Introduction
Numerous types of cancer develop as a result of
dysregulated gene expression derived from epigenetic modifications
such as methylation or acetylation (1,2). In
particular, DNA-bound core histones regulate gene activation and
inactivation via the opposing activities of histone
acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes.
HDACs play crucial roles in epigenetic modulation by
deacetylating histone and non-histone substrates that are involved
in processes critical for both normal development and cancer
(3). HDACs have been classified
into four classes based on structural homologies between humans and
yeast. Class I HDACs are ubiquitously expressed in the nuclei of
all cells (4), whereas HDACs 4, 5,
7 and 9, which are members of class II, are shuttled between the
nucleus and the cytoplasm and are specifically expressed in heart
tissue, bone, the nervous system and skeletal muscle (5). In several types of cancer, aberrant
expression of HDAC family members has been proposed as a hallmark
of multiple tumorigenic processes, including proliferation,
apoptosis, angiogenesis and metastasis (6). Moreover, a number of researchers have
suggested that HDACs are associated with chemotherapy resistance in
several types of cancer (7–11). Chemoresistance to effective
treatments for many tumor types presents a major obstacle, leading
researchers to study the use of molecularly targeted therapies as a
new class of chemotherapeutic agents. HDAC inhibitors, which target
HDAC proteins, have a wide range of effects, including cell cycle
arrest, apoptosis, anti-angiogenic effects, and autophagy, and are
utilized clinically as chemotherapeutic agents in several types of
cancer (5).
We previously reported that HDAC expression in
metastatic and non-metastatic breast cancer cells differs and that
high HDAC expression levels, with the exception of HDAC4, are
associated with invasiveness, which is of concern due to the role
in invasion of cancer metastasis (12). Furthermore, Stronach et
al(13) recently demonstrated
that HDAC4 may be a therapeutic target for platinum resistance in
ovarian cancer.
SMAD family member 4 (SMAD4) has also been reported
to be a gene that promotes 5-fluorouracil (5-FU) resistance in
colon cancer (14). SMAD4 is known
to be a mediator of the transforming growth factor (TGF) β
signaling pathway and has been shown to act as a suppressor of
tumor progression and to decrease tumor growth by inducing
apoptosis. SMAD4 inactivation by genetic abnormalities, including
deletion or mutation, has been identified in several types of
cancer, including colorectal, pancreatic, breast, ovarian, lung and
gastric cancer (15–19). Furthermore, reduction of SMAD4
expression by methylation of the SMAD4 promoter has been correlated
with advanced prostate cancer (20). However, the correlation between
HDAC4 and SMAD4 has not been elucidated.
In the present study, we investigated the
correlation between high HDAC4 expression and chemoresistance as
well as the molecular targets that are associated with HDAC4
overexpression-induced chemoresistance in breast cancer. Based on
these results, we suggest that SMAD4 in the TGFβ signaling pathway
is regulated by HDAC4 and is associated with 5-FU resistance in
breast cancer cells.
Materials and methods
Cell culture and transfection
MDA-MB-231 (HTB-26) and MCF-7 (HTB-22) breast cancer
cells that had been obtained from the American Type Culture
Collection (ATCC) were cultured in Dulbecco’s Modified Eagle’s
Medium (DMEM; Lonza) supplemented with 10% fetal bovine serum (FBS;
Gibco) and 1% penicillin/streptomycin (Lonza). The cells were
maintained at 37ºC in a humidified atmosphere containing 5%
CO2.
To obtain stable expression of the HDAC4 protein,
MCF-7 cells were transfected with Lipofectamine® 2000
(Invitrogen) using a human HDAC4 full-length cDNA vector
constructed in a pcDNA3-EGFP Plasmid (Addgene) carrying the
neomycin resistance gene. Transfected clones were cultured in
selective media supplemented with 600 μg/ml of G418 (Sigma).
Transient transfection was performed using
Lipofectamine® 2000 in order to overexpress hHDAC4 and
the Lipofectamine® RNAiMAX Reagent (Invitrogen) was used
to knockdown hHDAC4 and hSMAD4 expression according to the
manufacturer’s recommended protocol.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using a Nucleospin RNAII kit
(Macherey-Nagel, Germany) and cDNAs were synthesized using M-MLV
Reverse Transcriptase (Promega). qRT-PCR reactions were carried out
in triplicate using iQ™ SYBR-Green Supermix and a CFX96 qPCR
machine (BioRad). The primers used to detect HDAC4, SMAD4 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: HDAC4
forward, 5′-agcgtgagcaagatcctc-3′ and reverse,
5′-gccaagtactcagcgtctcc-3′; SMAD4 forward,
5′-cccaggatcagtaggtggaa-3′ and reverse, 5′-cccagcctttcacaaaactc-3′;
and GAPDH forward, 5′-acagtcagccgcatcttctt-3′ and reverse,
5′-acgaccaaatccgttgactc-3′. The amplification conditions were: a
predenaturation step at 95ºC for 3 min, followed by 40 cycles of
denaturation at 95ºC for 15 sec, annealing at 60ºC for 15 sec and
extension at 72ºC for 15 sec. The comparative threshold cycle (Ct)
method, 2−ΔΔCt, was used to calculate fold
amplification.
Western blot analysis
The cells were lysed with RIPA buffer containing 50
mM Tris-HCl at pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM
NaCl, 1 mM EDTA, 1 mM Na3Vo4, 1 mM NaF and
proteinase inhibitors. Proteins were separated using 8% SDS-PAGE
gels and transferred to polyvinylidene difluoride membranes (PVDF;
Millipore, Germany). Blots were then incubated in 5% skim milk
(Difco) for 1 h and probed with anti-HDAC4 (Abcam) and anti-GAPDH
(Santa Cruz Biotechnology, Inc.) primary antibodies overnight,
followed by incubation with a horseradish peroxidase
(HRP)-conjugated anti-rabbit secondary antibody. Immunoreactive
proteins were detected using an enhanced chemiluminescence kit
(Thermo).
In vitro cytotoxicity assay
To identify differences in the cytotoxicity of
anticancer drugs, including cisplatin (Sigma), paclitaxel
(Bristol-Myers Squibb SRL, Italy), gemcitabine (Lilly, Korea) and
5-FU (Sigma), cells were plated in 96-well plates at a density of
5×103 cells/well. Twenty-four hours later, the cells
were treated with anticancer drugs and incubated for 24, 48 and 72
h. The surviving cells were treated with 500 μg/ml of MTT solution
for 2 h, after which point the absorbance was measured at 540 nm.
The survival rate was calculated as the ratio of the absorbance of
the treated wells to that of the control wells.
Microarray analysis
Biotinylated cRNA was produced according to the
manufacturer’s protocol. Briefly, total RNA was reverse-transcribed
to cDNA using T7 oligo(dT) primers and second-strand cDNAs were
synthesized through in vitro transcription and labeled with
biotin-NTP. The amounts of the labeled cDNAs were quantified using
an ND-1000 Spectrophotometer (NanoDrop).
Labeled cRNA samples were hybridized to each human
HT-12 v4 Expression Bead Array (Illumina, Inc.) for 16–18 h at
58ºC. Amersham fluorolink streptavidin-Cy3 (GE Healthcare
Bio-Sciences, UK) and an Illumina Bead Array Reader confocal
scanner were used to detect the array signal according to the
manufacturer’s instructions. The microarray experiments on
independent samples were performed in four times replicate.
For data analysis, raw data were extracted from the
software provided by the manufacturer (GenomeStudio v2011.1;
Illumina, Inc.). The raw data were then filtered using a detection
P-value of <0.05 in ≥ 50% of the samples. Selected gene signal
values were transformed and normalized using the logarithmic and
quantile methods, respectively. Significant statistical values for
the expression data were determined using fold-change and the local
pooled error (LPE) test. The false discovery rate (FDR) was
controlled by adjusting the P-values using the Benjamini-Hochberg
algorithm.
Functional annotation analysis of the significant
probe list was performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/home.jsp).
The data discussed in this publication have been
deposited in the NCBI gene expression omnibus (GEO) and are
accessible through the GEO series accession number GSE42242
(http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42242).
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed using a ChIP Assay kit
(Millipore) according to the manufacturer’s protocol. Briefly,
cells were cross-linked with 1% formaldehyde and sonicated on ice
in order to fragment the chromatin. The lysates were diluted with
chromatin-dilution buffer and the soluble fractions were
immunoprecipitated using anti-acetyl-histone H3 (Millipore) at 4ºC
overnight. Protein-A Sepharose beads were complexed with
anti-acetyl-histone H3 and washed to remove nonspecific binding.
The antibody-bound chromatin was eluted using elution buffer (1%
SDS and 0.1 M NaHCO3). The eluted chromatin samples were
treated with proteinase K at 45ºC for 1 h and the DNA was extracted
using the phenol chloroform method. Finally, PCR was performed on
the eluted DNA using four primer sets that were targeted to the
SMAD4 promoter region. The sequences of the primers targeting the
SMAD4 promoter were: P1 forward, 5′-gtggaaggaggagcagtgtc-3′ and
reverse, 5′-tgtcacctttgccatacattg-3′; P2 forward,
5′-tgtgtgtttccttccccttc-3′ and reverse, 5′-tccttgcaggctacaggact-3′;
P3 forward, 5′-tcctttgttccagcctcact-3′ and reverse, 5′-aaa
ctgaaggaagatctgtcagc-3′; P4 forward, 5′-tgaaattacccggatgt ggt-3′
and reverse, 5′-ctaggggagagcaggaagg-3′; and P5 forward,
5′-gctcgtgggagaatcaagtt-3′ and reverse, 5′-caaaacagaaattgg
ctgga-3′. The PCR products were analyzed on 2% standard
Tris/Acetate/EDTA (TAE) agarose gels that had been stained with
GelRed™ Nucleic Acid Gel Staining solution (Biotium).
Statistical analyses
All graphed data are presented as the means ±
standard deviation (SD). The results were analyzed using analysis
of variance (ANOVA) and the Student’s t-test. P-values <0.05 or
0.01 were considered to indicate statistically significant
differences.
Results
HDAC4 expression affects 5-FU
chemoresistance in breast cancer cells
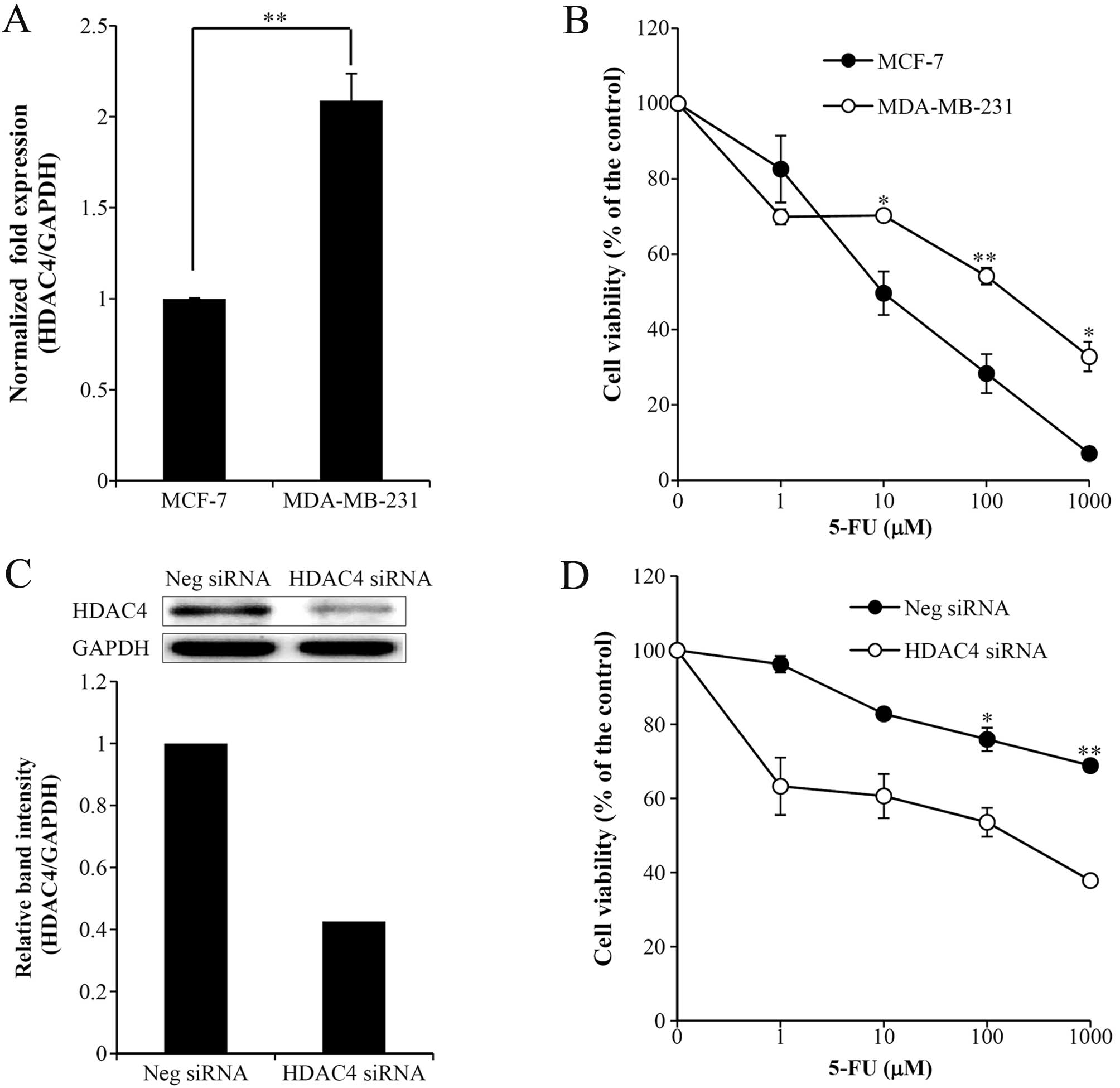
We previously reported that HDAC expression was
associated with cancer progression in metastatic MDA-MB-231 and
non-metastatic MCF-7 cells. As a result, the increased expression
of HDAC1, 6 and 8 in MDA-MB-231 cells as compared to MCF-7 cells
was correlated with tumor cell invasiveness. Although the
expression of HDAC4 was increased in MDA-MB-231 cells, HDAC4 did
not increase tumor cell invasion (12). To investigate whether increased
HDAC4 expression may be associated with chemoresistance to
anticancer drugs, we verified the cytotoxicity of various
anticancer drugs, including cisplatin, paclitaxel, gemcitabine and
5-FU, in MCF-7 and MDA-MB-231 cells (data not shown). Significant
chemoresistance was demonstrated only for 5-FU. As shown in
Fig. 1B, more pronounced 5-FU
chemoresistance was observed in MDA-MB-231 cells than in MCF-7
cells, confirming that the decreased HDAC4 expression in MDA-MB-231
cells transfected with siRNA against HDAC4 was associated with
reduced 5-FU resistance (Fig. 1C and
D). Therefore, we proposed that HDAC4 expression may be
associated with 5-FU chemoresistance. To test the direct
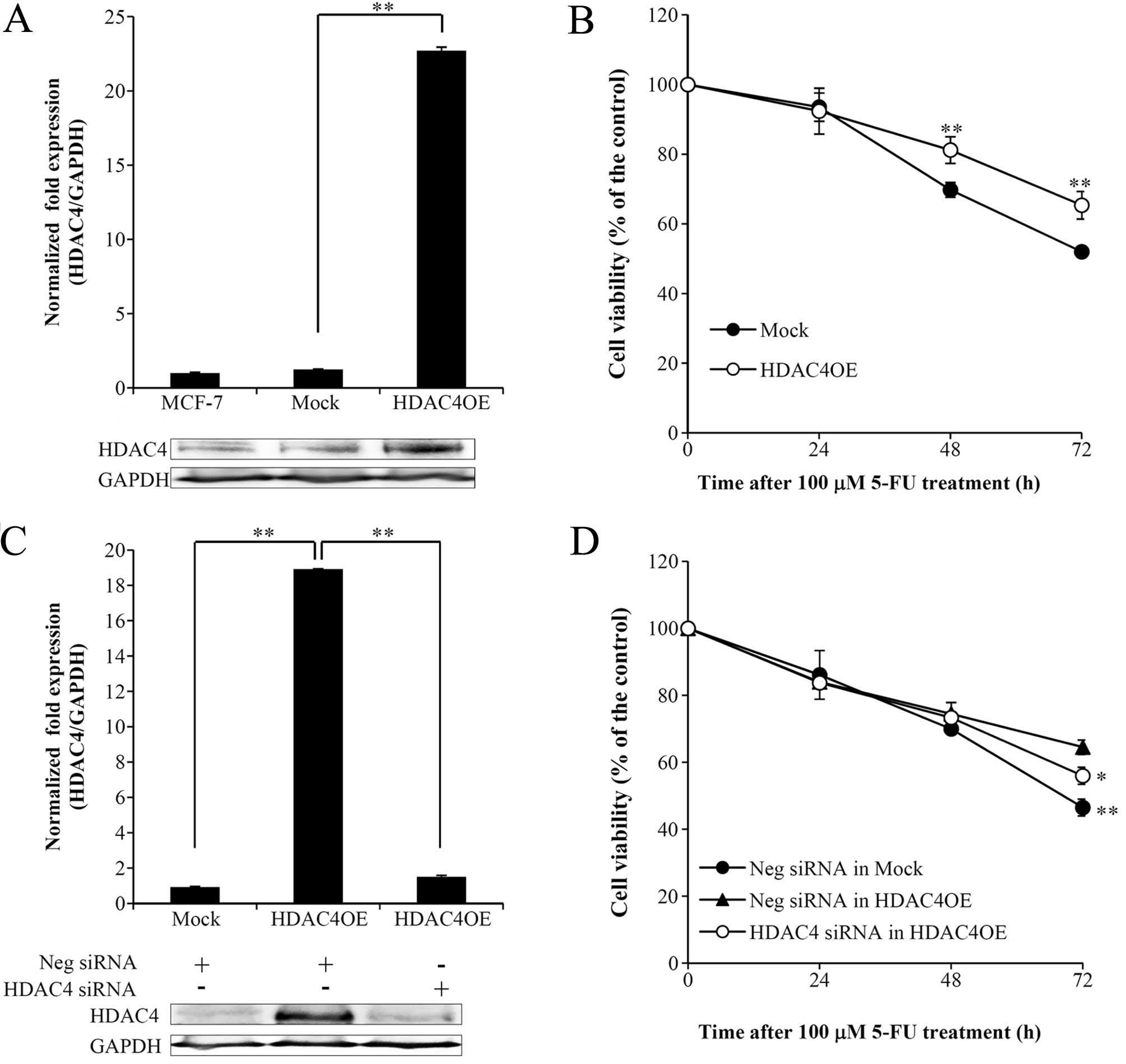
relationship between HDAC4 and 5-FU resistance, we established
MCF-7 cells stably overexpressing HDAC4 (HDAC4OE). MCF-7 cells with
elevated HDAC4 expression were shown to be more resistant to 5-FU
than control cells (Fig. 2A and B).
Moreover, we found that HDAC4 knockdown with siRNA in HDAC4OE cells
reversed 5-FU chemoresistance (Fig. 2C
and D). These observations suggested that HDAC4 participates in
5-FU chemoresistance.
Identification of significant HDAC4
overexpression-mediated alterations in the TGFβ signaling pathway
in MCF-7 cells
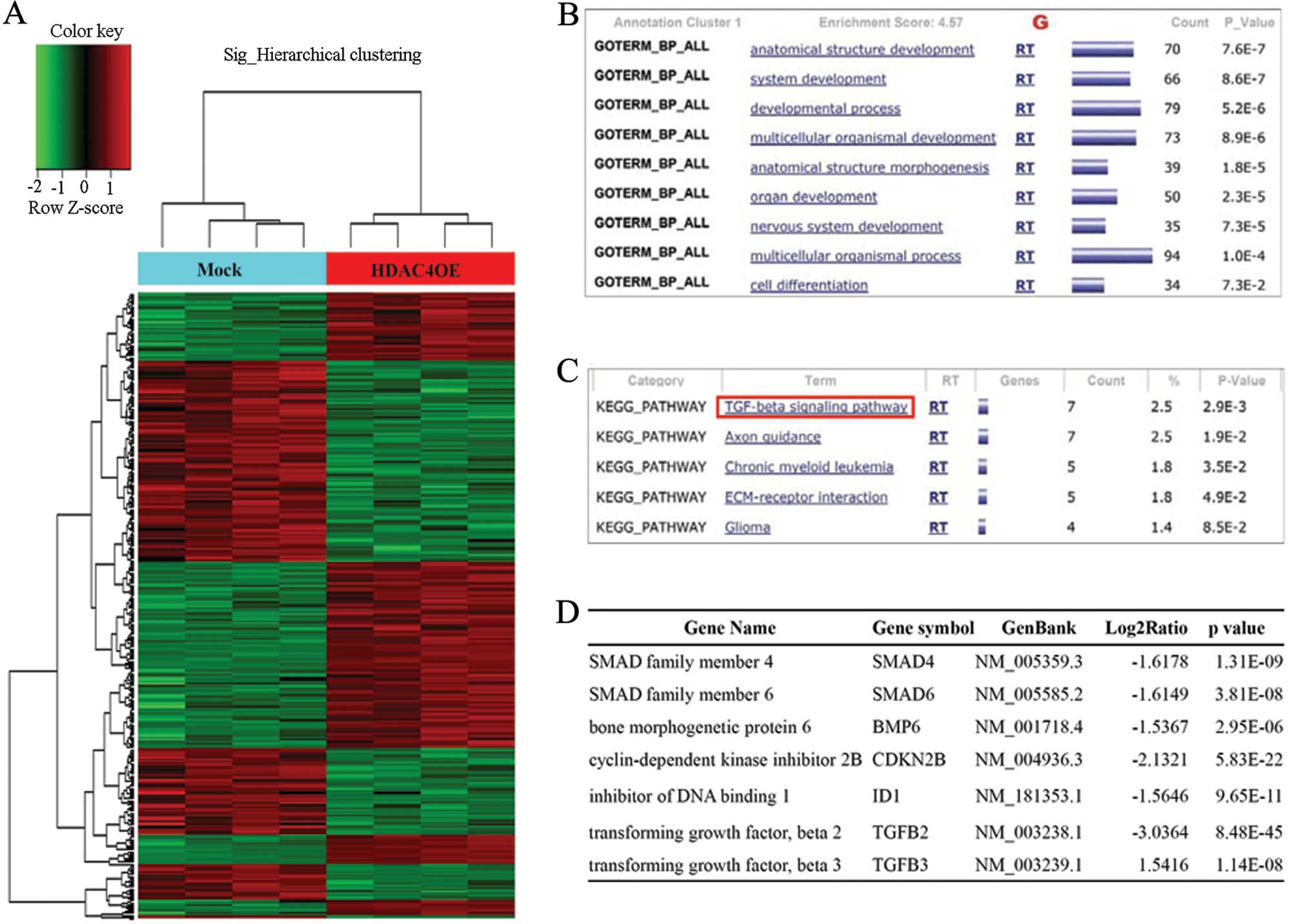
To identify differentially expressed genes in
HDAC4OE cells, we performed microarray analysis in mock-transfected
and HDAC4OE cells. Among the genes regulated by HDAC4
overexpression, 355 genes were shown to have significantly
different expression levels (the expression of 185 genes was
increased, and the expression of 170 genes was decreased) based on
the criteria of a 1.5-fold difference with a P-value <0.05.
Hierarchical clustering analysis was performed using differentially
expressed genes (Fig. 3A). To
investigate alternative pathways of differentially expressed genes,
the functional gene ontology of each gene was analyzed against the
DAVID. As a result, 285 genes were defined (Fig. 3B). We identified 84 genes, including
SMAD4, SMAD6, bone morphogenetic protein 6 (BMP6), inhibitor of DNA
binding 1 (ID1), TGFβ2 and TGFβ3, involved in the TGFβ signaling
pathway from the most significant cluster (P<0.01) (Fig. 3C and D). Recently, Papageorgis et
al(14) reported that SMAD4
inactivation induced 5-FU resistance in colon cancer. Therefore, we
selected SMAD4 as a candidate gene that may regulate 5-FU
chemoresistance in HDAC4OE cells.
HDAC4-mediated regulation of the SMAD4
gene impacts 5-FU chemoresistance
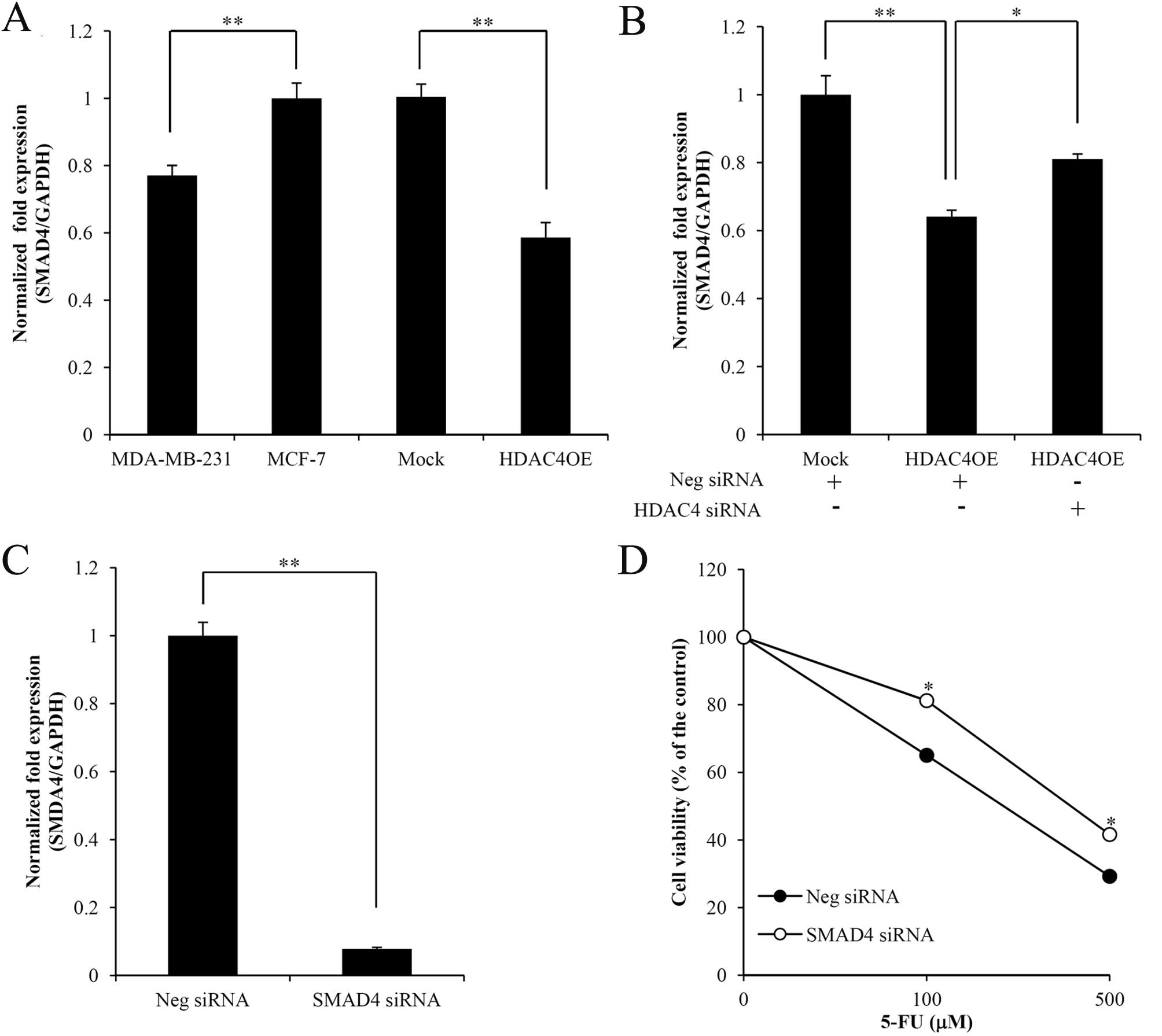
To confirm the correlation between SMAD4 and HDAC4,
the pattern of SMAD4 expression in HDAC4OE and MDA-MB-231 cells was
characterized using qRT-PCR. SMAD4 expression was downregulated in
cells in which HDAC4 was highly expressed (Fig. 4A), and SMAD4 expression was
upregulated when HDAC4 siRNA was transduced into HDAC4OE cells
(Fig. 4B). Consequently, we
hypothesized that HDAC4 may regulate SMAD4 expression in breast
cancer cells. To investigate alterations in chemoresistance
mediated by SMAD4 expression due to HDAC4 overexpression in MCF-7
cells, we performed cytotoxicity assays comparing SMAD4 knockdown
and control MCF-7 cells. As a result, the cytotoxicity of 5-FU in
SMAD4 knockdown cells was elevated (Fig. 4C and D). Therefore, SMAD4 can be
reliably associated with 5-FU resistance in breast cancer
cells.
HDAC4 regulates SMAD4 gene expression via
deacetylation of the SMAD4 promoter
To determine whether SMAD4 transcriptional
repression by HDAC4 is mediated through a modification of histones
in the SAMD4 promoter region, we compared the SMAD4 promoter region
to the UCSC genome browser (http://genome.ucsc.edu/) and found that histone
acetylation was enriched. We also identified a putative binding
site for yin-yang 1 (YY1), which is known to transcriptionally
repress several genes by recruiting HDAC4 (21–24)
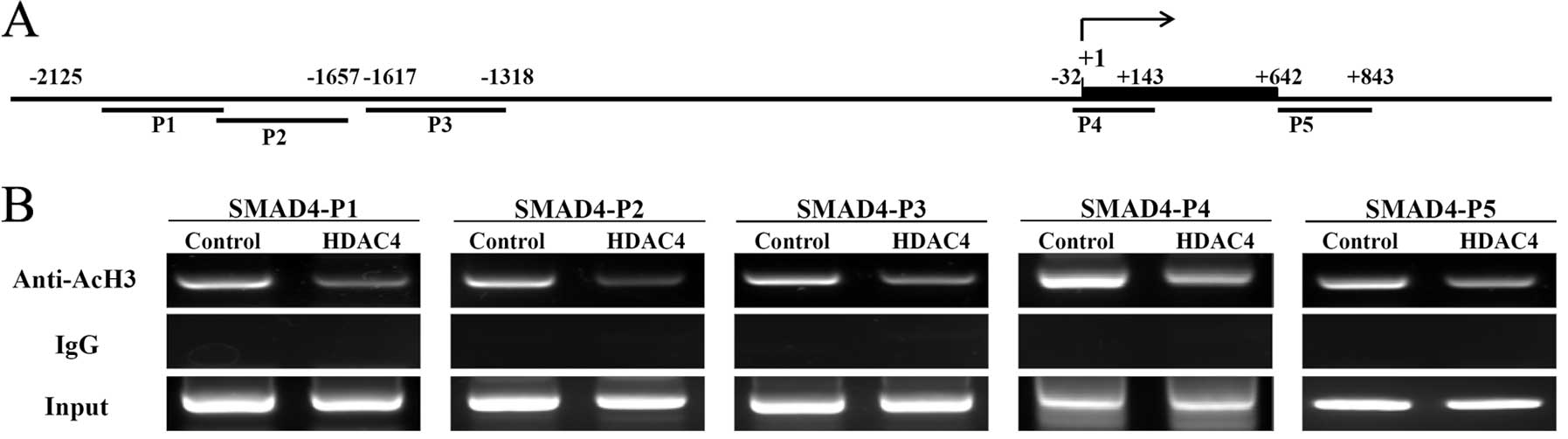
(Fig. 5A). The acetylation status
of histones in defined regions of SMAD4 was investigated using ChIP
assays comparing acetylation between control and HDAC4OE cells. The
ChIP results clearly confirmed that acetylation of histone H3 was
reduced at five regions within the SMAD4 promoter following
overexpression of HDAC4 (Fig. 5B).
Furthermore, ChIP assays using a HDAC4 antibody were unable to
detect the SMAD4 band, which indicated that HDAC4 does not bind
directly to the SMAD4 promoter region (data not shown).
Consequently, we hypothesized that HDAC4 may form a complex with a
transcription factor bound to the SMAD4 promoter.
Discussion
Alterations in the global pattern of histone
acetylation have been observed in several types of cancer. For
instance, Fraga et al(25)
reported that loss of acetylation at lysine 16 of histone 4 (H4K16)
is a common characteristic of cancer cells, and global
hypomethylation of lysine 8 of histone 3 (H3K8) has been suggested
to be a prognostic indicator of tumorigenesis in breast cancer
(26). The relationship between
hypomodified and hypermodified histones has also been reported to
be associated with mortality in prostate and lung cancer (27,28).
In addition, we previously identified differences in HDAC
expression between metastatic and non-metastatic breast cancer
cells, with high HDAC expression in metastatic breast cancer cells
related to invasiveness. However, despite the high expression of
HDAC4, it was not shown to participate in invasion (12). Overexpression of HDAC4 has been
reported to cause gene-induced resistance to platinum chemotherapy
by modulating the acetylation of signal transducer and activator of
transcription (STAT) 1 in ovarian cancer (13). In addition, HDAC4 was shown to
modulate resistance to docetaxel under hypoxic conditions and the
acetylation of HIF1α in hepatoma cells (9). Therefore, we hypothesized that HDAC4
expression was related to chemoresistance in breast cancer and
sought to identify the relationship between HDAC4 expression and
5-FU resistance in breast cancer (Fig.
1B and D). We confirmed that HDAC4 overexpression in MCF-7
cells induced 5-FU resistance, whereas HDAC4 knockdown in HDAC4OE
cells restored sensitivity to the drug (Fig. 2B and D), suggesting that HDAC4
expression induces 5-FU resistance in breast cancer. Both the
14-3-3σ and JAK/STAT pathways have recently been shown to be
associated with 5-FU resistance in MCF-7 cells (29,30).
We profiled genes that were altered by HDAC4
overexpression as HDAC4 is known to regulate the transcription and
stability of other genes and pathways related to acquired
resistance through HDAC4 overexpression (Fig. 3A–C). The results of our studies
indicated that genes related to the TGFβ signaling pathway,
including SMAD4, SMAD6, BMP6, ID1, TGFβ2 and TGFβ3, were most
significantly affected by altered HDAC4 expression (Fig. 3D). Among these genes, SMAD4 has been
most commonly reported as downregulated in numerous types of
cancer. Papageorgis et al(14) previously suggested that SMAD4
inactivation induces malignancy in colon cancer and 5-FU
chemoresistance, and the results of a study conducted by Shi et
al(31) indicated that
reductions in SMAD4 expression promoted cell proliferation and
invasion in colorectal carcinomas. In addition, advanced gastric
cancer displays lower SMAD4 expression than early gastric cancer
(32). In breast cancer, Stuelten
et al(33) found decreased
levels of SMAD4 expression in breast cancer tissue and demonstrated
that low SMAD4 expression was correlated with poor prognosis
(34). He et al(35) reported that SMAD4 mRNA and protein
levels were associated with poor outcome in glioma patients. Of
note, our study confirmed the HDAC4-mediated downregulation of
SMAD4 in HDAC4OE cells (Fig. 4A and
B). Moreover, we verified that SMAD4 mediated 5-FU resistance
in MCF-7 cells (Fig. 4C and D). As
SMAD4 has been shown to suppress tumor growth by inducing apoptosis
in MCF-7 cells (36),
HDAC4-mediated regulation of SMAD4 transcriptional expression may
be one of the mechanisms underlying 5-FU resistance.
Most genes develop transcriptional regulation
through epigenetic modifications, such as methylation and
acetylation, without alteration to their DNA sequences. For
example, HDAC4 was shown to regulate histone H3 acetylation in the
p21 proximal promoter in colon, ovarian cancer, and osteosarcoma
cells (37,38). In addition, Reddy et
al(24) suggested that HDAC4
forms a nucleosome remodeling and deacetylating (NuRD) complex in
MCF-7 cells and that this complex represses tumor-suppressing gene
transcription. In our study, the SMAD4 promoter appeared to have
reduced histone H3 acetylation following HDAC4 overexpression
(Fig. 5B), and based on these
findings, we suggest that increased deacetylation of the SMAD4
promoter due to HDAC4-mediated negative regulation of SMAD4 gene
expression may lead to chemoresistance.
In conclusion, we provide evidence supporting the
relevance of HDAC4 expression for 5-FU resistance in breast cancer
as well as the molecular basis for HDAC4-mediated gene regulation.
We also suggest that HDAC4 regulates SMAD4 expression and modifies
histone H3 in the SMAD4 promoter region.
Acknowledgements
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF), funded by the Ministry of Education, Science and Technology
(2010-0007110).
References
|
1
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellis L, Atadja PW and Johnstone RW:
Epigenetics in cancer: targeting chromatin modifications. Mol
Cancer Ther. 8:1409–1420. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sandoval J and Esteller M: Cancer
epigenomics: beyond genomics. Curr Opin Genet Dev. 22:50–55. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakagawa M, Oda Y, Eguchi T, et al:
Expression profile of class I histone deacetylases in human cancer
tissues. Oncol Rep. 18:769–774. 2007.PubMed/NCBI
|
|
5
|
Khan O and La Thangue NB: HDAC inhibitors
in cancer biology: emerging mechanisms and clinical applications.
Immunol Cell Biol. 90:85–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dell’Aversana C, Lepore I and Altucci L:
HDAC modulation and cell death in the clinic. Exp Cell Res.
318:1229–1244. 2012.PubMed/NCBI
|
|
7
|
Song B, Wang Y, Xi Y, et al: Mechanism of
chemoresistance mediated by miR-140 in human osteosarcoma and colon
cancer cells. Oncogene. 28:4065–4074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Crea F, Nobili S, Paolicchi E, et al:
Epigenetics and chemoresistance in colorectal cancer: an
opportunity for treatment tailoring and novel therapeutic
strategies. Drug Resist Updat. 14:280–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geng H, Harvey CT, Pittsenbarger J, et al:
HDAC4 protein regulates HIF1α protein lysine acetylation and cancer
cell response to hypoxia. J Biol Chem. 286:38095–38102. 2011.
|
|
10
|
Tinari N, De Tursi M, Grassadonia A, et
al: An epigenetic approach to pancreatic cancer treatment: the
prospective role of histone deacetylase inhibitors. Curr Cancer
Drug Targets. 12:439–452. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim MG, Pak JH, Choi WH, Park JY, Nam JH
and Kim JH: The relationship between cisplatin resistance and
histone deacetylase isoform overexpression in epithelial ovarian
cancer cell lines. J Gynecol Oncol. 23:182–189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park SY, Jun JA, Jeong KJ, et al: Histone
deacetylases 1, 6 and 8 are critical for invasion in breast cancer.
Oncol Rep. 25:1677–1681. 2011.PubMed/NCBI
|
|
13
|
Stronach EA, Alfraidi A, Rama N, et al:
HDAC4-regulated STAT1 activation mediates platinum resistance in
ovarian cancer. Cancer Res. 71:4412–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Papageorgis P, Cheng K, Ozturk S, et al:
Smad4 inactivation promotes malignancy and drug resistance of colon
cancer. Cancer Res. 71:998–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takagi Y, Kohmura H, Futamura M, et al:
Somatic alterations of the DPC4 gene in human colorectal cancers in
vivo. Gastroenterology. 111:1369–1372. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang LH, Kim SH, Lee JH, et al:
Inactivation of SMAD4 tumor suppressor gene during gastric
carcinoma progression. Clin Cancer Res. 13:102–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hahn SA, Schutte M, Hoque AT, et al: DPC4,
a candidate tumor suppressor gene at human chromosome 18q21.1.
Science. 271:350–353. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schutte M, Hruban RH, Hedrick L, et al:
DPC4 gene in various tumor types. Cancer Res. 56:2527–2530.
1996.PubMed/NCBI
|
|
19
|
Nagatake M, Takagi Y, Osada H, et al:
Somatic in vivo alterations of the DPC4 gene at 18q21 in human lung
cancers. Cancer Res. 56:2718–2720. 1996.PubMed/NCBI
|
|
20
|
Aitchison AA, Veerakumarasivam A, Vias M,
et al: Promoter methylation correlates with reduced Smad4
expression in advanced prostate cancer. Prostate. 68:661–674. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han S, Lu J, Zhang Y, et al: Recruitment
of histone deacetylase 4 by transcription factors represses
interleukin-5 transcription. Biochemical J. 400:439–448. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Feng Y, Xu L, et al: YY1
restrained cell senescence through repressing the transcription of
p16. Biochim Biophys Acta. 1783:1876–1883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren G, Zhang G, Dong Z, et al: Recruitment
of HDAC4 by transcription factor YY1 represses HOXB13 to affect
cell growth in AR-negative prostate cancers. Int J Biochem Cell
Biol. 41:1094–1101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reddy SD, Pakala SB, Molli PR, et al:
Metastasis-associated protein 1/histone deacetylase 4-nucleosome
remodeling and deacetylase complex regulates phosphatase and tensin
homolog gene expression and function. J Biol Chem. 287:27843–27850.
2012. View Article : Google Scholar
|
|
25
|
Fraga MF, Ballestar E, Villar-Garea A, et
al: Loss of acetylation at Lys16 and trimethylation at Lys20 of
histone H4 is a common hallmark of human cancer. Nat Genet.
37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elsheikh SE, Green AR, Rakha EA, et al:
Global histone modifications in breast cancer correlate with tumor
phenotypes, prognostic factors, and patient outcome. Cancer Res.
69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seligson DB, Horvath S, Shi T, et al:
Global histone modification patterns predict risk of prostate
cancer recurrence. Nature. 435:1262–1266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barlési F, Giaccone G, Gallegos-Ruiz MI,
et al: Global histone modifications predict prognosis of resected
non small-cell lung cancer. J Clin Oncol. 25:4358–4364.
2007.PubMed/NCBI
|
|
29
|
Uluer ET, Aydemir I, Inan S, Ozbilgin K
and Vatansever HS: Effects of 5-fluorouracil and gemcitabine on a
breast cancer cell line (MCF-7) via the JAK/STAT pathway. Acta
Histochem. 114:641–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng G, Xiong Y, Yi S, et al: 14-3-3σ
regulation by p53 mediates a chemotherapy response to
5-fluorouracil in MCF-7 breast cancer cells via Akt inactivation.
FEBS Lett. 586:163–168. 2012.
|
|
31
|
Shi Q, Zhong YS, Yao LQ, et al:
Down-regulation of Smad4 enhances proliferation and invasion of
colorectal carcinoma HCT116 cells and up-regulates Id2. Mol Med
Rep. 5:89–95. 2012.PubMed/NCBI
|
|
32
|
Kim JY, Park DY, Kim GH, et al: Smad4
expression in gastric adenoma and adenocarcinoma: frequent loss of
expression in diffuse type of gastric adenocarcinoma. Histol
Histopathol. 20:543–549. 2005.PubMed/NCBI
|
|
33
|
Stuelten CH, Buck MB, Dippon J, Roberts
AB, Fritz P and Knabbe C: Smad4-expression is decreased in breast
cancer tissues: a retrospective study. BMC cancer. 6:252006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Kruijf EM, Dekker TJ, Hawinkels LJ, et
al: The prognostic role of TGF-β signaling pathway in breast cancer
patients. Ann Oncol. 24:384–390. 2013.
|
|
35
|
He SM, Zhao ZW, Wang Y, et al: Reduced
expression of SMAD4 in gliomas correlates with progression and
survival of patients. J Exp Clin Cancer Res. 30:702011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Q, Wu L, Oelschlager DK, et al: Smad4
inhibits tumor growth by inducing apoptosis in estrogen
receptor-alpha-positive breast cancer cells. J Biol Chem.
280:27022–27028. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wilson AJ, Byun DS, Nasser S, et al: HDAC4
promotes growth of colon cancer cells via repression of p21. Mol
Biol Cell. 19:4062–4075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mottet D, Pirotte S, Lamour V, et al:
HDAC4 represses p21 (WAF1/Cip1) expression in human cancer cells
through a Sp1-dependent, p53-independent mechanism. Oncogene.
28:243–256. 2009. View Article : Google Scholar : PubMed/NCBI
|