Introduction
Hepatocellular carcinoma (HCC) is a common, highly
invasive malignant tumor associated with a high mortality rate. It
is the third leading cause of cancer-related mortality worldwide
(1) and the highest incidence rates
are reported in East Asia (2,3).
Recurrence, metastasis and the development of new primary tumors
are the most common causes of mortality among patients with HCC
(4). The mainstay of therapy is
surgical resection, however, only 10–20% of patients are suitable
for surgical treatment (5). In
addition, recent developments in genomic technologies have enabled
the study of molecular aberrations and deregulations directly from
patient specimens in a comprehensive manner (6). However, the target gene locus
associated with genomic therapy remains unelucidated. Thus, given
the single means of treatment and the lack of a targeted locus,
there is an urgent need for developing novel treatments that can
prevent the early invasion and metastasis of HCC and for surveying
new molecular-targeted sites that offer the possibility for the
effective development of gene therapy.
Epithelial to mesenchymal transition (EMT), which
converts epithelial cells into migratory mesenchymal cells, is
crucial for embryonic development. It is characterized by
upregulation of extracellular matrix components, loss of
intercellular cohesion, an increased rate of cellular migration and
invasion, as well as increased resistance to apoptosis. Recently,
EMT has emerged as a pivotal event in the development of invasion
and metastasis associated with cancer progression (7). This type of transition involves a
decrease in E-cadherin expression and gain of vimentin expression,
leading to increased cell migration, invasion and tumorigenicity
(8). Downregulation of E-cadherin
is a major event in EMT. E-cadherin, which is a key cadherin, plays
a major role in the establishment and maintenance of intercellular
adhesion, cell polarity and tissue architecture (9). Currently, there is substantial
evidence that functional perturbation of E-cadherin expression
during the process of EMT occurs at the transcriptional level.
According to studies, numerous transcription factors have been
associated with the regulation of EMT, including SNAIL (10), Zeb1 (11), Zeb (2), Twist1 (12) and Slug (13). Among these regulators, SNAIL is a
prominent inducer of EMT which strongly suppresses E-cadherin
expression (14).
SNAIL is a well-known zinc finger (ZF)
transcriptional repressor which is essential during embryonic
development (15). Numerous studies
have shown that SNAIL is significantly upregulated in a variety of
malignant tumors. There is also evidence that SNAIL enhances
invasive and metastatic potential, and the malignant tumors
exibiting SNAIL upregulation are prone to distant metastasis, deep
invasion and poor prognosis. Conversely, in vitro
experiments have shown that inhibition of SNAIL mRNA expression
significantly reduces cell motility and invasion capacity and
decreases invasion to the extracellular matrix (16). Briefly, SNAIL-induced EMT converts
epithelial cells into mesenchymal cells with migratory properties
contributing to the acquisition of invasive and metastatic
potential of tumors. In addition, the induction is largely due to
the repression of E-cadherin. Thus, E-cadherin repressors are
regarded as markers of malignancy and metastasis and provide hope
for gene therapy of HCC. However, the molecular mechanism of SNAIL
in regulating HCC carcinogenesis and progression remains unclear.
In the present study, we verify the relationship of SNAIL and
E-cadherin in vitro by the RNA interference (siRNA)
method.
Small hairpin RNA (shRNA) expression vector systems
have been established to induce RNA interference (RNAi) in
mammalian cells (17). RNAi has
emerged as a powerful technique through which to downregulate the
expression of specific genes in cells and in animals. Among the
shRNA delivery mechanisms, viral vectors, and in particular
lentiviral vectors, are capable of specific, highly stable and
functional silencing of gene expression in a variety of human cells
including primary non-dividing cells (18). In the present study, we used a
lentiviral vector, carrying SNAIL shRNA for transfection into HepG2
cells in order to knock down the expression of the target gene
SNAIL. Through gene expression analysis at the mRNA and protein
levels, we observed that knockdown of SNAIL led to the activation
of E-cadherin expression, resulting in reduced cell proliferation
and viability of HCC cells. Thus, targeted inactivation of SNAIL
may provide a novel therapeutic strategy for HCC patients
expressing this gene.
Materials and methods
Construction of the pGenesil-SNAIL
siRNA
Three pairs of shRNAs were used for screening the
most effective downregulation of the gene fragment. In the enzyme
site of the vector pGenesil-1, three coding regions corresponding
to the target human SNAIL (NM_005985), starting at positions 122,
150 and 518 in the sequence were selected as siRNA target sequences
under the guide of the siRNA designing software. The target shRNA
oligonucleotides were annealed and ligated with the siRNAs, and the
pGenesil-1 vector was cleaved by BamHI and HindIII,
and then the products were recovered and purified. shRNA
oligonucleotide fragment and the pGenesil-1 vector were ligated,
and the recombinant plasmids were named pGenesil-SNAIL shRNA-1–3.
Next, pGenesil-GFP-SNAIL shRNAs were transfected into HepG2 cells,
respectively, using the Lipofectamine 2000 reagent (Invitrogen,
Carlsbad, CA, USA). The most effective siRNA, by screening using
analysis of the SNAIL mRNA and protein levels in HepG2 cells, was
detected and used for subsequent experiments.
Construction and production of lentiviral
vectors (Fig. 1)
The most effective shRNA expression cassette was
selected and cut off by BamHI and MluI from the
pGenesil-1 vector, and ligated into pLV-mCMV-ZsGreen-PGK-puro
shuttle vector, which was called pLV-SNAIL shRNA. Plasmids were
purified with a MaxPrep kit and successful ligations were verified
by sequencing (Sango, China). Recombinant lentiviral vectors were
produced by co-transfecting HEK-293T cells with the lentiviral
expression plasmid and packaging plasmids using the calcium
phosphate method. Briefly, 8 μg shRNA plasmid DNA, 5 μg lentiviral
helper1 and 6 μg lentiviral helper 2 plasmids were mixed with
sterile ddH2O to a final volume of 450 μl and mixed with
50 μl of 2.5 M CaCl2. After transfection, infectious
media, containing shRNA lentiviral vectors, were harvested at 48
and 72 h.
Cell culture and infection
The human hepatocellular cancer HepG2 cell line and
HEK293T cells were purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA). Cells were grown in 5%
CO2 at a saturated humidity, at 37°C and cultured as a
monolayer in RPMI-1640 medium supplemented with 100 U/ml penicillin
and 100 μg/ml streptomycin, 2 mmol/l glutamine and 10% FBS.
HepG2 cells were passaged to 40% confluence the
following day. The viral media were added to HepG2 cells with 8
μg/ml Polybrene 4 times over two days. After infection for 72 h,
viral media were removed and cells were collected for further
experiments.
Cell proliferation assay
Cell viability was assessed by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
method. HepG2 cells (the LV-SNAIL siRNA, LV-nonsense RNA and
parental cell groups) were seeded in 96-well plates in culture
medium at an optimal density (5×103 cells/well) in
triplicate wells. After growing for 12, 24 and 48 h, 20 μl of 5
mg/ml MTT (in PBS) was added to each well and continually incubated
for 4 h at 37°C in a CO2 incubator. The formazan
granules obtained from the cells were dissolved in 150 μl dimethyl
sulfoxide (DMSO) for 10 min. Cell viability was then measured in
terms of the optical density (OD) at a wavelength of 490 nm. Each
cell viability assay was performed and repeated three times.
Quantification by real-time PCR
Transfected and untreated cells were collected and
washed with phosphate-buffered saline (PBS). Total RNA was
extracted from the cells of the three groups, and cDNA was
generated with a PrimeScript® RT reagent kit (Takara,
Chiga, Japan) at a total volume of 20 μl according to the
manufacturer’s instructions. cDNA was then used in each
amplification reaction. Reactions were performed using the
SYBR® Premix Ex Taq™ (Takara), under the following PCR
conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5
sec and 60°C for 30 sec. Each sample was also subjected to melting
curve analysis to confirm amplification specificity. GAPDH was used
as a control housekeeping gene. The expression of SNAIL was
assessed by normalization of the cycle threshold (Ct) of these
genes to that of GAPDH. A Ct value was obtained from each
amplification curve by using the software provided by the
manufacturer (Roche, Mannheim, Germany).
Western blot analysis
Cell lysates of the lentiviral-infected HepG2 cell
clones and the non-transfected HepG2 cell controls were harvested
and subjected to SDS-PAGE in 10% polyacrylamide gel. Bands were
probed for SNAIL protein expression using rabbit polyclonal
anti-SNAIL and secondary goat anti-rabbit alkaline phosphatase
antibody (Abcam, Cambridge, MA, USA). GAPDH detection was performed
using rabbit polyclonal anti-GAPDH and mouse monoclonal anti-GAPDH
(Abcam). Blots were incubated with appropriate HRP-conjugated
secondary antibodies and specific binding was detected by ECL
(Pierce, Rockford, IL, USA).
Results
Selection of the most effective
SNAIL-specific shRNA
Expression levels of SNAIL mRNA and protein in the
HepG2 cell line were detected in previously published data by our
laboratory (19). The levels of
SNAIL mRNA and protein were were previously found to be increased
in metastatic tumor samples in comparison to non-metastatic tumor
samples and the same phenomena were detected in HepG2 cells
(19). We determined the effects of
SNAIL inhibition on the viability and growth of HepG2 cells. For
screening the most effective shRNA fragment, three plasmids
containing shSNAIL (pGenesil-SNAIL shRNA) were constructed and
transfected into HepG2 cells using the Lipofectamine 2000 reagent,
respectively. GFP expression in the HepG2 cells was observed under
a fluorescence microscope 36 h after transfection with the pGenesil
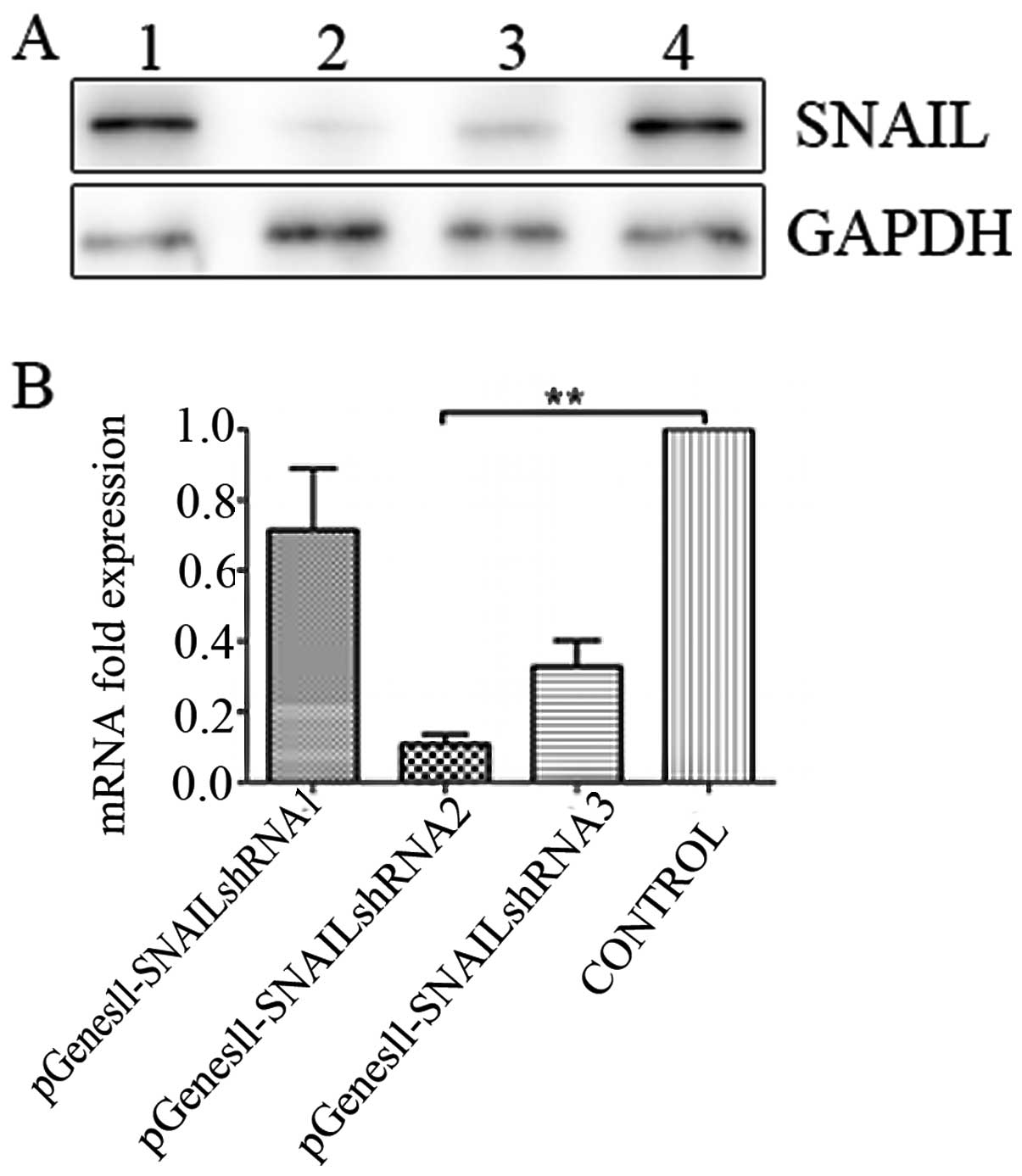
vectors. Results of the western blot assay showed that
pGenesil-SNAIL shRNA2 significantly suppressed the expression of
SNAIL at the protein level in HepG2 cells, while real-time PCR
determined expression at the mRNA level. According to the results
of the real-time PCR and western blot assay, SNAIL shRNA2 was the
most effective and, thus, was used in the subsequent experiments
(Fig. 2).
Construction and analysis of the LV-SNAIL
shRNA
The most efficient shRNA expression cassette was
selected and constructed into the lentiviral vector, named LV-SNAIL
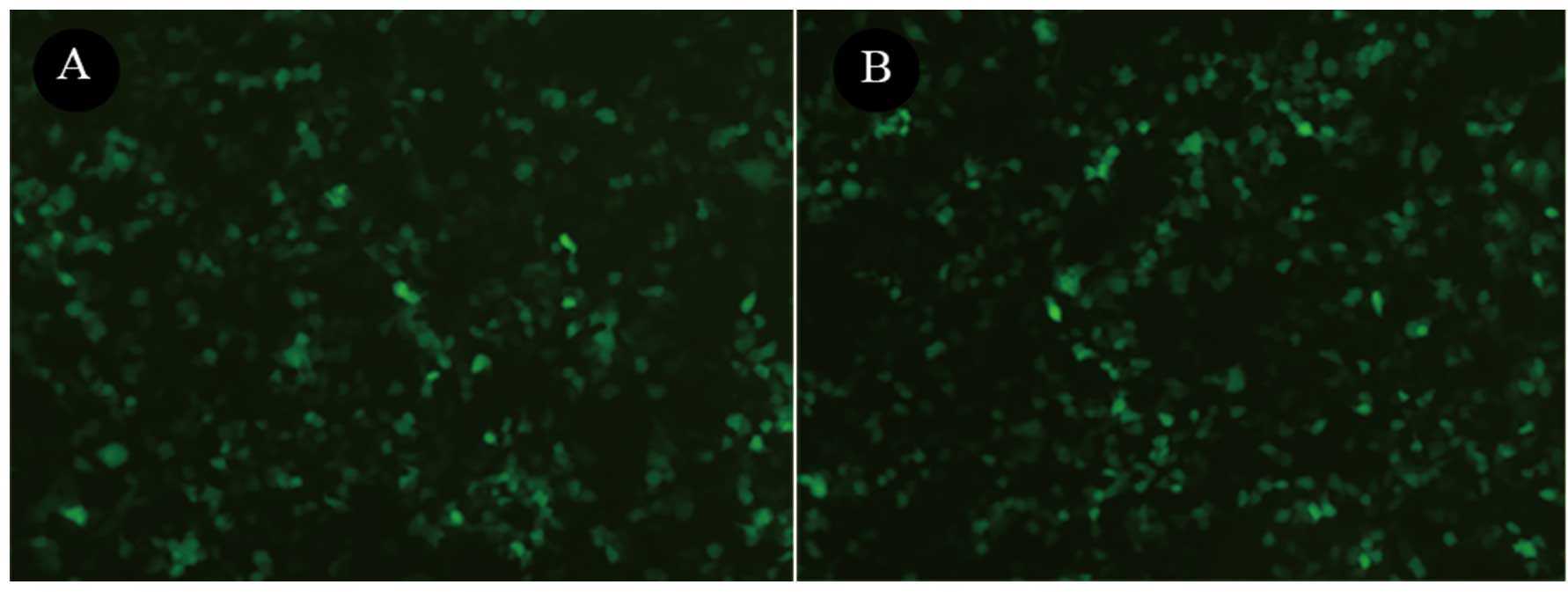
shRNA. To determine the effect of LV-SNAIL shRNA on the expression
of SNAIL, GFP expression was observed under a fluorescence
microscope in the HepG2 cells 72 h after infection with LV-SNAIL
shRNA and LV-CONTROL-GFP (Fig. 3).
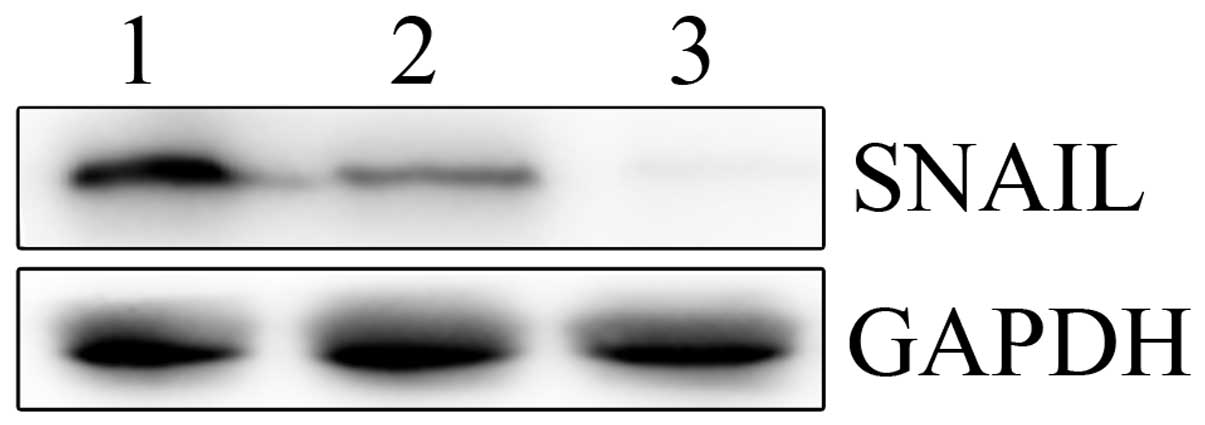
Next, real-time PCR and western blotting were performed to
determine the mRNA and protein levels of SNAIL in the LV-SNAIL
shRNA, LV-CONTROL-GFP and parental cell groups. As shown in
Fig. 4, LV-SNAIL shRNA
significantly inhibited expression of SNAIL protein when compared
with the levels in the HepG2 cells and the LV-CONTROL-GFP
group.
Cell proliferation analysis in HepG2
cells
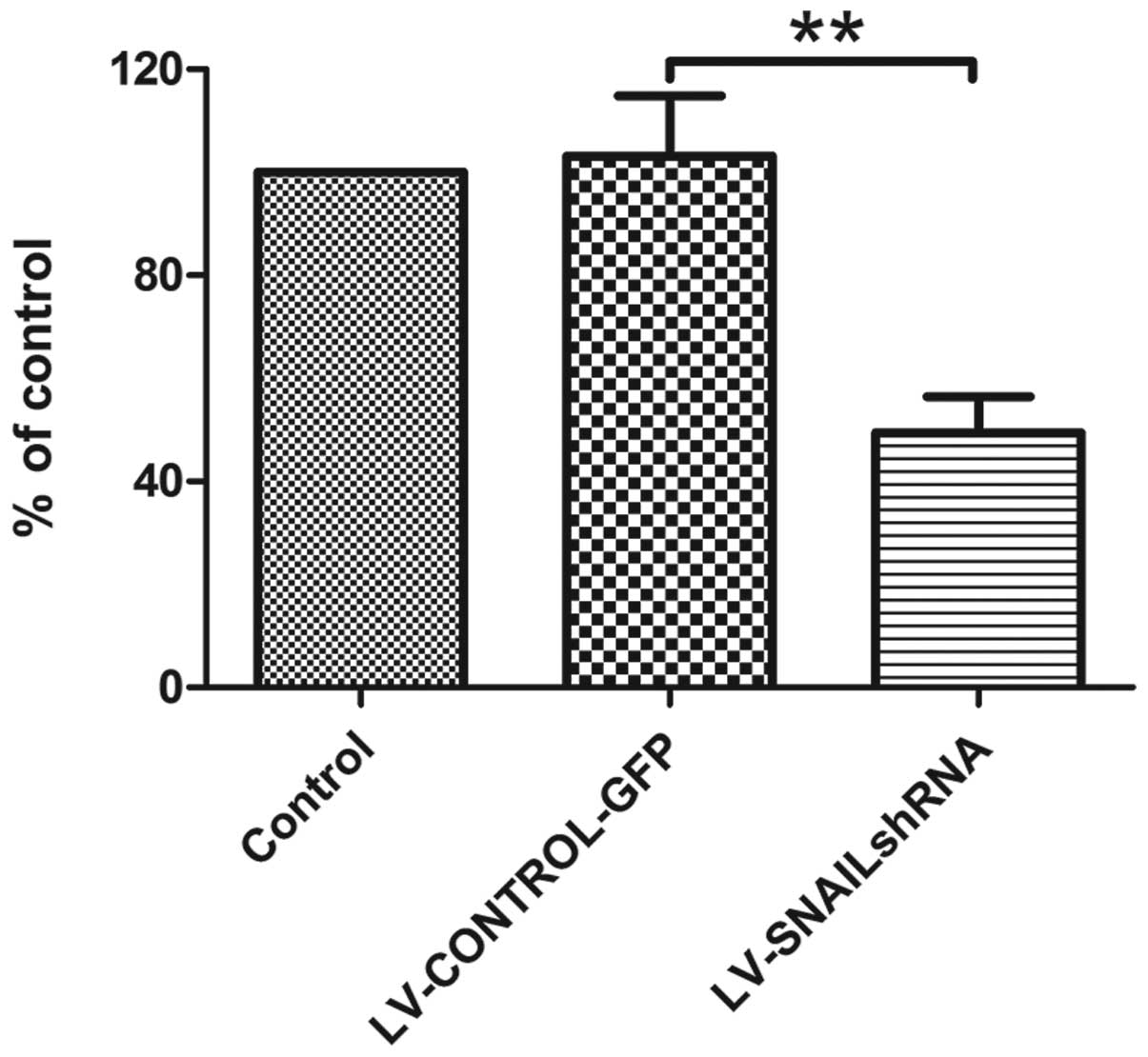
Cell proliferation was monitored for 4 days after
HepG2 cells were infected with LV-SNAIL shRNA, LV-CONTROL-GFP and
in parental cells without infection. MTT assay was used to evaluate
cell viability. The absorption value at 490 nm was determined. As a
result, we found that transfection with LV-CONTROL-GFP had no
effect on the cell proliferation and viability in the cell groups
(Fig. 5) while transfection with
LV-SNAIL shRNA caused a dramatic reduction in proliferation when
compared with the proliferation rate in the parental HepG2
cells.
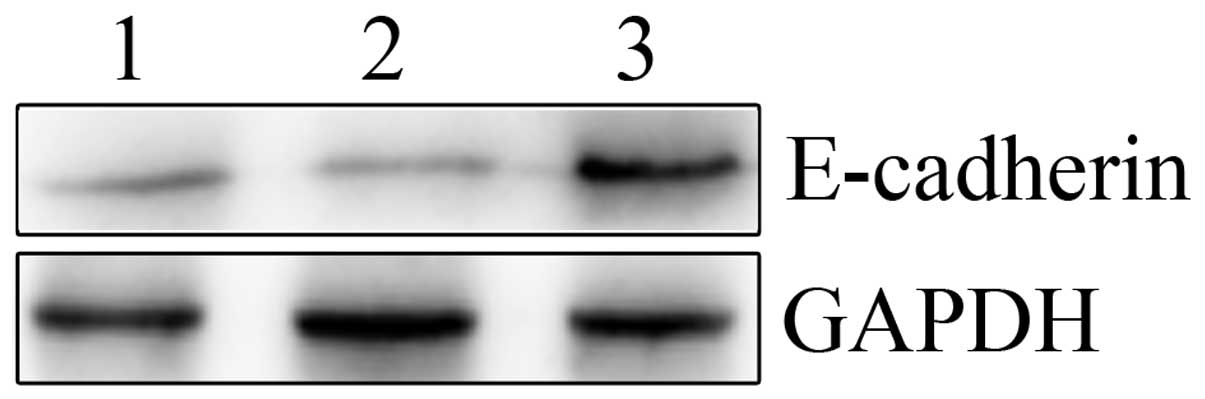
E-cadherin expression assay
As shown in Fig. 6,
E-cadherin protein expression was evaluated by western blotting in
the human HepG2 cell lines. Silencing of SNAIL expression by
LV-SNAIL shRNA significantly increased the expression of E-cadherin
when compared with the LV-SNAIL shRNA and parent groups. LV-SNAIL
shRNA markedly downregulated the protein level of SNAIL and
upregulated the protein level of E-cadherin in HepG2 cells when
compared with these levels in the other two cell groups.
Discussion
The SNAIL signaling pathway plays a significant role
in various physiological processes, including cell growth, cell
cycle regulation (20) and
embryonic development (15).
However, SNAIL has been implicated in cell proliferation, invasion
and metastasis in a diverse group of human cancers, including liver
cancer. Recent studies have also revealed that the promotion of
SNAIL activation contributes to oncogenesis and the invasion and
metastasis of cancers (21,22). Thus, the purpose of the present
study was to assess the changes in the mRNA and protein expression
of SNAIL and E-cadherin involved in EMT in HepG2 cell lines.
Epithelial to mesenchymal transition (EMT), which
converts epithelial cells into migratory mesenchymal cells, is
crucial for embryonic development. It is characterized by
upregulation of extracellular matrix components, loss of
intercellular cohesion, increased rate of cellular migration and
invasion, as well as increased resistance to apoptosis. Recently,
EMT has emerged as a pivotal event in the development of the
invasive and metastatic potential associated with cancer
progression (7). This type of
transition occurs through a decrease in E-cadherin expression and
gain of vimentin expression, leading to increased cell migration,
invasion and tumorigenicity (8).
Downregulation of E-cadherin is a major event in EMT. E-cadherin,
which is a key cadherin, plays a major role in the establishment
and maintenance of intercellular adhesion, cell polarity and tissue
architecture (9). At present, there
is substantial evidence that functional perturbation of E-cadherin
expression during the process of EMT occurs at the transcriptional
level. According to various studies, numerous transcription factors
have been associated with regulation of EMT, including SNAIL
(10), Zeb1 (11), Zeb (2), Twist1 (12) and Slug (13). Among these regulators, SNAIL is a
prominent inducer of EMT which strongly suppresses E-cadherin
expression (14).
In order to elucidate the role played by SNAIL in
HCC, we suppressed SNAIL in HCC cells using the RNAi method by a
lentiviral vector. RNAi is the process by which double-strand RNA
induces potent and specific inhibition of eukaryotic gene
expression through the degradation of complementary messenger RNA,
and is functionally similar to the process of post-transcriptional
gene silencing. The inhibitory potency and the method of
transferring siRNA into HCC cells are two critical factors for
successful application of the RNAi method. In recent years, shRNAs
have been proven to provide long-lasting silencing and maximal
inhibition of gene expression at lower concentration (23). However, the inhibitory potency of
shRNA is related to specificity of the target sequence, thus we
used three pairs of shRNAs for screening the most effective
downregulated gene fragment. Thus, we chose SNAIL siRNA2 which was
able to knock out the expression of SNAIL efficiently. In addition,
in order to identify the target gene or the vector fragment, we
used EGFP siRNA as the control group and parental cells as the
blank group. In the present study, we found that LV-SNAIL siRNA2
suppressed the expression of SNAIL at both the mRNA and protein
levels in HCC. In the control group of HepG2 cells, however, the
expression SNAIL was significantly increased compared with the
experimental group. We concluded that LV-shSNAIL had a high
specificity for SNAIL.
In the present study, a lentiviral vector was
selected as our shRNA delivery vehicle since lentiviruses can
transfect both dividing and non-dividing cells at a high efficiency
and sustain long-term gene expression by integration into the host
genome. What is more, lentiviral vectors are safe for use in
humans, and they have the potential to become the means for human
gene therapy. At present, an increasing number of studies suggest
that activation of SNAIL plays an important role in the early
metastasis of carcinomas (24).
Furthermore, disruption of SNAIL expression has been reported to
suppress cell metastasis by decreasing EMT and increasing
epithelium-specific genes, such as E-cadherin in other carcinomas
(25). In our study, the expression
of SNAIL and E-cadherin exhibited a negative correlation, which may
be another major verification for our findings. Thus, our results
indicate that blocking SNAIL expression upregulates E-cadherin
expression directly or indirectly in HepG2 cells.
An increasing number of studies suggest that
activation of SNAIL plays an important role in many carcinomas
(26,27). Furthermore, disruption of SNAIL at
the transcriptional level has been reported to suppress cell
invasion by decreasing cell-cell homotypic adhesion and increasing
cell motility (14). Moreover,
inappropriate and constitutive activation of SNAIL may be
responsible for HCC progression by regulating expression of target
genes, such as cadherin and MMP-2 (9). In the present study, the survival of
cells using a cell proliferation analysis was examined and we found
that silencing of SNAIL by RNAi in HepG2 cells resulted in reduced
proliferation and survival. Conversely, the expression of
E-cadherin in the SNAIL-silenced cell group was upregulated, which
indicates that the reduced proliferation and survival which
resulted from the silencing of SNAIL by RNAi was caused by the
upregulation of the expression of E-cadherin.
Overall, our study indicates that siRNA targeting of
SNAIL mRNA via a lentiviral vector system effectively sustains
knockdown of the SNAIL gene expression in HepG2 cells. In the
present study, we describe the successful construction of a
lentiviral RNAi vector targeting SNAIL that may provide a useful
tool with which to study the function of the SNAIL gene in liver
cancer cells. Our findings strongly suggest that SNAIL and
E-cadherin play a significant role in HCC progression, and exhibit
a negative correlation. Furthermore, the expression of E-cadherin
may be responsible for the reduced proliferation and survival of
HepG2 cells. Targeting SNAIL activation may prove to be an
effective approach for controlling HCC progression. Thus, the SNAIL
signaling pathway may provide a novel therapeutic target for the
treatment of HCC.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Comijn J, Berx G, Vermassen P, et al: The
two-handed E box binding zinc finger protein SIP1 downregulates
E-cadherin and induces invasion. Mol Cell. 7:1267–1278. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Osada T, Sakamoto M, Ino Y, Iwamatsu A,
Matsuno Y, Muto T and Hirohashi S: E-cadherin is involved in the
intrahepatic metastasis of hepatocellular carcinoma. Hepatology.
24:1460–1467. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tung-Ping Poon R, Fan ST and Wong J: Risk
factors, prevention, and management of postoperative recurrence
after resection of hepatocellular carcinoma. Ann Surg. 232:10–24.
2000.PubMed/NCBI
|
|
5
|
Clark HP, Carson WF, Kavanagh PV, Ho CP,
Shen P and Zaqoria RJ: Staging and current treatment of
hepatocellular carcinoma. Radiographics. 25(Suppl 1): S3–S23. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoshida Y, Moeini A, Alsinet C, Kojima K
and Villanueva A: Gene signatures in the management of
hepatocellular carcinoma. Semin Oncol. 39:473–485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hugo H, Ackland ML, Blick T, Lawrence MG,
Clements JA, Williams ED and Thompson EW: Epithelial-mesenchymal
and mesenchymal-epithelial transitions in carcinoma progression. J
Cell Physiol. 213:374–383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Neal CL, McKeithen D and Odero-Marah VA:
Snail negatively regulates cell adhesion to extracellular matrix
and integrin expression via the MAPK pathway in prostate cancer
cells. Cell Adh Migr. 5:249–257. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyoshi A, Kitajima Y, Sumi K, Sato K,
Haqiwara A, Koqa Y and Mivazaki K: Snail and SIP1 increase cancer
invasion by upregulating MMP family in hepatocellular carcinoma
cells. Br J Cancer. 90:1265–1273. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cano A, Perez-Moreno MA, Rodriqo I, et al:
The transcription factor Snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eger A, Aiqner K, Sondereqqer, et al:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Mani SA, Donaher JL, et al: Twist,
a master regulator of morphogenesis, plays an essential role in
tumor metastasis. Cell. 117:927–939. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolos V, Peinado H, Perez-Moreno MA, Fraga
MF, Esteller M and Cana A: The transcription factor Slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: a comparison with Snail and E47 repressors. J Cell
Sci. 116:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haraguchi M: The role of the
transcriptional regulator snail in cell detachment, reattachment
and migration. Cell Adh Migr. 3:259–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: an alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Palmer MB, Majumder P, Green MR, Wade PA
and Boss JM: A 3′ enhancer controls Snail expression in melanoma
cells. Cancer Res. 67:6113–6120. 2007.
|
|
17
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishitsuji H, Ikeda T, Miyoshi H, Ohashi
T, Kannaqi M and Masuda T: Expression of small hairpin RNA by
lentivirus-based vector confers efficient and stable
gene-suppression of HIV-1 on human cells including primary
non-dividing cells. Microbes Infect. 6:76–85. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen D, Zheng X, Jiao X, Gao Y, Zhang K
and Liang J: Transcriptional repressor snail and metastasis in
hepatocellular carcinoma. Hepatogastroenterology. 59:1359–1365.
2012.PubMed/NCBI
|
|
20
|
Park JH, Sung IJ, Lee SW, Kim KW, Kim YS
and Yoo MA: The zinc-finger transcription factor Snail
downregulates proliferating cell nuclear antigen expression in
colorectal carcinoma cells. Int J Oncol. 26:1541–1547. 2005.
|
|
21
|
Fan XJ, Wan XB, Yang ZL, et al: Snail
promotes lymph node metastasis and Twist enhances tumor deposit
formation through epithelial-mesenchymal transition in colorectal
cancer. Hum Pathol. 44:173–180. 2012. View Article : Google Scholar
|
|
22
|
Sun M, Guo X, Qian X, et al: Activation of
the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell
Biol. 4:304–315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim DH, Behlke MA, Rose SD, Chang MS, Choi
S and Rossi JJ: Synthetic dsRNA Dicer substrates enhance RNAi
potency and efficacy. Nat Biotechnol. 23:222–226. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Emadi Baygi M, Soheili ZS, Schmitz I,
Sameie S and Schulz WA: Snail regulates cell survival and inhibits
cellular senescence in human metastatic prostate cancer cell lines.
Cell Biol Toxicol. 26:553–567. 2010.PubMed/NCBI
|
|
25
|
Becker KF, Rosivatz E, Blechschmidt K,
Kremmer E, Sarbia M and Hofler H: Analysis of the E-cadherin
repressor Snail in primary human cancers. Cells Tissues Organs.
185:204–212. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Merikallio H, Turpeenniemi-Hujanen T,
Paakko P, et al: Snail promotes an invasive phenotype in lung
carcinoma. Respir Res. 13:1042012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boutet A, Esteban M, Maxwell P and Nieto
M: Reactivation of Snail genes in renal fibrosis and carcinomas: a
process of reversed embryogenesis? Cell Cycle. 6:638–642. 2007.
View Article : Google Scholar : PubMed/NCBI
|