Introduction
Breast cancer is the most common cancer among women
worldwide and is a highly heterogeneous disease. Therefore, there
is a pressing need for methods with which to stratify patients into
the different risk groups more accurately than the current
clinicopathological classifications (1–4). A
molecular-based approach to classify breast tumours was first
proposed by Sørlie et al(5).
In this study, breast carcinomas were clustered based on gene
expression profiles determined using DNA microarrays. Breast
tumours were divided into luminal A [estrogen receptor-positive
(ER+) and/or progesterone receptor-positive
(PR+)/human epidermal growth factor receptor 2-negative
(HER2−)], luminal B (ER+ and/or
PR+/HER2+), basal-like, HER2+ and
normal-like breast cancer. These subtypes are associated with
distinct prognosis and treatment options. Immunohistochemical-based
molecular classifications have also been proposed alternatively
(6–8). Immunohistochemistry (IHC) has been
demonstrated to be an efficient and acceptable surrogate of gene
expression analysis (6,9–11).
Several lines of evidence have confirmed that the subclassification
of ER+ cancers and the prognostic value of gene
signatures is largely driven by the expression levels of
proliferation-related genes and that proliferation markers, such as
Ki67, may provide equivalent prognostic information to that
provided by gene signatures. In particular, according to the new
St. Gallen consensus recommendations, Ki67 is one of the prognostic
markers that is considered important to subclassify luminal A and
luminal B, together with HER2 expression (12). The absence of ER, PR and HER2 is
used to define the triple-negative subtype, which represents ~15%
of breast tumours and is not a homogeneous entity (13–15).
Thus, new prognostic and/or predictive factors may provide
additional risk stratifications to better guide treatment decisions
in these different subtypes of breast cancer.
Survivin (also called baculoviral inhibitor of
apoptosis repeat-containing 5, BIRC5) is a member of the family of
inhibitor of apoptosis proteins (IAP) and is a multifunctional
protein implicated in a number of cellular processes, including
apoptosis, mitosis and angiogenesis (16). Survivin is present during fetal
development and is rarely detectable, but is sometimes present in
terminally differentiated normal adult tissues (17–19).
Importantly, survivin is abundantly expressed in most types of
cancers, including breast, colorectal, lung, gastric, bladder and
liver cancer, melanoma and malignant lymphoma (20). The incidence of survivin expression
in cancer is reported to range from 30 up to 100% (21). High survivin expression is
associated with poor prognosis in most human cancers. Although it
exhibits a high degree of tumour-specific expression and is one of
the 16 cancer-related genes included in the Oncotype DX assay
(22), the role of survivin as a
breast cancer biomarker has remained the subject of much debate.
Previous studies using quantitative reverse
transcription-polymerase chain reaction (qRT-PCR) and IHC have
reported that survivin is either prognostically irrelevant or
associated with improved or adverse outcome in primary breast
cancer patients (23–25). Such discordant results could perhaps
be explained by the fact that these studies did not account for
subcellular localization of survivin, which can be present in both
nuclear and cytoplasmatic pools. These different pools are
immunochemically and functionally different and are independently
modulated during the cell cycle (26). Furthermore, recent studies have
demonstrated that increased expression of nuclear, as opposed to
cytoplasmic, survivin was associated with decreased overall
survival (OS) and breast cancer-specific survival (BCSS) (27,28).
Testin (TES) is a putative tumour suppressor. The
human TES gene is localised to the fragile site FRA7G at 7q31.2,
and downregulation of TES has been reported in many human
malignancies (29–33). In addition, a profound reduction in
growth potential was detected in different cancer cell lines in
which TES was overexpressed (34,35).
TES is a highly conserved protein of 421 amino acids containing
three C-terminal LIM domains, which are responsible for
protein-protein interactions coordinating intracellular and
extracellular pathways. In particular, TES is a component of the
focal adhesion complex, which is important in the regulation of
epithelial physiology and localises to cell-matrix adhesions,
cell-cell contacts and actin stress fibres. In mice, TES interacts
and colocalises with a variety of cytoskeletal proteins, including
zyxin, mena, VASP, talin and actin (36,37).
Overexpression of TES decreased cell motility (36–38).
Moreover, restoration of TES expression in breast cancer and
uterine sarcoma cell lines inhibited their growth by induction of
apoptosis (34). In association
with alterations of cell adhesion and motility, TES expression
resulted in activation of caspase-dependent and -independent
apoptosis in the absence or with a reduced level of survivin
(34).
Expression of TES and its relationship with survivin
have never been evaluated in a large series of human breast
tumours. The aim of this study was to determine whether TES and
survivin expression could characterise the different breast cancer
subtypes and their correlation with clinicopathological
parameters.
Materials and methods
Patient samples
The study was carried out on 242 consecutive cases
of breast carcinomas that were obtained from the Cantonal Institute
of Pathology (Locarno, Switzerland). The study was approved by the
Cantone Ticino Ethics Committee. All cases were diagnosed during
the period from January 1981 to December 2009 with a median
follow-up time of 5.2 years (SD, 3.4 years). The median age at
diagnosis was 54.4 (SD, 12.0). The histological diagnosis was
determined during routine pathological assessment. The tumours were
graded according to the Scarff-Bloom Richardson classification as
modified by Elston and Ellis (39).
Staging at the time of diagnosis was based on the TNM system
(40). The clinicopathological
characteristics of the patients are listed in Table I. All patients underwent surgery ±
radiotherapy and systemic standard treatment. Survival data,
including disease-free survival (DFS) and BCSS, were maintained on
a prospective basis. DFS was defined as the interval (in months)
from the date of the primary surgical treatment to the first
loco-regional or distant recurrence and BCSS was defined as the
time (in months) from the date of the primary surgical treatment to
the time of death from breast cancer.
 | Table IClinicopathological data of the
patient cohort. |
Table I
Clinicopathological data of the
patient cohort.
| Variables | Total (n=242)
(%) |
|---|
| Age (years, mean ±
SD) | 54.6±12.0 |
| Histologic
subtype |
| Ductal | 219 (90.5) |
| Lobular | 23 (9.5) |
| Lymph node
status |
| Negative | 134 (55.4) |
| Positive | 104 (42.9) |
| Unknown | 4 (1.7) |
| Histologic
grade |
| I | 32 (13.2) |
| II | 122 (50.4) |
| III | 87 (36.0) |
| Unknown | 1 (0.4) |
| Tumour stage |
| 0 | 7 (2.9) |
| I | 92 (38.0) |
| II | 111 (45.9) |
| III | 28 (11.6) |
| Unknown | 4 (1.7) |
| ER |
| <10% | 22 (9.1) |
| ≥10% | 220 (90.9) |
| PR |
| <10% | 85 (35.1) |
| ≥10% | 157 (64.9) |
| Ki67 |
| <10% | 66 (27.3) |
| ≥10% | 172 (71.1) |
| Unknown | 4 (1.7) |
| HER2 IHC |
| Negative/moderate
(0–2+) | 208 (85.9) |
| Strong (3+) | 27 (11.2) |
| Unknown | 7 (2.9) |
| Event |
| NED | 197 (81.4) |
| Local
recurrence | 10 (4.1) |
| Metastasis | 15 (6.2) |
| Contralateral
tumour | 12 (5.0) |
| Death | 8 (3.3) |
Tumour classification
Tumours were classified according to standard
molecular subtypes as follows: luminal A type (154/242, 63.6%),
luminal B type (39/242, 16.1%; in particular 24/242, 9.9%, with
both ER and PR positivity), HER2 overexpression type (7/242, 2.9%),
and triple-negative type (35/242, 14.5%).
Immunohistochemistry
Sections (3-μm) were cut from formalin-fixed
paraffin-embedded (FFPE) blocks and mounted on positive-charged
slides. Immunostaining was performed using anti-survivin rabbit
polyclonal antibody (1:250, cat. ab469) and anti-TES mouse
monoclonal antibody (1:200, cat. ab57292; both from Abcam,
Cambridge, MA, USA). The specificity of both antibodies was
previously confirmed by western blot analysis. Cell nuclei were
counterstained with hematoxylin solution. Slides were evaluated by
at least two investigators in a blinded manner. Positive samples
for each antibody and negative samples, in which the primary
antibody was omitted, were used as controls. Adjacent normal breast
tissues in most samples served as the internal negative or positive
control depending on the protein tested.
Data analysis
Immunostaining for survivin was recorded according
to staining intensity, distribution in the cytoplasm and/or nucleus
and the percentage of positive tumour cells. In cases where
staining was heterogeneous in the slide, examined fields included
those with the highest and lowest percentage of stained cells. A
mean percentage of positive tumour cells was determined in at least
five areas at a magnification of ×400. A tumour was assessed as
survivin-positive if the staining was positive-cytoplasmatic,
positive-nuclear or both. Staining was scored as follows: score 0,
no staining or staining in <5% of cells; score 1, weak staining
in 6–19% of cells; score 2, moderate staining in 20–40% of cells;
score 3, strong staining in >40% of cells. For statistical
analysis, scores 0 and 1 were considered negative, and scores 2 and
3 were considered positive for both cytoplasmatic and nuclear
staining. Immunostaining for cytoplasmatic TES was scored as
follows: score 0, no staining or staining in 2% of cells; score 1,
3–40% positively stained cells (weak staining); score 2, 41–65%
positively stained cells (moderate staining); score 3, >65%
positively stained cells (strong staining).
Data on ER, PR, Ki67 and HER2/neu were obtained
through standard clinical testing, using IHC for ER and PR and the
Hercep Test™ (Dako, Glostrup, Denmark) for HER2/neu. HER2 staining
was divided into two groups, with negative to moderate (0–2+) HER2
expression and strong (3+) overexpression. Cases scoring 2+ for
HER2 immunostaining were subsequently assessed by fluorescence
in situ hybridisation study (FISH).
Statistical analysis
The statistical analysis was carried out using the
semi-quantitative results of the immunohistochemical staining. A
univariate statistical analysis was carried out using Chi-Square
test and odds ratios (ORs) to analyse the categorical variables and
the Student’s t-test for independent samples to analyse continuous
variables. The relationship between ‘survivin overexpression and
TES downregulation’ with ‘triple-negative phenotype’ was also
studied in the multivariate analysis, taking into account the
effect of the confounding variable ‘histological grade’, using a
binary logistic model. Continuous data were tested for normality.
The analysis of time to event was performed using Kaplan-Meier
methodology. Statistical significance was defined as a value of
P=0.05, two-tailed. All statistical analyses were performed using
PASW Statistics 19 (formerly SPSS 19).
Results
Expression of survivin and TES in breast
tissues
Cytoplasmatic or nuclear survivin expression was
detected in 97% of the breast carcinomas. No expression of survivin
was observed in the adjacent normal breast tissues, with the
exception of a few samples where the normal breast tissues showed
cytoplasmic positivity. The above percentage was within the range
of previously published studies (17,18,41).
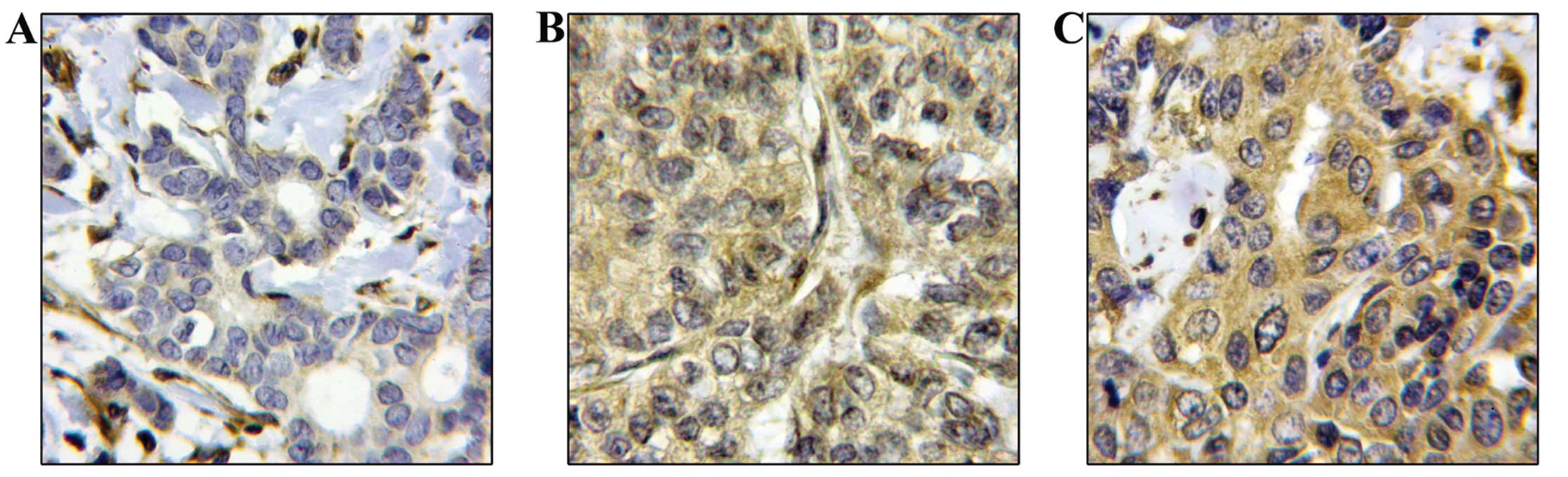
Positive nuclear staining of survivin was found in 9/242 of the
breast tumours (3.7%; score 2 and 3), while positive cytoplasmic
staining was present in 22.3% of the cases (54/242). In 172 out of
242 (71%) patients there was positive staining both in the nucleus
and in the cytoplasm (Fig. 1).
These results are in accordance with previous studies (42).
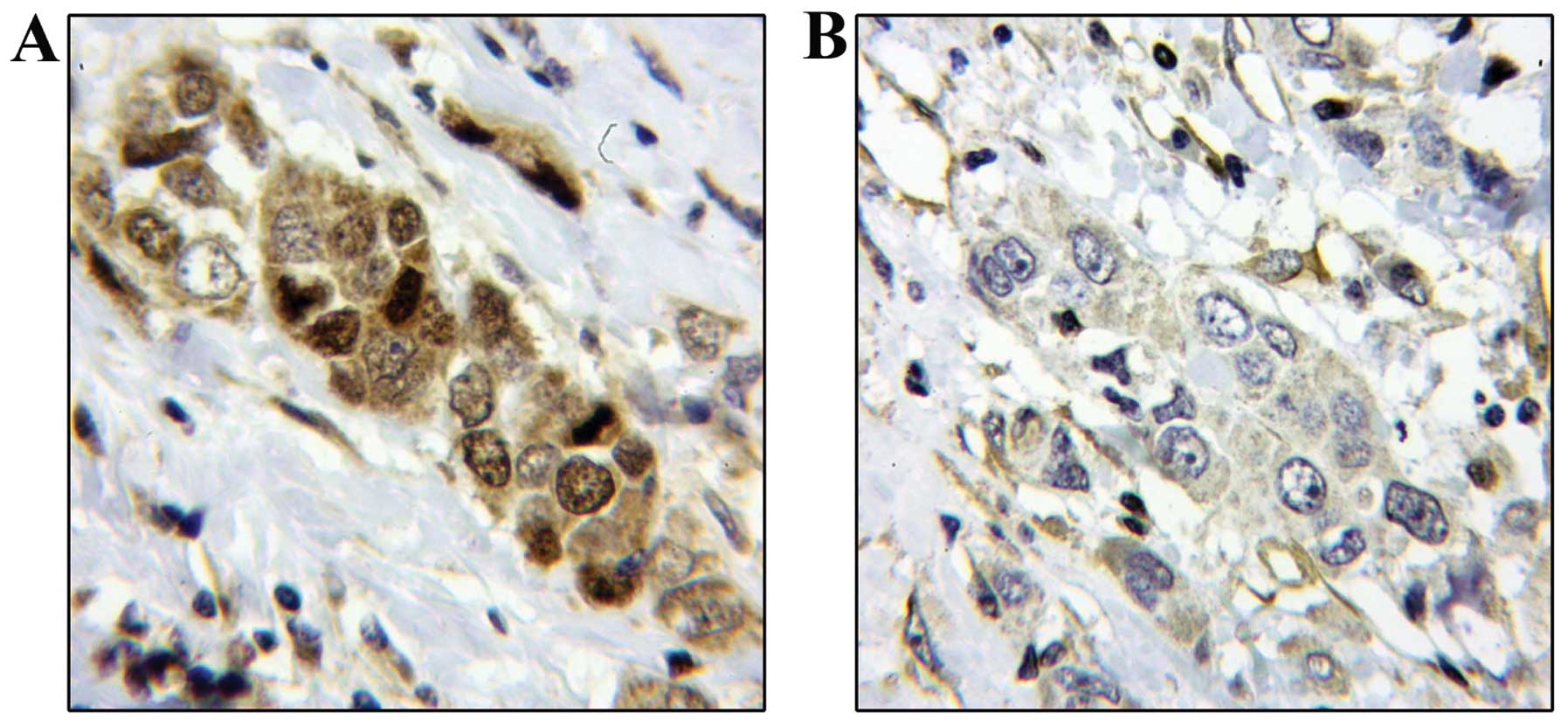
Reduced TES expression was found in 74.7% of the
cases (181/242). Tumours were divided into three different
categories: no staining, weak staining and moderate staining (score
0, 1 and 2, respectively) (Fig. 2).
The adjacent normal breast tissues with surrounding mesenchymal and
endothelial cells showed specific immunoreactivity and represented
an internal positive control for TES antibody specificity.
Correlation between survivin and TES
expression and clinical outcome of breast cancer patients
From a clinical perspective, we assessed the
correlation between survivin and TES expression and
clinicopathological parameters in the primary breast cancers
(Table II). TES status, alone and
in association with nuclear expression of survivin, was
significantly correlated with increased histological grade, being
predominantly present in grade III tumours (P=0.004 and 0.003,
respectively). There was also a statistically significant
correlation between high nuclear survivin expression (>40% of
tumour cells) and the presence of lymph node metastases (P=0.09).
There was no significant association between nuclear and/or
cytoplasmic survivin expression, TES expression and age of
patients, histological type (ductal or lobular) and HER2 status.
However, there was a trend toward a significant association between
absent or weak expression of TES and Ki67 expression (P=0.055). The
results of the χ2 analysis of these data are summarised
in Table II.
 | Table IIRelationship between survivin and TES
expression and standard clinicopathological and immunohistochemical
markers. |
Table II
Relationship between survivin and TES
expression and standard clinicopathological and immunohistochemical
markers.
| Parameter | Nuclear survivin
high expression (score 2–3)
n (%) | Chi-square
P-value | TES
negative/low/reduced expression (score 0–2)
n (%) | Chi-square
P-value | Association of
survivin (score 2–3) and TES (score 0–2)
n (%) | Chi-square
P-value |
|---|
| Grade | | | | | | |
| 1 | 15/32 (46.9) | 0.097 NS | 19/32 (59.4) | 0.004 | 13/32 (40.6) | 0.003 |
| 2 | 60/122 (49.2) | | 87/122 (71.3) | | 67/122 (54.9) | |
| 3 | 45/87 (51.7) | | 75/87 (86.2) | | 63/87 (72.4) | |
| Histological nodal
status | | | | | | |
| Negative | 57/134 (42.5) | 0.009 | 54/134 (40.3) | 0.224 NS | 78/134 (58.2) | 0.814 NS |
| Positive | 62/104 (59.6) | | 51/104 (49) | | 63/104 (60.6) | |
| Ki67 | | | | | | |
| <10 | 45/66 (68.2) | 0.146 NS | 22/66 (33.3) | 0.055 NS | 33/66 (50) | 0.087 NS |
| ≥10 | 133/172 (77.3) | | 81/172 (47.1) | | 107/172 (62.2) | |
On account of the short median follow-up time of 5.2
years, Kaplan-Meier survival analysis did not reveal any
association between survivin and TES expression and BCSS or DFS.
However, we found a higher percentage of events (i.e. local
recurrence, distant metastases and death) in cases with positive
nuclear survivin expression (22.2 vs. 11.3%, P=0.039) and low TES
expression (23.8 vs. 14.6%, P=0.068) (Tables I and III).
 | Table IIICorrelation of survivin and TES
expression with event incidence. |
Table III
Correlation of survivin and TES
expression with event incidence.
| Survivin high
expression (score 2–3)
n (%) | Survivin negative
or low expression (score 0–1)
n (%) | Chi-square
P-value | TES negative/low
expression (score 0–1)
n (%) | TES moderate/strong
expression (score 2–3)
n (%) | Chi-square
P-value |
|---|
| Event
incidence | 36/162 (22.2) | 9/80 (11.3) | 0.039 | 25/105 (23.8) | 20/137 (14.6) | 0.068 NS |
| NED | 126/162 (77.8) | 71/80 (88.8) | | 80/105 (76.2) | 117/137 (85.4) | |
Association of survivin and TES
expression with breast cancer subtypes
The prognostic implications of breast cancer
subtypes have been described in several reports. We found a
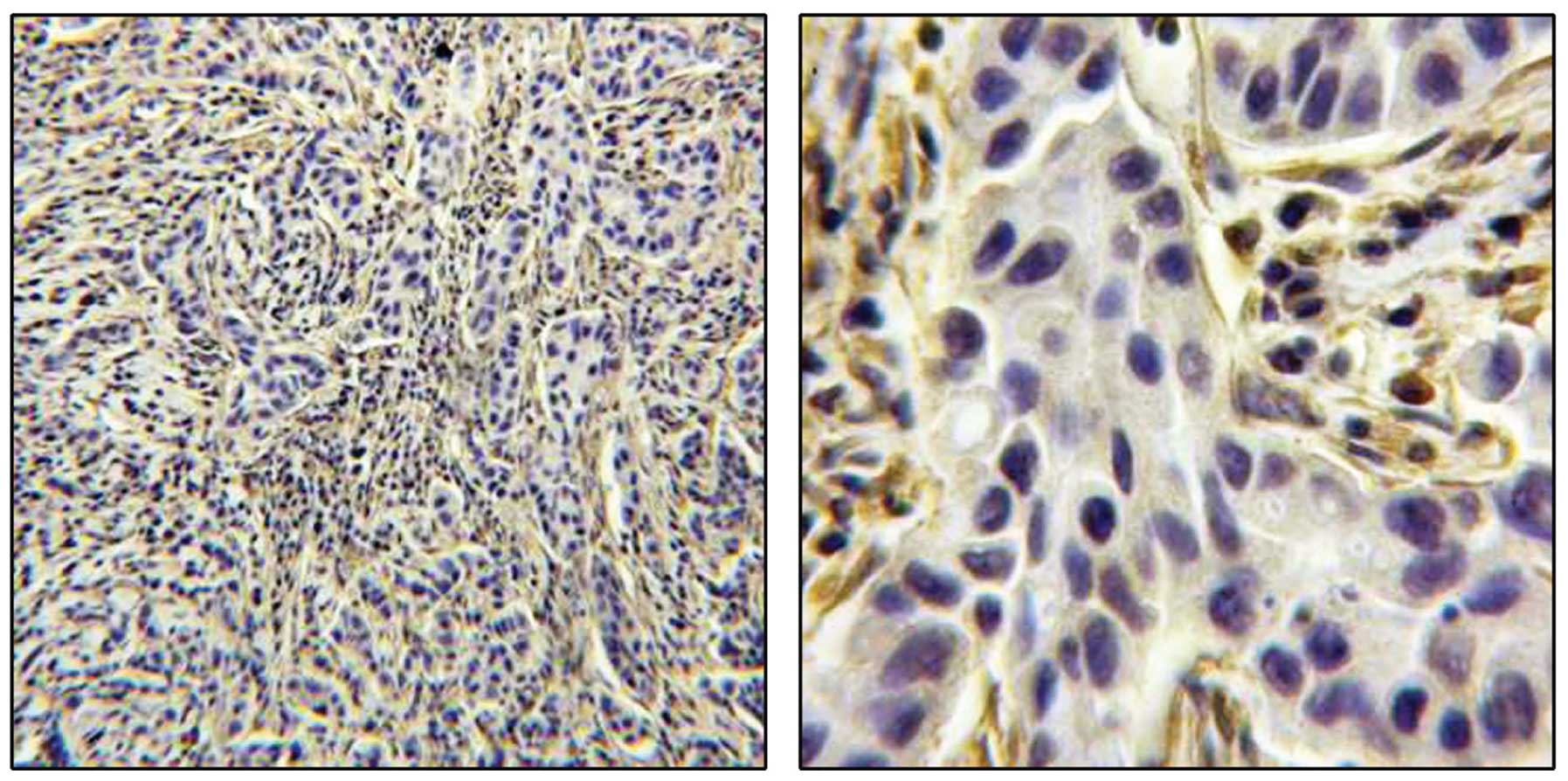
statistically significant association between the subcellular
localization of survivin (moderate/strong nuclear staining with or
without cytoplasmic staining), reduced TES expression (score 0–2)
and the triple-negative breast cancer subtype (P=0.009) (univariate
OR, 3.20; 95% CI, 1.34–7.66) (Fig.
3). There was also a significant association between nuclear
survivin expression and the triple-negative subtype (P=0.022)
(univariate OR, 4.15; 95% CI, 1.22–14.1). A multivariate analysis
based on a binary logistic model was carried out, using the
variable ‘survivin overexpression and TES downregulation’ as the
dependent one and ‘histological grade’ as well as ‘triple-negative
phenotype’ as covariates. The histological grade was dichotomised
into high (grade 2 and 3) and low (grade 1). Triple-negative
phenotype exhibited a statistically significant result [P=0.018;
OR, 2.90; 95% CI, 1.2–6.97 (reference group = no triple
phenotype)], while the histological grade exhibited borderline
significance [P=0.051; OR, 2.15; 95% CI, 0.997–4.62 (reference
group = low histological grade)]. These results showed a
significant association between the triple-negative phenotype and
survivin overexpression and TES downregulation, independently of
the histological grade. Furthermore, there was a significant
correlation between the absence or low expression of TES
(immunohistochemical score 0–1) and the luminal B subtype with
ER+ and PR+ expression (P=0.019) (univariate
OR, 2.9; 95% CI, 1.19–7.06), independently of the histological
grade (adjusted multivariate OR, 2.67; 95% CI, 1.09–6.65; P=0.032)
(Fig. 4). These data are summarised
in Table IV. Instead, there was no
significant association between cytoplasmic and/or nuclear survivin
expression, absence or downregulation of TES and the luminal A and
HER2 subtypes.
 | Table IVCorrelation between survivin and TES
expression and breast cancer molecular subtypes. |
Table IV
Correlation between survivin and TES
expression and breast cancer molecular subtypes.
Subtype
n (%) | Nuclear survivin
high expression (score 2–3)
n (%) | Fisher test
P-value | TES
negative/low/reduced expression (score 0–2)
n (%) | Fisher test
P-value | Association of
survivin (score 2–3) and TES (score 0–2)
n (%) | Fisher test
P-value |
|---|
Triple-negative
35/242 (14.89) | 32/35
(91.4)
vs.
149/207 (72.0) | 0.012 | 29/35
(82.9)
vs.
152/207 (73.4) | 0.295 NS | 28/35
(80.0)
vs.
115/207 (55.6) | 0.009 |
Univariate
OR=4.15
95% CI, 1.22–14.1 | 0.022 |
Univariate
OR=1.75
95% CI, 0.69–4.44 | 0.24 NS |
Univariate
OR=3.20
95% CI, 1.34–7.66 | 0.009 |
Adjusteda
OR=4.07
95% CI, 1.19–13.88 | 0.025 | Adjusteda
OR=1.54
95% CI, 0.60–3.95 | 0.37 NS | Adjusteda
OR=2.90
95% CI, 1.20–6.97 | 0.018 |
Luminal B
24/242 (9.9) | 17/24
(70.8)
vs.
164/218 (75.2) | 0.63 NS | 16/24
(66.6)
vs.
89/218 (40.8) | 0.018 | 15/24
(62.5)
vs.
128/218 (58.7) | 0.83 NS |
Univariate
OR=0.80
95% CI, 0.32–2.03 | 0.64 NS |
Univariate
OR=2.90
95% CI, 1.19–7.06 | 0.019 |
Univariate
OR=1.17
95% CI, 0.49–2.80 | 0.72 NS |
Adjusteda
OR=0.77
95% CI, 0.30–1.98 | 0.59 NS | Adjusteda
OR=2.67
95% CI, 1.09–6.65 | 0.032 | Adjusteda
OR=1.06
95% CI, 0.44–2.55 | 0.90 NS |
Discussion
Advances in high-throughput methodologies have
revolutionised the scientific approach to highly complex diseases.
Breast cancer subtypes have been extensively characterised by gene
expression analysis using microarrays. However, this approach is
not feasible for large-scale clinical applications or retrospective
studies using FFPE tissue samples. In these situations
immunohistochemical staining for specific biomarkers provides a
useful alterative. In the present study, we showed that decreased
expression of TES and increased levels of nuclear survivin were
preferentially associated with the triple-negative subtype. This
subtype generally presents high histological grade, Ki67
overexpression and unfavourable prognosis.
Survivin is a bifunctional protein, which is both an
integral component of the chromosome passenger complex and a
negative regulator of apoptosis. Survivin exists in distinct
intracellular pools. The predominant cytosolic fraction and a
smaller nuclear pool are independently modulated during cell cycle
progression and control the assembly of a normal bipolar mitotic
apparatus (43). More importantly,
cytoplasmic localisation of survivin in non-malignant cells
suppresses apoptosis, while nuclear translocation may be important
to regulate proliferation (44).
Survivin intracellular localisation is regulated by an active and
evolutionarily conserved, Crm1-dependent nuclear export signal,
which appears to be essential for survivin tumour-promoting
functions. In particular, inhibition of this signal abrogates the
anti-apoptotic effect of survivin, while maintains its mitotic
activity. This suggests that increased levels of nuclear survivin
lead to a proliferative aggressive phenotype (28,45,46).
However, the exact prognostic and clinical implications of the
nuclear or cytoplasmatic localisation of survivin remain
controversial. Here, we found that nuclear survivin is a predictor
of worse outcome in breast cancer and a strong association between
nuclear survivin and the triple-negative breast cancer subtype.
Based on the results of our previous study on TES in
breast cancer cell lines (34), we
assessed the pattern of TES expression in breast tumours to
determine whether reduced expression of TES would be preferentially
associated with specific tumour subgroups. We found that TES was
expressed in ducts and lobules of normal breast. In particular, TES
was present in epithelial, myoepithelial and basal cells in normal
tissues. TES was significantly reduced in tumour cells in a large
fraction of the breast cancers. In some breast tumours with
adjacent normal tissues with hyperplastic foci, TES was detected
only in the normal myoepithelial and basal cells. Negative
expression of TES in the columnar cells is likely to be the result
of dysplastic transformation of the breast epithelium. However, the
significance of this particular localisation warrants
investigation, as it suggests that the pattern of expression of TES
is more complex than originally believed. TES was also abundantly
expressed in the stroma and endothelial cells (47) surrounding both normal and tumour
tissues. Remarkably, we found a significant correlation between TES
downregulation, together with nuclear survivin expression, and the
triple-negative subtype. In contrast, regardless of survivin,
negative or very weak expression of TES was strongly correlated
with the luminal B subtype, ER+ and PR+
tumours.
Tumourigenesis is a multistep process resulting from
the accumulation of genetic and epigenetic changes (48,49).
DNA methylation, a major epigenetic modification, leads to gene
silencing. The frequency of hypermethylation of CpG dinucleotides
varies significantly between breast cancer subtypes (50). CpG islands were found to be more
frequently methylated in luminal B tumours than in the other tumour
subtypes (50). Furthermore,
depending on ER status and irrespective of the molecular subtype, a
higher methylation frequency was observed in ER+ and
PR+ tumours (50). The
human TES gene is located in the fragile chromosomal region FRA7G.
Common fragile sites are regions in mammalian chromosomes prone to
breakage and rearrangements. This genetic instability can lead to
disease manifestations and may play a role in oncogenesis (51). FRA7G is a locus of 300 kb, localised
between markers D7S486 and D7S522, which shows loss of
heterozygosity in many human malignancies (52,53).
This region is known to encompass several genes, in addition to
TES, including caveolin-1, caveolin-2 (54) and MET (55). The methylation of CpG in the TES
promoter is a frequent event in gastric tumours (32). In previous studies, methylation of
the TES promoter was also shown to be common in breast cancer
(30,34) and may be involved in TES
downregulation. The different correlation of TES in regards to
triple-negative and luminal B subtypes could be linked to a
different grade of hypermethylation of CpG islands in the TES
promoter region (50).
TES is an important structural protein and may serve
as a platform to integrate multiple signal transduction events.
Current data suggest the possibility that downregulation of TES is
associated with alterations in cell adhesion and motility and
therefore can lead to development of tumours with an aggressive
phenotype. The reduced expression of TES in tumours of the
basal-like/triple-negative subtype, along with its expression in
myoepithelial/basal cells of the normal breast, can lead to
speculate a possible role in epithelial-to-mesenchymal transition
(EMT). EMT is an important process associated with the ability of
epithelial cells to detach from a primary tumour and metastasise.
It is also possible that the tumour-suppressive function of TES may
reside within alternative, yet unknown functions. A possible
function of TES in survivin-dependent pathways may stem from
maintenance of a basal/myoepithelial phenotype in
basal-like/triple-negative breast cancer, as it has been noted for
caveolin 1 and 2 (56,57).
Reduced expression of TES characterises breast
cancer subtypes with particularly poor outcome such as
triple-negative and luminal B tumours, and therefore can be
considered an important marker to aid in predicting the course of
disease, either by itself or in association with established
markers, such as survivin. Further studies generating long-term
follow-up data are warranted to confirm the usefulness of TES as a
biomarker in breast cancer. Furthermore, a greater understanding of
the molecular and functional role of TES in aggressive types of
breast cancer may lead to more selective and effective treatment
for breast cancer patients.
Acknowledgements
We thank Professor Luca Mazzucchelli, Dr Milo
Frattini and Dr Jessica Barizzi for the FFPE tissue sections of
human breast cancer and for their advice in the immunohistochemical
analysis. This study was supported by the Fondazione Ticinese per
la Ricerca sul Cancro (Lugano, Switzerland).
References
|
1
|
Span PN, Sweep FC, Wiegerinck ET,
Tjan-Heijnen VC, Manders P, Beex LV and deKok JB: Survivin is an
independent prognostic marker for risk stratification of breast
cancer patients. Clin Chem. 50:1986–1993. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
The International Agency for Research on
Cancer (IARC). Pathology and Genetics: Tumours of the Breast and
Female Genital Organs. World Health Organization Classification of
Tumours. 4. 3rd edition. Tavassoli FA and Devilee P: IARC Press;
Lyon: 2003
|
|
3
|
Page DL: Special types of invasive breast
cancer, with clinical implications. Am J Surg Pathol. 27:832–835.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weigelt B, Horlings HM, Kreike B, Hayes
MM, Hauptmann M, Wessels LF, de Jong D, Van de Vijver MJ, Van’t
Veer LJ and Peterse JL: Refinement of breast cancer classification
by molecular characterization of histological special types. J
Pathol. 216:141–150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE
and Børresen-Dale AL: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001.PubMed/NCBI
|
|
6
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M and Perou
CM: Immunohistochemical and clinical characterization of the
basal-like of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Banerjee S, Reis-Filho JS, Ashley S,
Steele D, Ashworth A, Lakhani SR and Smith IE: Basal-like breast
carcinomas: clinical outcome and response to chemotherapy. J Clin
Pathol. 59:729–735. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheang MC, Voduc D, Bajdik C, Leung S,
McKinney S, Chia SK, Perou CM and Nielsen TO: Basal-like breast
cancer defined by five biomarkers has superior prognostic value
than triple-negative phenotype. Clin Cancer Res. 14:1368–1376.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Laurinavicius A, Laurinaviciene A,
Ostapenko V, Dasevicius D, Jarmalaite S and Lazutka J:
Immunohistochemistry profiles of breast ductal carcinoma: factor
analysis of digital image analysis data. Diagn Pathol. 7:272012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS
and Millikan RC: Race, breast cancer subtypes, and survival in the
Carolina Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morrison DH, Rahardja D, King E, Peng Y
and Sarode VR: Tumour biomarker expression relative to age and
molecular subtypes of invasive breast cancer. Br J Cancer.
107:382–387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Geyer FC, Rodrigues DN, Weigelt B and
Reis-Filho JS: Molecular classification of estrogen
receptor-positive/luminal breast cancer. Adv Anat Pathol. 19:39–53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bertucci F, Finetti P, Cervera N, Esterni
B, Hermitte F, Viens P and Birnbaum D: How basal are
triple-negative breast cancers? Int J Cancer. 123:236–240. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de Ruijter TC, Veeck J, de Hoon JP, van
Engeland M and Tjan-Heijnen VC: Characteristics of triple-negative
breast cancer. J Cancer Res Clin Oncol. 137:183–192.
2011.PubMed/NCBI
|
|
15
|
Nakagawa M, Bando Y, Nagao T, Takai C,
Ohnishi T, Honda J, Moriya T, Izumi K, Takahashi M, Tangoku A and
Sasa M: Among triple-negative breast cancers, HER2 (0) breast
cancer shows a strong tendency to be basal-like compared with HER2
(1+) breast cancer: preliminary results. Breast Cancer. 19:54–59.
2012.PubMed/NCBI
|
|
16
|
Duffy MJ, O’Donovan N, Brennan DJ,
Gallagher WM and Ryan BM: Survivin: a promising tumor biomarker.
Cancer Lett. 249:49–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adamkov M, Halasova E, Kajo K, Machalekova
K, Vybohova D, Varga I and Rajcany J: Survivin: a promising
biomarker in breast carcinoma. Neoplasma. 57:572–577. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nassar A, Sexton D, Cotsonis G and Cohen
C: Survivin expression in breast carcinoma: correlation with
apoptosis and prognosis. Appl Immunohistochem Mol Morphol.
16:221–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dallaglio K, Marconi A and Pincelli C:
Survivin: a dual player in healthy and diseased skin. J Invest
Dermatol. 132:18–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, espressed in cancer and
lynphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nassar A, Lawson D, Cotsonis G and Cohen
C: Survivin and caspase-3 expression in breast cancer: correlation
with prognostic parameters, proliferation, angiogenesis, and
outcome. Appl Immunohistochem Mol Morphol. 16:113–120. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paik S, Shak S, Tang G, Kim C, Baker J,
Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher
ER, Wickerham DL, Bryant J and Wolmark N: A multigene assay to
predict recurrence of tamoxifen-treated, node-negative breast
cancer. N Engl J Med. 351:2817–2826. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chu JS, Shew JY and Huang CS:
Immunohistochemical analysis of survivin espression in primary
breast cancers. J Formos Med Assoc. 103:925–931. 2004.PubMed/NCBI
|
|
24
|
Kennedy SM, O’Driscoll L, Purcell R,
Fitz-Simons N, McDermott EW, Hill AD, O’Higgins NJ, Parkinson M,
Linehan R and Clynes M: Prognostic importance of survivin in breast
cancer. Br J Cancer. 88:1077–1083. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hinnis AR, Luckett JC and Walker RA:
Survivin is an independent predictor of short-term survival in poor
prognostic breast cancer patients. Br J Cancer. 96:639–645. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Caldas H, Jiang Y, Holloway MP, Fangusaro
J, Mahotka C, Conway EM and Altura RA: Survivin splice variants
regulate the balance between proliferation and cell death.
Oncogene. 24:1994–2007. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brennan DJ, Rexhepaj E, O’Brien SL,
McSherry E, O’Connor DP, Fagan A, Culhane AC, Higgins DG, Jirstrom
K, Millikan RC, Landberg G, Duffy MJ, Hewitt SM and Gallagher WM:
Altered cytoplasmic-to-nuclear ratio of survivin is a prognostic
indicator in breast cancer. Clin Cancer Res. 14:2681–2689. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rexhepaj E, Jirstrom K, O’Connor DP,
O’Brien SL, Landberg G, Duffy MJ, Brennan DJ and Gallagher WM:
Validation of cytoplasmic-to-nuclear ratio of survivin as an
indicator of improved prognosis in breast cancer. BMC Cancer.
10:6392010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gunduz E, Gunduz M, Beder L, Nagatsuka H,
Fukushima K, Sutcu R, Delibas N, Yamanaka N, Shimizu K and Nagai N:
Downregulation of TESTIN and its association with cancer history
and a tendency toward poor survival in head and neck squamous cell
carcinoma. Arch Otolaryngol Head Neck Surg. 135:254–260. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tatarelli C, Linnenbach A, Minori K and
Croce CM: Characterization of the human TESTIN gene localized in
the FRA7G region at 7q31.2. Genomics. 68:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weeks RJ, Kees UR, Song S and Morison IM:
Silencing of TESTIN by dense biallelic promoter methylation is the
most common molecular event in childhood acute lymphoblastic
leukaemia. Mol Cancer. 9:1632010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma H, Weng D, Chen Y, Huang W, Pan K, Wang
H, Sun J, Wang Q, Zhou Z, Wang H and Xia J: Extensive analysis of
D7S486 in primary gastric cancer supports TESTIN as a candidate
tumor suppressor gene. Mol Cancer. 9:1902010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Cicek MS, Plummer SJ, Jorgenson E,
Casey G and Witte JS: Association of testis derived transcript gene
variants and prostate cancer risk. J Urol. 177:894–898. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sarti M, Sevignani C, Calin GA, Aqeilan R,
Shimizu M, Pentimalli F, Picchio MC, Godwin A, Rosenberg A, Drusco
A, Negrini M and Croce CM: Adenoviral transduction of TESTIN gene
into breast and uterine cancer cell lines promotes apoptosis and
tumor reduction in vivo. Clin Cancer Res. 11:806–813.
2005.PubMed/NCBI
|
|
35
|
Tobias ES, Hurlstone AF, Mackenzie E,
McFarlane R and Black DM: The TES gene at 7q31.1 is methylated in
tumours and encodes a novel growth-suppressing LIM domain protein.
Oncogene. 20:2844–2853. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Coutts AS, MacKenzie E, Griffith E and
Black DM: TES is a novel focal adhesion protein with a role in cell
spreading. J Cell Sci. 116:897–906. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Griffith E, Coutts AS and Black DM: RNAi
knockdown of the focal adhesion protein TES reveals its role in
actin stress fibre organisation. Cell Motil Cytoskeleton.
60:140–152. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Griffith E, Coutts AS and Black DM:
Characterisation of chicken TES and its role in cell spreading and
motility. Cell Motil Cytoskeleton. 57:133–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I The value of histological
grade in breast cancer: experience from a large study with
long-term follow-up. Histopathology. 41:154–161. 2002.
|
|
40
|
Union Internationale Contre Cancer;
International Union Against Cancer. TNM Atlas. 5th edition.
Wittekind CH, Hutter R, Greene FL, Klimpfinger M and Sobin LH:
Springer-Verlag; Berlin: 2005, View
Article : Google Scholar
|
|
41
|
Jha K, Shukla M and Pandey M: Survivin
expression and targeting in breast cancer. Surg Oncol. 21:125–131.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Al-Joudi FS, Iskandar ZA, Hasnan J, Rusli
J, Kamal Y, Imran AK, Ahmed M and Zakaria J: Expression of survivin
and its clinicopathological correlations in invasive ductal
carcinoma of the breast. Singapore Med J. 48:607–614.
2007.PubMed/NCBI
|
|
43
|
Fortugno P, Wall NR, Giodini A, O’Connor
DS, Plescia J, Padgett KM, Tognin S, Marchisio PC and Altieri DC:
Survivin exists in immunochemically distinct subcellular pools and
is involved in spindle microtubule function. J Cell Sci.
115:575–585. 2002.PubMed/NCBI
|
|
44
|
Moon WS and Tarnawski AS: Nuclear
translocation of survivin in hepatocellular carcinoma: a key to
cancer cell growth? Hum Pathol. 34:1119–1126. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Knauer SK, Krämer OH, Knösel T, Engels K,
Rödel F, Kovács AF, Dietmaier W, Klein-Hitpass L, Habtemichael N,
Schweitzer A, Brieger J, Rödel C, Mann W, Petersen I, Heinzel T and
Stauber RH: Nuclear export is essential for the tumor-promoting
activity of survivin. FASEB J. 21:207–216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stauber RH, Mann W and Knauer SK: Nuclear
and cytoplasmic survivin: molecular mechanism, prognostic, and
therapeutic potential. Cancer Res. 67:5999–6002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Archacki SR, Angheloiu G, Moravec CS, Liu
H, Topol EJ and Wang QK: Comparative gene expression analysis
between coronary arteries and internal mammary arteries identifies
a role for the TES gene in endothelial cell functions relevant to
coronary artery disease. Hum Mol Genet. 21:1364–1373. 2012.
View Article : Google Scholar
|
|
48
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33:245–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dworkin AM, Huang TH and Toland AE:
Epigenetic alterations in breast: implications for breast cancer
detection, prognosis and treatment. Semin Cancer Biol. 19:165–171.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Holm K, Hegardt C, Staaf J,
Vallon-Christersson J, Jönsson G, Olsson H, Borg A and Ringnér M:
Molecular subtypes of breast cancer are associated with
characteristic DNA methylation patterns. Breast Cancer Res.
12:R362010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hellman A, Zlotorynski E, Scherer SW,
Cheung J, Vincent JB, Smith DI, Trakhtenbrot L and Kerem B: A role
for common fragile site induction in amplification of human
oncogenes. Cancer Cell. 1:89–97. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang H, Qian J, Proffit J, Wilber K,
Jenkins R and Smith DI: FRA7G extends over a broad region:
coincidence of human endogenous retroviral sequences (HERV-H) and
small polydispersed circular DNAs (spcDNA) and fragile sites.
Oncogene. 16:2311–2319. 1998. View Article : Google Scholar
|
|
53
|
Huang H, Qian C, Jenkins RB and Smith DI:
Fish mapping of YAC clones at human chromosomal band 7q31.2:
identification of YACS spanning FRA7G within the common region of
LOH in breast and prostate cancer. Genes Chromosomes Cancer.
21:152–159. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Elgelmann JA, Zhang XL and Lisanti MP:
Genes encoding human caveolin-1 and -2 are co-localized to the
D7S522 locus (7q31.1), a known fragile site (FRA7G) that is
frequently deleted in human cancers. FEBS Lett. 436:403–410. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lin JC, Scherer SW, Tougas L, Traverso G,
Tsui LC, Andrulis IL, Jothy S and Park M: Detailed deletion mapping
with a refined physical map of 7q31 localizes a putative tumor
suppressor gene for breast cancer in the region of MET. Oncogene.
13:2001–2008. 1996.PubMed/NCBI
|
|
56
|
Elsheikh SE, Green AR, Rakha EA, Samaka
RM, Ammar AA, Powe D, Reis-Filho JS and Ellis IO: Caveolin 1 and
Caveolin 2 are associated with breast cancer basal-like and
triple-negative immunophenotype. Br J Cancer. 99:327–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mercier I, Bryant KG, Sotgia F, Bonuccelli
G, Witkiewicz AK, Dasgupta A, Jasmin JF, Pestell RG and Lisanti MP:
Using Caveolin-1 epithelial immunostaining patterns to stratify
human breast cancer patients and predict the Caveolin-1 (P132L)
mutation. Cell Cycle. 8:1396–1401. 2009. View Article : Google Scholar
|