultured in RPMI-1640 (Gibco,
Grand Island, NY, USA) supplemented with 10% fetal bovine serum
(Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin in a
humidified incubator at 37°C with 5% CO2.
STR analysis
DNA was extracted from SACC-83 and SACC-LM cells as
previously described (10). STR
profiling PCR was carried out in the ABI 3100 genetic analyzer
machine by the use of 10 ng of DNA as a template and the
Goldeneye™16C STR detection kit.
Immunostaining
SACC-83 and SACC-LM cells were plated into 24-well
plates containing round coverslides in RPMI-1640 culture medium for
24 h, then fixed with 4% paraformaldehyde for 10 min, and subjected
to immunostaining assay to identify the cell type of SACC-83 and
SACC-LM. Briefly, the glass coverlids were rinsed twice with PBS,
and endogenous peroxidase was blocked by the use of 3% hydrogen
peroxide in PBS for 10 min. The samples were blocked with normal
goat serum for 1 h, incubated with anti-human pan-cytokeratin
(pan-CK), cytokeratin (AE1), CK8/18 and S100P (Zhongshan Golden
Bridge Biotechnology Co., Ltd., Beijing, China) overnight at 4°C,
followed by horseradish peroxidase-conjugated immunoglobulin for 30
min, developed for color with peroxidase substrate
3,3′-diaminobenzidine (DAB), counterstained with hematoxylin, and
recorded using an Olympus DP controller (Olympus, Tokyo,
Japan).
Transwell invasion assay
Cell invasion assays were performed using transwell
chambers with a polycarbonate membrane (Millipore, Bedford, MA,
USA) coated with 40 μl diluted matrix gel (BD Biosciences). Cells
were trypsinized and seeded at 1×105 cells/well/0.1 ml
serum-free RPMI-1640 medium in the upper chambers, and 0.5 ml of
RPMI-1640 medium supplemented with 20% FBS was added into each
lower chamber. 48 h after incubation, cells on the surface of the
membrane were wiped off, and the membranes were fixed with 95%
ethanol and stained with 1% crystal violet (Sigma-Aldrich). The
invaded cells clinging to the bottom of the membrane were evaluated
by light microscopy at 10× magnification, and by measurement of
optical density (OD)570 for the solution derived from
the crystal violet discolored from the invaded cells by using 300
μl of 33% glacial acetic acid. All experiments were performed in
triplicate and similar results were obtained from three independent
experiments.
Quantitative PCR (qPCR) analysis
Total RNA was extracted from SACC-83 or SACC-LM cell
lines using the TRIzol reagent according to the manufacturer’s
instructions (Invitrogen, Carlsbad, CA, USA). Complementary DNA was
reverse transcribed by the use of 2.5 μg of RNA as a template. qPCR
was performed using the ABI 7500 real-time PCR machine coupled with
SYBR Green chemistry (Applied Biosystems, Foster City, CA, USA).
All PCR reactions were in 20 μl of total volume containing 10 μl of
SYBR Green PCR master mix, 50 ng cDNA, 250 nM of each primer. All
amplifications were performed in triplicate for each sample and
repeated three times. The thermal cycling was 10 min at 95°C,
followed by 40 cycles at 95°C for 15 sec, at 60°C for 60 sec. The
specificity of amplification was monitored using the dissociation
curve of the amplified product. Relative expression of the target
genes was calculated using the 2-ΔΔCt method.
Western blotting
Cells were harvested and lysed in RIPA buffer with
protease inhibitors (Applygen Technologies, Inc., Beijing, China).
Protein concentration was determined using the BCA Protein Assay
(Thermo Fisher Scientific, Waltham, MA, USA), and 40 μg of protein
was loaded for each sample. Proteins were separated on a sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to a polyvinylidene difluoride membrane. The membranes
were blocked in 5% non-fat dry milk for 1 h and probed with
antibodies against S100P (Sino Biological Inc., Beijing, China),
S100A4 (BD Biosciences), matrix metalloproteinase (MMP)-1, MMP-2,
MMP-9, MMP-12, MMP-14, MMP-17 (Epitomics Inc., Burlingame, CA,
USA), cyclooxygenase (COX)-2, phospho-Akt (p-Akt) (Cell Signaling
Technology, Danvers, MA, USA), and β-actin (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) separately at 4°C
overnight. Following incubation with peroxidase-linked secondary
antibodies, immunoreactive proteins were visualized by enhanced
chemiluminescence (ECL) reagent (Applygen Technology, Inc.).
Statistical analysis
Quantitative data are expressed as the means ± SD
and compared using a t-test through SPSS software, version 13.0
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
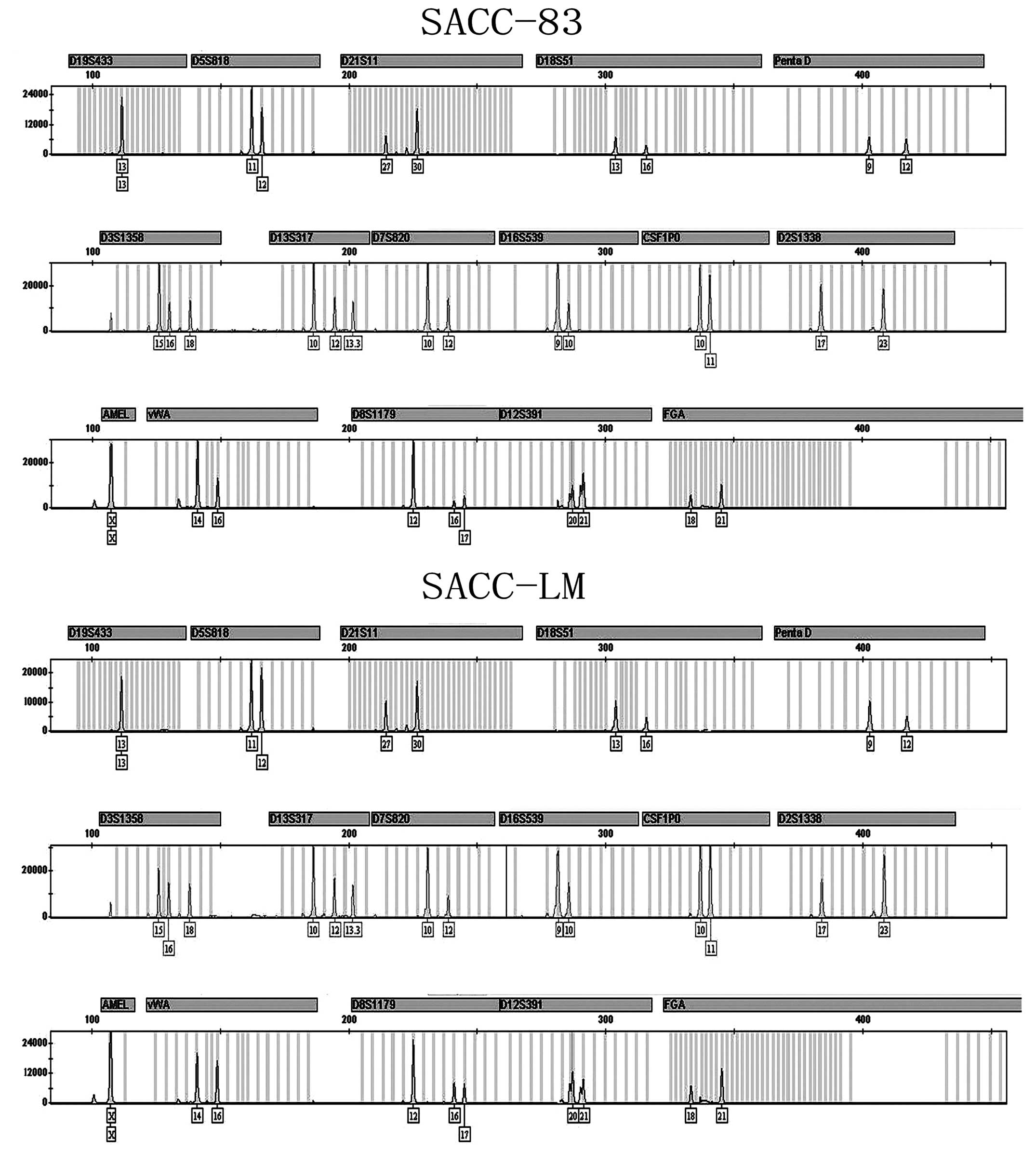
STR profiling analysis
To investigate whether SACC-83 and SACC-LM cell
lines originated from the same patient, STR assay was performed
using Goldeneye™16C STR detection kit. The results are shown in
Fig. 1. SACC-83 and SACC-LM had the
identical STR profiling in 16 gene foci [D19S433 (13, 13);
D5S818 (11, 12); D21S11 (27, 30); D18S51 (13, 16);
PentaD (9, 12); D3S1358 (15, 16,
18); D13S317 (10, 12, 13.3);
D2S1338 (17, 23), D7S820 (10, 12),
D16S539 (9, 10), CSF1PO (10, 11);
vWA (14, 16); D8S1179 (12, 16,
17); D12S391 (20, 21);
FGA (18, 21) and amelogenin (X, X)]. Compared with
HeLa cell STR profiling [D19S433 (13, 14);
D5S818 (11, 12); D21S11 (27, 28); D18S51 (16, 16);
PentaD (8, 15); D3S1358 (15, 18);
D13S317 (12, 14); D2S1338 (17, 17),
D7S820 (8, 12), D16S539 (9, 10),
CSF1PO (9, 10); vWA (16, 18);
D8S1179 (12, 13); D12S391 (20, 25);
FGA (21, 21) and amelogenin (X, X)] only 3 of the
16 STR foci (D5S818, D16S539 and amelogenin) were the same as those
in SACC-83 and SACC-LM. The remaining 13 STR foci listed above were
different from SACC-83 and SACC-LM. The results indicated that the
SACC-83 and SACC-LM cell lines are derived from the same patient
and, most importantly, they have thus far not been contaminated by
HeLa cells (Fig. 1).
SACC-83 and SACC-LM derive from
adenoepithelial cells
To verify the cell type of SACC-83 and SACC-LM,
immunostaining was performed by using the epithelial markers
pan-cytokeratin and cytokeratin AE1, and the luminal markers CK8/18
and S100P. Immunostaining results showed that both SACC-83 and
SACC-LM cells were positive not only for epithelial, but also for
luminal markers (Fig. 2). This
indicated that both SACC-83 and SACC-LM originated from oral
adenoepithelial cells. However, S100P expression in SACC-LM was
higher than that in SACC-83, which indicated SACC-LM had different
molecular features compared with SACC-83.
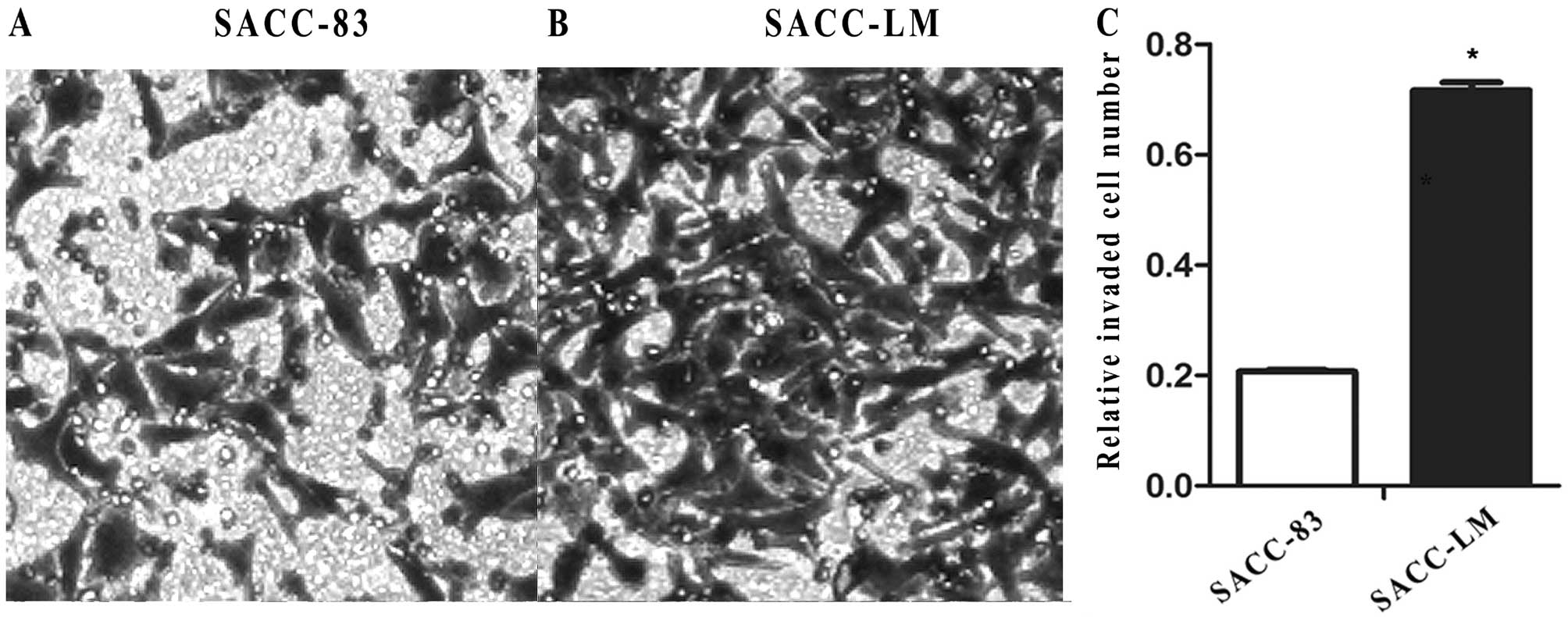
Invasion ability of SACC-83 and
SACC-LM
Prior to the investigation of the molecular
differences between SACC-83 and SACC-LM, the invasion ability was
explored based on the results in the previous studies. The
transwell assay showed that the invaded SACC-83 cells were fewer
than the invaded SACC-LM cells (Fig. 3A
and B). The results were further confirmed by measurement of
OD570 for the solution derived from the crystal violet
discolored from the invaded cells by using 300 μl of 33% glacial
acetic acid (0.207±0.055 in SACC-83 vs. 0.717±0.001 in SACC-LM;
P<0.01) (Fig. 3C). These results
indicate that SACC-LM has increased invasion ability compared to
SACC-83.
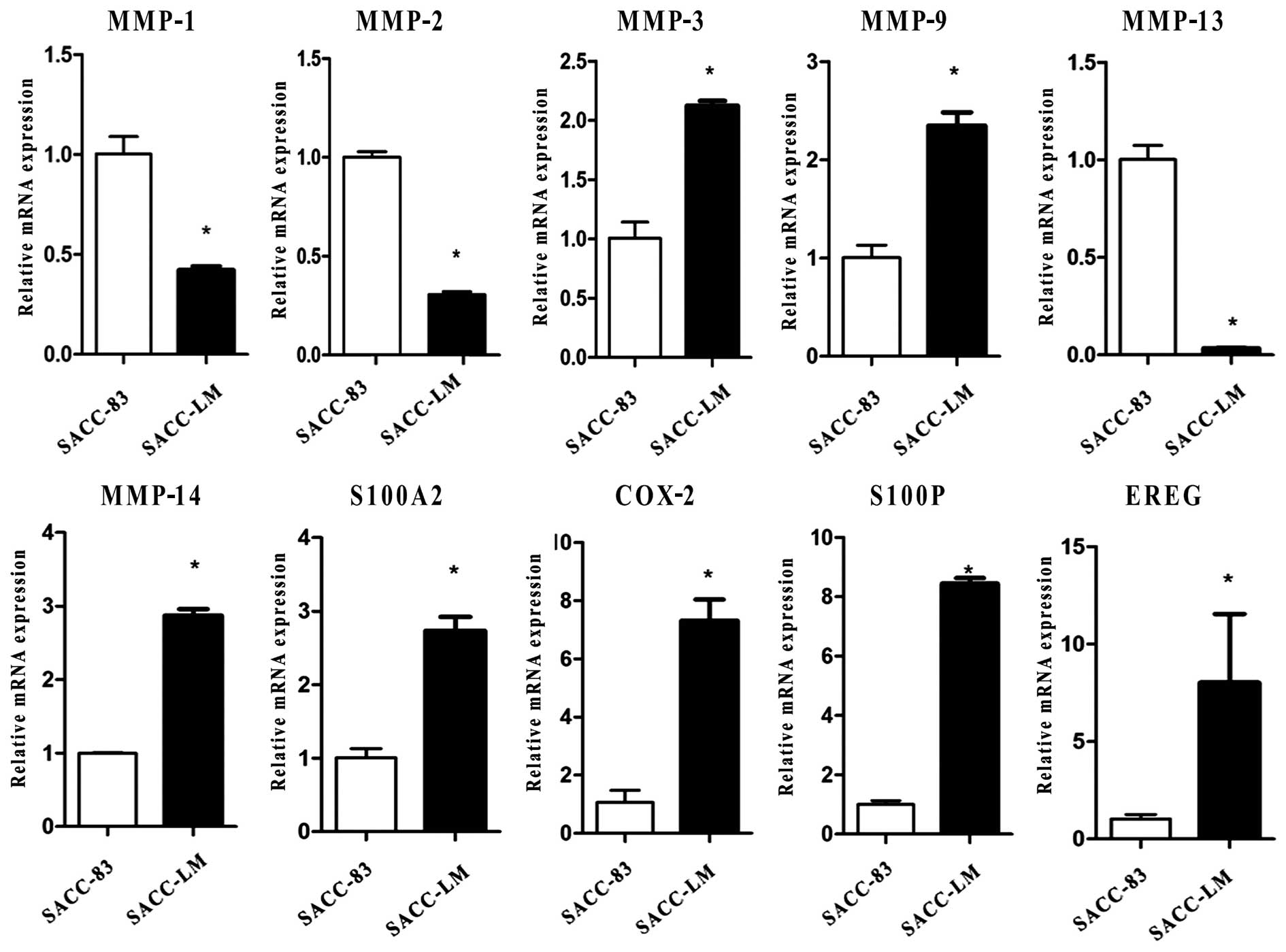
Identification of molecular markers
through qPCR and western blotting
To identify the molecular markers of SACC-83 and
SACC-LM, we screened 136 genes which included 12 genes in the MMP
family, 8 genes in Smads, 7 genes in BMPs, 13 genes in the S100
family, 17 genes in the cytokeratin family and 79 genes related to
cancer cell proliferation and metastasis. With the exception of
some genes that were expressed unstably in the two cell lines or
showed no significant difference between SACC-83 and SACC-LM, 29 of
the 136 genes showed significant differential expression in the two
cell lines (Table I). Herein we
presented 10 of 29 genes at the mRNA level, and 6 of 10 genes
further confirmed by western blotting at the protein level.
Real-time PCR results showed that compared with SACC-83, SACC-LM
expressed MMP-3, MMP-9, MMP-14, S100A2, S100P, COX-2, EREG at high
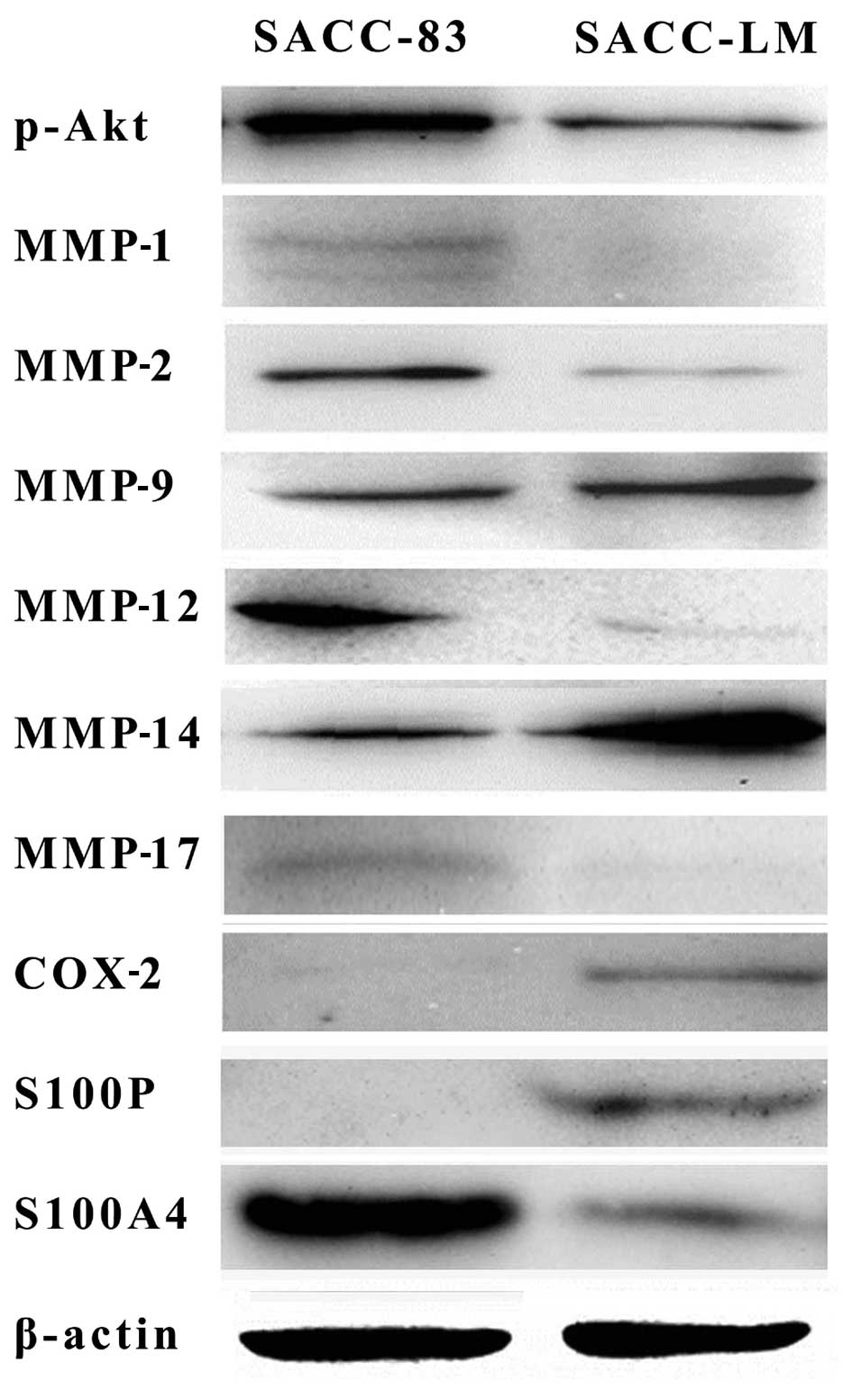
levels and MMP-1, MMP-2, MMP-13 at low levels (Fig. 4). Western blotting of MMP-1, MMP-2,
MMP-9, MMP-14, COX-2, S100P further confirmed the qPCR results. In
addition, western blotting results also showed that p-Akt, S100A4,
MMP-12, MMP-17 were highly expressed in SACC-83 compared to SACC-LM
(Fig. 5).
 | Table IThe list of the 29 differentially
expressed genes in SACC-LM compared to SACC-83. |
Table I
The list of the 29 differentially
expressed genes in SACC-LM compared to SACC-83.
| Abbreviation | GenBank accession
no. | Fold-change | P-value |
|---|
| Increased |
| COX-2 | NM_000963 | 6.9 | 0.001 |
| MMP-3 | NM_002422 | 2.1 | 0.001 |
| MMP-9 | NM_004994 | 2.3 | 0.001 |
| MMP-14 | NM_004995 | 2.9 | 0.002 |
| BMP7 | NM_001719 | 66.8 | 0.032 |
| S100P | NM_005980 | 8.4 | 0.001 |
| S100A2 | NM_005978 | 2.7 | 0.001 |
| S100A13 | NM_001024210 | 1.6 | 0.039 |
| S100A16 | NM_080388 | 2.1 | 0.047 |
| KRT15 | NM_002275 | 8.4 | 0.031 |
| EREG | NM_001432 | 9.8 | 0.002 |
| WNT2 | NM_003391 | 2.3 | 0.04 |
| WNT3 | NM_030753 | 1.8 | 0.005 |
| Decreased |
| MMP-1 | NM_001145938 | −2.4 | 0.006 |
| MMP-2 | NM_001127891 | −3.2 | 0.006 |
| uPA | NM_001145031 | −6.9 | 0.033 |
| Trail | NM_001190942 | −2.6 | 0.046 |
| FXYD5 | NM_001164605 | −1.5 | 0.031 |
| KRT7 | NM_005556 | −2.7 | 0.019 |
| KRT16 | NM_005557 | −2.2 | 0.012 |
| MCAM | NM_006500 | −2.5 | 0.008 |
| NME4 | NM_005009 | −2.3 | 0.035 |
| NOTCH1 | NM_017617 | −2.4 | 0.036 |
| CCL3 | NM_002983 | −2.4 | 0.006 |
| CCL15 | NM_032965 | −3.4 | 0.049 |
| CCL19 | NM_006274 | −2.6 | 0.036 |
| CXCL5 | NM_002994 | −2.7 | 0.045 |
| BMP3 | NM_001201 | −2.4 | 0.039 |
| BMP5 | NM_021073 | −2.3 | 0.022 |
Discussion
Cell lines are useful for a wide array of
experiments in the life sciences. However, cell cross-contamination
is increasing and requires further study (13–15).
To date, false cell lines are widely distributed and used in the
laboratories and cell banks worldwide. The German Cell Bank
reported that nearly 20% of cell lines submitted to them were
contaminated by different species or individuals (16). Several of the problems are related
to the HeLa cell line which is cross-contaminated with 106 cell
lines (3). Therefore, it is crucial
and necessary that researchers know the cell lines they are
studying and make scientific results reliable and reproducible.
A variety of methods are available for identifying
cross-contamination including isozyme typing, HLA typing,
karyotyping, DNA fingerprinting, short tandem repeat (STR). STR
profiling is an inexpensive and reliable method to confirm cell
line identity (17–20). In this study, SACC-83 and SACC-LM
cells had identical STR profiling, which indicated that the SACC-83
and SACC-LM cell lines derived from the same patient. Compared with
the HeLa STR profiling, SACC-83 and SACC-LM had 81.25% difference
from HeLa in the 16 foci of STR profiling, indicating that SACC-83
and SACC-LM have as yet not been contaminated by HeLa cells.
Pan-cytokeratin and cytokeratin AE1 are the
classical epithelial markers, which react specifically with a wide
variety of normal and neoplastic epithelial tissues. Cytokeratin
8/18 and placental S100 (S100P) are the luminal markers which
express in duct cells and duct carcinoma (21–24).
SACC-83 and SACC-LM cells were positive for epithelial cells as
well as for luminal marker CK8/18 and S100P. These indicated that
both SACC-83 and SACC-LM originated from oral adenoepithelial
cells.
However, it remains unclear whether SACC-83 and
SACC-LM cross-contaminated each other. Owing to increasing research
using SACC-83 and SACC-LM as models to explore the underlying
mechanism of SACC, it is necessary to find the molecular features
for identifying and avoiding cross-contamination and misuse of the
two cell lines. Herein, we further investigated the invasion
abilities of the two SACC cell lines. The transwell assay results
showed the invasion ability of the cells was stable in these two
cell lines. SACC-LM had increased invasion ability compared to
SACC-83, which is consistent with previous studies (8). This indicated that SACC-83 and SACC-LM
had different biological characteristics. It is well known that the
gene determines the phenotype and the biological behavior of cells.
Therefore, we next explored the expression of 136 genes at the mRNA
level and examined the biomarkers to identify the two cell lines.
Finally, 29 of 136 differential expression genes were identified
via real-time PCR and western blotting.
Western blotting results showed that p-Akt
expression is higher in SACC-83 than in SACC-LM, which further
confirmed that SACC-LM cells were more vulnerable than SACC-83
under anoikis conditions (data not shown). In addition, MMP
protein, particularly MMP-2 and MMP-9 are critical for cancer
metastasis (25,26). In general, concurrent overexpression
of MMP-2 and MMP-9 contribute to invasion and metastasis of tumor
cells by degrading extracellular matrix as well as promoting
angiogenesis. Of note, compared with the parent cell line SACC-83,
MMP-1, MMP-2, MMP-12 and MMP-13 were downregulated in the daughter
cell line SACC-LM. However, other MMPs such as MMP-9, MMP-14 were
upregulated in SACC-LM, which may contribute to the high invasion
and lung metastasis ability. Other metastasis-related genes
including COX-2, S100P and EREG expression were higher in SACC-LM
than in SACC-83, which indicated these genes could promote cancer
spread. However, further investigations are required to elucidate
the precise underlying mechanisms related to SACC metastasis.
In conclusion, SACC-83 and SACC-LM are homologous
cancer cell lines, which are not contaminated by each other or
other cancer cell lines. Compared with SACC-83, SACC-LM presents
higher expression of COX-2, S100P, and lower expression of MMP-2,
p-Akt, which could be the candidates for identifying the homologous
cell lines SACC-83 and SACC-LM.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (No. 81172556).
References
|
1
|
Lara-Padilla E and Caceres-Cortes JR: On
the nature of the tumor-initiating cell. Curr Stem Cell Res Ther.
7:26–35. 2012. View Article : Google Scholar
|
|
2
|
Chatterjee R: Cell biology. Cases of
mistaken identity. Science. 315:928–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Capes-Davis A, Theodosopoulos G, Atkin I,
et al: Check your cultures! A list of cross-contaminated or
misidentified cell lines. Int J Cancer. 127:1–8. 2010.
|
|
4
|
Stacey GN: Cell contamination leads to
inaccurate data: we must take action now. Nature. 403:3562000.
View Article : Google Scholar
|
|
5
|
Parson W, Kirchebner R, Muhlmann R, et al:
Cancer cell line identification by short tandem repeat profiling:
power and limitations. FASEB J. 19:434–436. 2005.PubMed/NCBI
|
|
6
|
Capes-Davis A, Reid YA, Kline MC, et al:
Match criteria for human cell line authentication: Where do we draw
the line? Int J Cancer. 132:2510–2519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li SL: Establishment of a human cancer
cell line from adenoid cystic carcinoma of the minor salivary
gland. Zhonghua Kou Qiang Yi Xue Za Zhi. 25:29–31. 621990.(In
Chinese).
|
|
8
|
Dong L, Wang YX, Li SL, et al: TGF-beta1
promotes migration and invasion of salivary adenoid cystic
carcinoma. J Dent Res. 90:804–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu K, Li SL, Gan YH, Wang CY and Yu GY:
Epiregulin promotes migration and invasion of salivary adenoid
cystic carcinoma cell line SACC-83 through activation of ERK and
Akt. Oral Oncol. 45:156–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu K, Gan YH, Li SL, et al: Relationship
of activated extracellular signal-regulated kinase 1/2 with lung
metastasis in salivary adenoid cystic carcinoma. Oncol Rep.
21:137–143. 2009.PubMed/NCBI
|
|
11
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang JP and Li CY: The effect of
transforming growth factor-beta1 on EDA region of fibronectin in
oral squamous cell carcinoma and adenoid cystic carcinoma cells.
Zhonghua Kou Qiang Yi Xue Za Zhi. 42:47–51. 2007.(In Chinese).
|
|
13
|
MacLeod RA, Dirks WG, Matsuo Y, Kaufmann
M, Milch H and Drexler HG: Widespread intraspecies
cross-contamination of human tumor cell lines arising at source.
Int J Cancer. 83:555–563. 1999.PubMed/NCBI
|
|
14
|
Lacroix M: Persistent use of ‘false’ cell
lines. Int J Cancer. 122:1–4. 2008.
|
|
15
|
Masters JR: Cell-line authentication: end
the scandal of false cell lines. Nature. 492:1862012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Masters J: False cell lines.
Carcinogenesis. 23:3712002. View Article : Google Scholar
|
|
17
|
Masters JR: False cell lines: the problem
and a solution. Cytotechnology. 39:69–74. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Azari S, Ahmadi N, Tehrani MJ and Shokri
F: Profiling and authentication of human cell lines using short
tandem repeat (STR) loci: Report from the National Cell Bank of
Iran. Biologicals. 35:195–202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
American Type Culture Collection Standards
Development Organization Workgroup ASN-0002. Cell line
misidentification: the beginning of the end. Nat Rev Cancer.
10:441–448. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barallon R, Bauer SR, Butler J, et al:
Recommendation of short tandem repeat profiling for authenticating
human cell lines, stem cells, and tissues. In Vitro Cell Dev Biol
Anim. 46:727–732. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shamloula MM, El-Shorbagy SH and Saied EM:
P63 and cytokeratin8/18 expression in breast, atypical ductal
hyperplasia, ductal carcinoma in situ and invasive duct carcinoma.
J Egypt Natl Cancer Inst. 19:202–210. 2007.PubMed/NCBI
|
|
22
|
Raspollini MR, Comin CE, Crisci A and
Chilosi M: The use of placental S100 (S100P), GATA3 and napsin A in
the differential diagnosis of primary adenocarcinoma of the bladder
and bladder metastasis from adenocarcinoma of the lung.
Pathologica. 102:33–35. 2010.PubMed/NCBI
|
|
23
|
Esheba GE, Longacre TA, Atkins KA and
Higgins JP: Expression of the urothelial differentiation markers
GATA3 and placental S100 (S100P) in female genital tract
transitional cell proliferations. Am J Surg Pathol. 33:347–353.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saghravanian N, Mohtasham N and Jafarzadeh
H: Comparison of immunohistochemical markers between adenoid cystic
carcinoma and polymorphous low-grade adenocarcinoma. J Oral Sci.
51:509–514. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Philip S, Bulbule A and Kundu GC: Matrix
metalloproteinase-2: mechanism and regulation of NF-kappaB-mediated
activation and its role in cell motility and ECM-invasion.
Glycoconj J. 21:429–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garavello W, Maggioni D, Nicolini G, Motta
L, Tredici G and Gaini R: Association between metalloproteinases 2
and 9 activity and ERK1/2 phosphorylation status in head and neck
cancers: An ex vivo study. Oncol Rep. 24:1073–1078.
2010.PubMed/NCBI
|