Introduction
Cancer initiation commonly occurs when the coding
region of a proto-oncogene is mutated to induce a gain of function
of the resulting gene product, such that it is excessively active,
or when the coding region of a tumor-suppressor gene suffers a
mutation that inactivates the resulting gene product. In either
case, the pathophysiologic consequence is excessive cell cycle
progression or defective programmed cell death. Cancer progression
and metastasis are also genetically programmed. However, whereas
mutations in the coding regions of critical genes underlie early
transformation, metastasis gene products are typically not mutated
in cancer. We previously demonstrated that aberrant expression or
splicing of metastasis-related genes underlie tumor progression
(1,2).
Osteopontin is a metastasis-related gene that
contributes to the progression of over 30 forms of cancer (3–5).
Aberrant splicing of osteopontin in cancers has been accounted for
by our identification of the variant form osteopontin-c, which is
selectively expressed in cancer cells but is absent from
non-transformed cells (6–8). Osteopontin-c supports
anchorage-independent survival and expansion, which are essential
components of tumor dissemination (9).
Although osteopontin (encoded by the gene spp1) has
been known to be produced at elevated levels by cancer cells
(10), the molecular underpinning
for its aberrant expression in cancer is incompletely accounted
for. Osteopontin may be induced as a downstream signal transduction
target of proto-oncogenic growth factors (11) or secondary to gain-of-function
events in transforming signaling pathways (12–14).
In either case, the binding of cognate transcription factors to
spp1 promoter regions is causative for the upregulated expression.
This opens the possibility that mutations or polymorphisms in the
promoter of the spp1 gene (Fig. 1)
may predispose to various levels of expression after
transformation, and hence to various levels of tumor
aggressiveness. Here, we investigated this hypothesis for breast
cancer.
 | Figure 1Spp1 polymorphisms. (A) Promoter. The
sequence is derived from NW_001838915.1 (whole genome shotgun
sequence) and NT_016354.19 (genomic contig) (the 160 bases proximal
to the transcription start site are also confirmed by GenBank nos.
NM_001040058.1, NM_001040060 and NM_000582; of note, the GenBank
sequences S78410.1 and D14813.1 contain a 60 nucleotide gap which
is likely a cloning artifact). The silent exon 1 is grey. The ‘A’
that starts the conventional numbering of the promoter sequence is
displayed in bold green font, as is the downstream ATG start site.
The literature has identified 12 polymorphic sites (red letters on
yellow background) in the spp1 promoter, of which 6 have rs or ss
numbers (positions −66, −156, −443, −616, −1748, −1776) (22,15,26,27,16).
Additional polymorphic sites have been reported, and comprise
−145/−146 (28), −302 (28), −593 (29), −655 (30), −1282 (31), −1625 (31) and −2053 (15). With the exception of one
insertion/deletion polymorphism at position −156, all are single
nucleotide exchanges. The position and nature of the polymorphism
is indicated above each site in blue. (B) Coding sequence. The
polymorphisms with assigned rs numbers are identified in the NCBI
SNP database. The protein sequence is shown under the nucleotide
sequence, and for non-synonymous base changes the amino acid
changes are depicted in red letters on yellow background. The
position and nature of the polymorphism is indicated above each
site in blue. |
Materials and methods
Patients
There were 4 sources of specimens. DNA from breast
cancer patients and healthy controls was obtained from the Division
of Human Genetics at The Ohio State University (50 breast cancers,
50 untransformed surrounding tissues, 50 healthy breasts). From
tumors previously analyzed for osteopontin RNA expression (6), DNA was obtained by phenol/chloroform
extraction (23 breast cancers, 11 surrounding tissues, 15 healthy
breasts). DNA from breast cancers and surrounding tissues was
purchased from Bioserve (86 tumors and 50 untransformed surrounding
tissues) and from Origene (82 tumors). The total number of samples
was 241 breast cancers, 111 surrounding tissues and 65 healthy
breast specimens.
DNA genotyping
Genotyping was carried out using ABI PRISM 7900HT
sequence detection system after performing polymerase chain
reaction (PCR) on the DNA samples. The PCR was performed as
directed by the ABI protocol for the TaqMan SNP genotyping assays
using TaqMan Universal PCR master mix, primer and TaqMan probe
(VIC/FAM) dye mix, and 5 ng/μl genomic DNA sample. The total
reaction volume was 5 μl. Then, post-PCR plate reads were performed
using the sequence detection system instrumentation to identify the
distinct alleles according to their fluorescent signals. One probe
set tested the spp1 polymorphic promoter sites −66 (rs28357094),
−443 (rs11730582), −1748 (rs2728127) and −1776 (rs29001511). We
also set out to investigate non-synonymous DNA sequence variations
in the coding region. The available probes for this comprised the
positions 305 (rs11544546), 367 (rs11544549), 794 (rs7435825) and
1025 (rs4660) and were all included in the present study.
RNA and real-time RT-PCR
Specimens of human breast tumors, non-transformed
surrounding tissue, as well as healthy breast tissue (obtained from
reduction mammoplasties) were provided by the tissue procurement
facility of the University of Cincinnati Medical Center/Children’s
Hospital (6). Total RNA was
extracted from specimens using TRIzol reagent (Invitrogen Life
Technologies). Total RNA was used for cDNA synthesis by reverse
transcription with Superscript II (Invitrogen Life Technologies)
according to the manufacturer’s protocol in a total volume of 20
μl.
All PCR reactions were performed on a Smart Cycler
(Cepheid, Sunnyvale, CA, USA) using SYBR-Green detection format.
cDNA (0.5 μl) was added to each PCR reaction in a total volume of
25 μl using the standard Invitrogen Life Technologies PCR buffer
system with optimized concentrations of MgCl2. For each
experiment a no-template reaction and cDNA from the reference cell
line MDA-MB−435 were included as negative and positive controls.
The conditions for PCR were a 94°C denaturation for 120 sec
followed by 40 cycles of: 94°C melting for 15 sec, a primer set
specific annealing temperature for 30 sec (6), extension at 72°C for 30 sec, and
ending with a melting curve program (60–95°C with a heating rate of
0.2°C and a continuous fluorescence measurement), and finally a
cooling step to 4°C. Product purity, product size, and absence of
primer dimers were confirmed by DNA melting curve analysis. Melt
curves yielded a single sharp peak for all template reactions, and
a minimal melt peak (resulting from primer dimers) or no melt peaks
for the no-template control reactions.
Statistics
We performed the Hardy-Weinberg exact test to
analyze deviations from equilibrium and association analysis to
evaluate genetic effects on phenotype using the statistical
software R (www.R-project.org). Single nucleotide
polymorphisms (SNPs) whose genotype frequencies departed from
Hardy-Weinberg equilibrium at p<0.01 were filtered out. Thus, we
evaluated associations among the 3 promoter SNPs rs11730582,
rs2728127 and rs29001511 in the promoter region with various breast
cancer characteristics. These statistical evaluations were carried
out using multivariate logistic regression under an additive
genetic model by χ2 test. The accepted significance
level for association analysis was 0.1.
The case-control haplotype analysis was performed
using Haploview v4.2 (http://www.broad.mit.edu/mpg/haploview). Similar to
the association analysis, the 3 SNPs, rs11730582, rs2728127 and
rs29001511 in the promoter region, were used to generate haplotype
frequencies, as the genotype data for these SNPs had a p-value
above the cut-off value of 0.01.
Results
Patient demographics
This study comprised 241 breast cancer specimens,
for which DNA from normal surrounding tissue was available for 111,
and 65 healthy breast samples. The entire cohort consisted
exclusively of women. In all groups, the mean age was close to 50
years. The demographic and cancer characteristics are specified in
Table I.
 | Table IPatient demographics. |
Table I
Patient demographics.
| Characteristics | Breast cancer
(241) | Normal surrounding
tissue (111) | Normal breasts
(71) |
|---|
| Cancer subtypes |
| Ductal | 212 | 98 | 0 |
| Lobular | 13 | 7 | 0 |
| Mucinous | 2 | 2 | 0 |
| Papillary | 0 | 0 | 0 |
| Age (years) (means ±
SEM) | 52.22±0.79 | 50.71±0.97 | 49.13±1.60 |
| Race |
| Caucasian | 85 | 49 | 53 |
| Asian | 87 | 51 | 1 |
| Black | 17 | 3 | 7 |
| Hispanic | 0 | 0 | 0 |
| Middle Eastern | 0 | 0 | 1 |
| Tumor size | 1.9±1.1 | 1.9±1.1 | na |
| Tumor grade |
| 1 | 13 | 6 | na |
| 2 | 63 | 34 | na |
| 3 | 70 | 19 | na |
| Tumor stage |
| T1 | 76 | 43 | na |
| T2 | 114 | 50 | na |
| T3 | 24 | 7 | na |
| T4 | 7 | 2 | na |
| N0 | 90 | 44 | na |
| N1 | 87 | 42 | na |
| N2 | 11 | 4 | na |
| N3 | 24 | 12 | na |
| N4 | 1 | 0 | na |
| M0 | 89 | 51 | na |
| M1 | 1 | 0 | na |
| Tumor stage |
| I | 42 | 22 | na |
| II | 130 | 60 | na |
| III | 52 | 20 | na |
| IV | 1 | 0 | na |
| ER status |
| + | 98 | 47 | na |
| − | 79 | 33 | na |
| PR status |
| + | 94 | 42 | na |
| − | 85 | 38 | na |
| HER2 status |
| + | 54 | 18 | na |
| − | 84 | 36 | na |
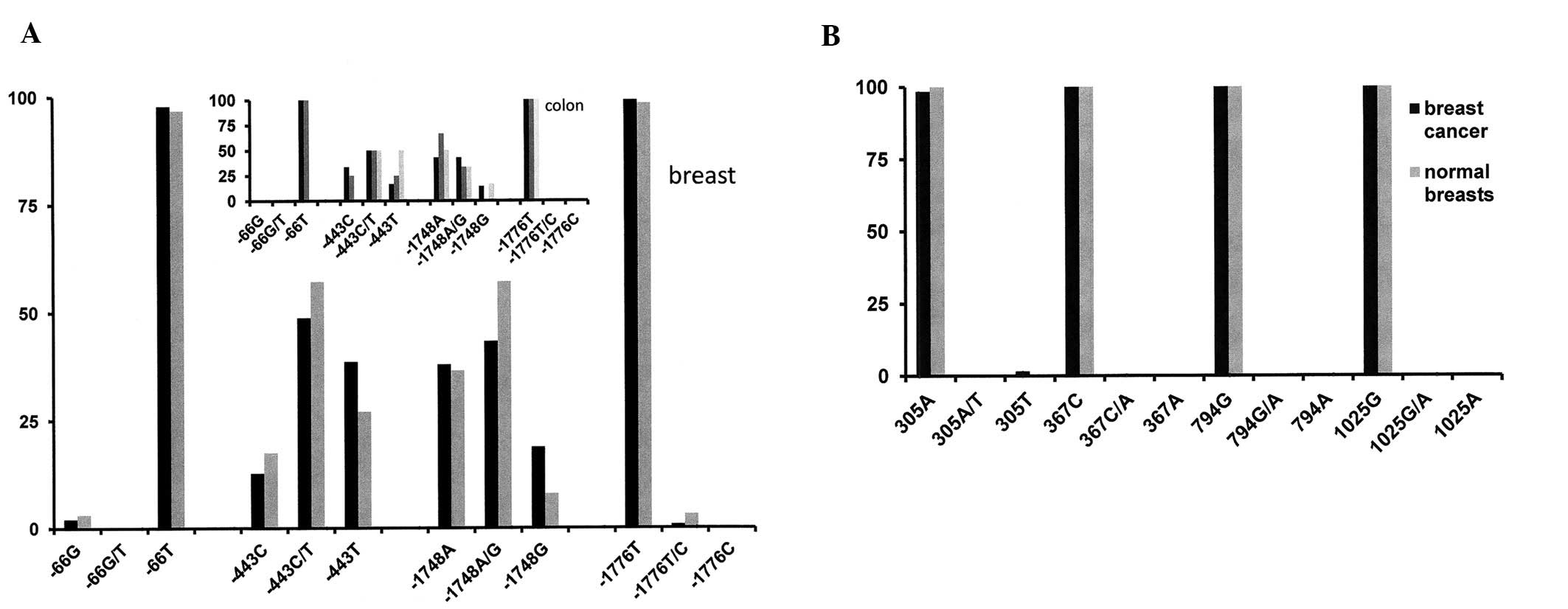
Individual polymorphic sites and
cancer
The polymorphic site in position −66 was not in
Hardy-Weinberg equilibrium and hence was not included in further
analyses. When comparing the other 3 promoter SNPs between cancers
and healthy controls, the association analysis by χ2
test using multivariate logistic regression under an additive
genetic model did not reveal significant differences between the
groups for positions −443, −1748 or −1776. However, a separate
analysis using the Cochran-Armitage trend test (CATT) and assuming
a recessive genetic model reached accepted levels of significance
for all three sites, implying the possibility of a weak association
of these polymorphisms with cancer. A small set of DNAs from colon
samples showed a distribution of SNPs very similar to breast cancer
(Fig. 2A). For studying the coding
region polymorphisms that are associated with amino acid changes, 4
probes were available. At SNP position 305, 3 tumors had a deviant
genotype from all other specimens. This SNP was in Hardy-Weinberg
dysequilibrium. The polymorphic sites in positions 367, 794 and
1025 showed one homozygous genotype for all specimens in this study
and therefore were not further analyzed (Fig. 2B).
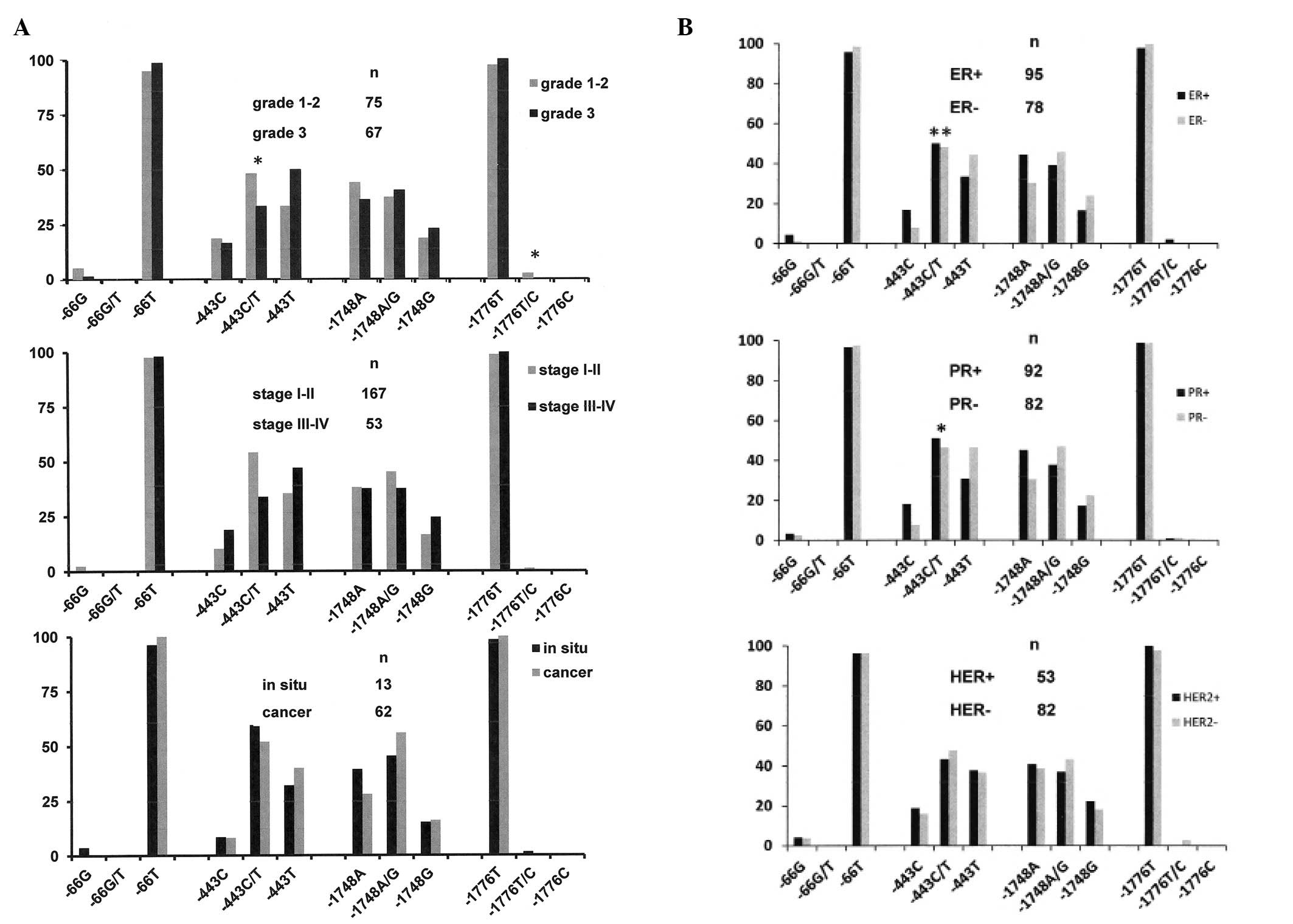
Individual polymorphic sites and clinical
measures of cancer
Within the cancer cohort, the polymorphisms in
position −443 was correlated with tumor grade (after combining
grades 1 plus 2 and comparing them to grade 3). The difference in
position −443 between low grade and high grade cancers was
confirmed by reanalysis with an allelic based test, with a
recessive genetic model, and an additive/multiplicative genetic
model using CATT. The polymorphic site in position −1776 just
barely reached a level of significance when grades 1 and 2 were
combined (grade 1 alone contained only 13 tumor samples) and
compared to grade 3. However a reanalysis of grade 1 vs. grade 2
and grade 2 vs. grade 3 did not reflect significant differences in
genotype at this position. There was no association between the
promoter SNPs and tumor stage (I–II vs. III–IV) or in situ
carcinoma vs. cancer (Fig. 3A). The
latter results were expected as stage and early transformation are
determined by the sampling time more than by tumor genetics.
Osteopontin expression has been associated with
breast cancer progression, regardless of the histologic subtype of
the cancer (4,6). Importantly, the polymorphic site at
−443, but not −1748 or −1776, showed differences between
ER-positive and ER-negative breast cancers and between PR-positive
and PR-negative breast cancers, but there was no association with
HER2 status. The −443 allele T was more common in the ER-negative
cancers and in the PR-negative cancers (Fig. 3B).
Somatic mutations
For 111 cancers, DNA from the matching untransformed
surrounding tissues was available. We tested it to detect possible
somatic mutations. None were found in position −66. In position
−443, 2 tumors were heterozygous in homozygous hosts, one with the
C/C genotype and the other with the T/T genotype. In position
−1748, 2 tumors had a heterozygous genotype, altered from the host
homozygous A/A. In position −1776, 1 tumor was heterozygous in a
homozygous T/T host. These results suggest that tumors may
encounter somatic mutations in the spp1 promoter that have the
potential to affect expression levels. For the coding region
polymorphic sites (positions 305, 367, 794 and 1025), there were no
differences between tumor and untransformed surrounding tissue.
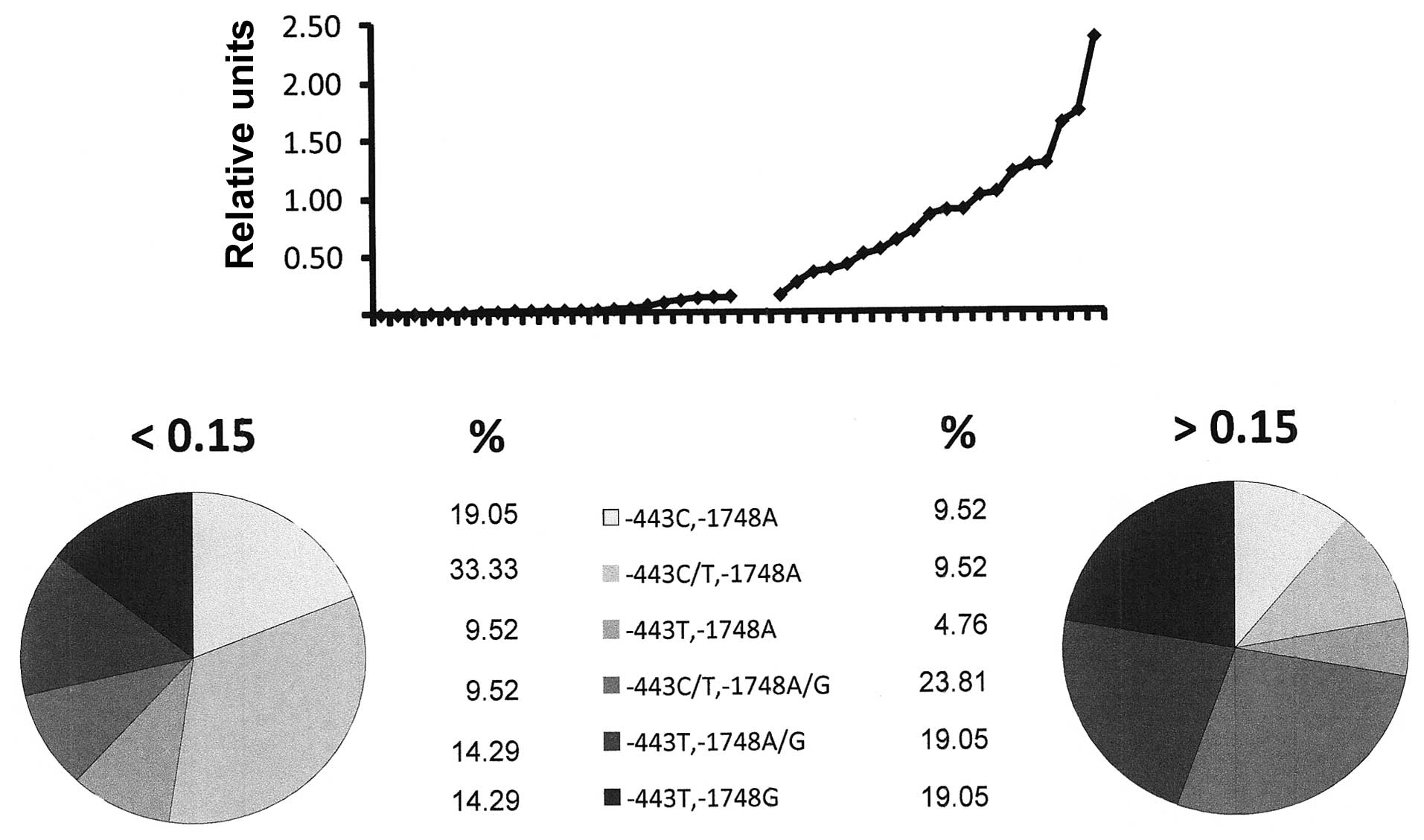
Associations of polymorphisms and
expression levels
For a subset of samples, information was available
concerning the expression levels of osteopontin according to
real-time RT-PCR analysis from breast tissue. We questioned whether
the promoter haplotype was correlated with expression, using 0.15
relative units as the cut-off between high and low osteopontin
expression. As the allelic distribution in positions −66 and −1776
was almost homogeneous in patients as well as in the normal
controls, we focused on −443 and −1748. The base G in position
−1748, on a homozygous or heterozygous background, was associated
with higher expression levels of osteopontin RNA in the breast
tissue (74% G in high expressors vs. 41% G in low expressors)
(Fig. 4). Of the 6 tumors with the
highest osteopontin expression (>1.2 relative units), 4 had a G
in position −1748. The polymorphic site in position −443 appeared
to have a lesser effect, but the fraction of T/T was increased and
the fraction of C/C was decreased in the population of high
expressors compared to the low expressors (47% T/T in high
expressors vs. 38% T/T in low expressors).
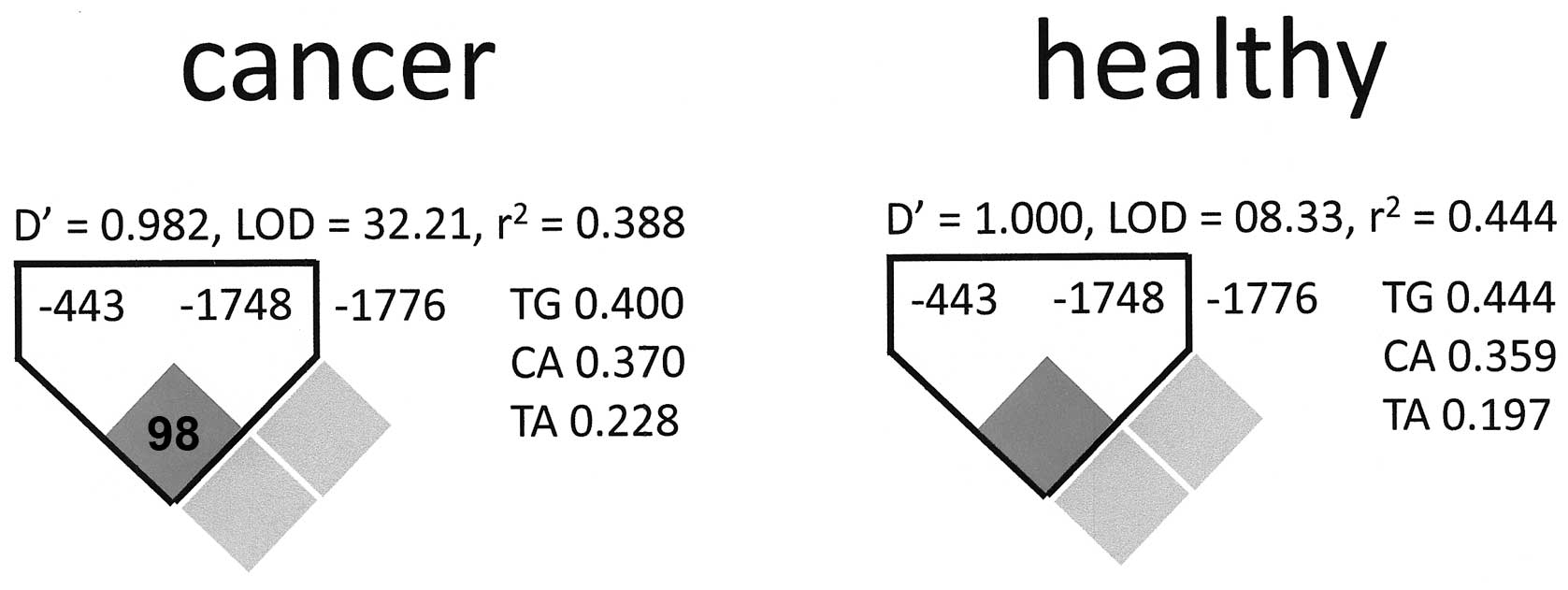
Haplotype analysis
We performed a haplotype analysis for the promoter
SNPs. The polymorphic site in position −66 was eliminated from this
evaluation as it was not in Hardy-Weinberg equilibrium. Among the
other three sites, there was an association between −443 and −1748
in the cancer patient group as well as in the healthy controls. No
association was found for SNP −1776 with either of the other
polymorphic sites (Fig. 5).
Discussion
Certain cancer-associated mutations may be
individually transforming, such as the chromosome translocation
that generates the chimeric kinase BCR-ABL in CML or the
loss-of-function mutations of RB that cause retinoblastoma. Other
mutations or polymorphisms have the nature of quantitative trait
loci that collectively affect the risk for transformation or
progression, with each individual site contributing only
moderately. It is safe to assume that many of the cancer-associated
genetic changes in the latter category have not been
identified.
The spp1 gene, which encodes osteopontin, is located
on chromosome 4q22.1. The very diverse roles of the gene product in
physiology and pathophysiology are regulated on the
post-transcriptional level (glycosylation, phosphorylation,
sulfation, calcium binding, heparin binding, proteolysis,
transglutamination), in RNA processing (3 alternative splice
variants, alternative translation from a non-canonical start site),
and genetically (polymorphisms in coding and non-coding regions).
Since 2004 (15,16), there has been increasing interest in
the biological roles for spp1 promoter polymorphisms in various
pathologies. Here, we report that promoter polymorphisms are also
relevant for breast cancer.
Abundant production of osteopontin is correlated
with aggressiveness (higher stage, higher grade and early
progression) in multiple forms of cancer (3). Known mechanisms for osteopontin
induction in cancer include the activation of gene expression due
to elevated signal transduction and the alternative splicing of the
spp1 transcript. It was possible that high expression polymorphisms
in the spp1 promoter may also contribute to an elevated risk for
tumor aggressiveness. Here, we tested this hypothesis. We found the
polymorphic site in position −443 of the promoter to be associated
with tumor grade, such that the allele T is more common in high
grade tumors. It is also more common in high expressors of
osteopontin compared to low expressors. Furthermore, this allele
occurs at a higher frequency in cancers that lack ER over cancers
that express this receptor and in cancers that lack PR over those
that express PR. We found that T in position −443 was associated
with higher aggressiveness of cancers, and consistently hormone
receptor-negative cancers tend to grow more rapidly and have a
worse prognosis than breast tumors that express ER or PR.
To assess our results against the existing base of
knowledge, we compared this study to the distribution of
polymorphic site frequencies according to public databases
(Table II). The polymorphism in
position −443 has been associated with various disease phenotypes
(Table III). A DNA sequence
similar to a c-MYB core binding motif, CAGTT, immediately precedes
the −443 polymorphic promoter position CAAGTT(C/T). However, the
canonical c-MYB site is 5′-(T/C)AAC(G/T)G-3′ (17), and transcription via c-MYB from the
non-canonical site in the spp1 promoter may be context-dependent.
While c-MYB causes higher transcription from the C allele, there is
evidence that under some circumstances the T allele may be
associated with higher levels of expression. In melanoma and
gastric cancer, the −443 allele C may have elevated transcription
over allele T or heterozygous C/T, causing an increased risk for
tumor progression and reduced survival rates (18,19).
In hepatitis C, the T/T genotype has been associated with an
increased anti-viral response to hepatitis C [which requires high
levels of osteopontin (20)],
however, the T allele may be more common in patients with high
hepatitis activity (reflective of a compromised antiviral response
due to low levels of osteopontin secretion) (21,22).
In diseases with autoimmune components, the published results
likewise point to a complex role for the SNP in position −443.
Nephropathy in diabetes is more common in carriers of the T allele
(23), which may reflect increased
inflammation due to high osteopontin expression. Conversely,
thrombocytopenia and hemolytic anemia in lupus have an
autoantibody-mediated pathogenesis, which is supported by
osteopontin, more strongly in carriers of the C allele (24). The polymorphic site −443 is
associated with osteoarthritis risk and severity. Thrombin-cleaved
osteopontin levels in the synovial fluid are lower in subjects with
the −443T/T genotype, resulting in milder disease (25). In the present study the T allele was
represented more frequently than the C allele at high tumor grade
and in tumors with high osteopontin RNA levels (of note, for a
subset of samples this result was confirmed by reanalysis in an
external core facility to exclude the possibility of an erroneous
data set). This implies an important role for c-MYB-independent
osteopontin expression in breast cancer.
 | Table IIFrequencies of the polymorphisms. |
Table II
Frequencies of the polymorphisms.
| Position | rs number | Alleles | HapMap ratio | ABI ratio | NCBI SNP ratio | MAF | Present study
ratio |
|---|
| −1776 | rs29001511 | C/T | 0.017/0.983 | 0.03/0.97
(Cauc)
0.00/1.00 (Black) | 0.017/0.983 | C=0.0059/13 | 0.007/0.993 |
| −1748 | rs2728127 | A/G | 0.5/0.5 | 0.66/0.34
(Cauc)
0.53/0.47 (Black) | 0.624/0.376 | G=0.3679/805 | 0.626/0.374 |
| −443 | rs11730582 | C/T | 0.26/0.74 | 0.44/0.56
(Cauc)
0.14/0.86 (Black)
0.305/0.695 (Asian) | 0.300/0.700 | C=0.3419/748 | 0.404/0.596 |
| −66 | rs28357094 | G/T | 0.5/0.5 | 0.26/0.74
(Cauc)
0.09/0.91 (Black) | 0.170/0.830 | G=0.1175/257 | 0.036/0.964 |
| 305 | rs11544546 | A/T | - | - | - | - | 0.98/0.02 |
| 367 | rs11544549 | C/A | - | - | - | - | 1.00/0.00 |
| 794 | rs7435825 | G/A | 1.00/0.00
(Cauc)
0.81/0.19 (Black)
1.00/0.00 (Asian) | 1.00/0.00
(Cauc)
0.88/0.14 (Black) | 1.00/0.00 | A=0.043/94 | 1.00/0.00 |
| 1025 | rs4660 | G/A | 1.00/0.00
(Cauc)
0.90/0.10 (Black)
1.00/0.00 (Asian) | 1.00/0.00
(Cauc)
0.88/0.12 (Black) | 0.951/0.049 | A=0.017/38 | 1.00/0.00 |
 | Table IIIFunctions of the polymorphic sites in
the spp1 promoter. |
Table III
Functions of the polymorphic sites in
the spp1 promoter.
| Position | Transcription
factor | Disease | Refs. |
|---|
| −66 | SP1/SP3 | Duchenne muscular
dystrophy | (32) |
| Osteoarthritis | (25) |
| Atherosclerosis
predisposition | (33) |
| Type 1
diabetes | (15,16,34) |
| −145/−146 | |
Nephrolithiasis | (28) |
| −156 | RUNX2 | Glioma | (15) |
| Pseudoxanthoma
elasticum | (31) |
| Systemic lupus
erythematosus | (35,36) |
| Systemic
sclerosis | (37) |
| Diastolic
dysfunction in hypertension | (38) |
| −443 | MYB | Hepatitis c | (21,22) |
| Melanoma | (18) |
| Gastric cancer | (19) |
| Diabetic
nephropathy | (23) |
| Thrombocytopenia,
anemia in SLE | (24) |
| Myocardial
infarction | (30) |
| Osteoarthritis | (25) |
| −593 | |
Nephrolithiasis | (29) |
| −1748 | | Pseudoxanthoma
elasticum | (31) |
The SNP frequency in the osteopontin promoter
(Fig. 1A) was roughly consistent
with the estimated variability in DNA sequence among humans which
is 0.3%. Notably, the coding region polymorphisms reported in the
NCBI SNP database was disproportionately higher. This is consistent
with the low evolutionary preservation of the osteopontin protein
sequence and with the low structural constraints of the molecule.
It may reflect an unstable chromosome locus. However, few of the
deposited SNPs are backed by larger population studies and those
located in critical functional sites (such as mutations of the RGD
motif) may be exceedingly rare. In this study, we assessed only
non-synonymous (i.e. amino acid-changing) polymorphic sites, for
which probes were available. The study population was entirely
homozygous for 3 of the 4. Further investigation is required to
assess the potential roles of coding region polymorphisms within
the spp1 gene in breast cancer. 56 of our specimens were assessed
with 2–6-fold coverage. For most of them, the reproducibility was
100%. Few samples with lower quality DNA had reproducibility in 4
of 6 repeats.
Acknowledgements
This research was supported by DOD grants BC095225
and PR094070 to G.F.W. Patient samples were obtained under IRB
protocol 04-01-29-01 from the University of Cincinnati.
References
|
1
|
Weber GF and Ashkar S: Stress response
genes: the genes that make cancer metastasize. J Mol Med.
78:404–408. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weber GF: Molecular mechanisms of
metastasis. Cancer Lett. 270:181–190. 2008. View Article : Google Scholar
|
|
3
|
Weber GF, Lett S and Haubein N:
Osteopontin is a marker for cancer aggressiveness and patient
survival. Br J Cancer. 103:861–869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weber GF, Lett GS and Haubein NC:
Categorical meta-analysis of osteopontin as a clinical cancer
marker. Oncol Rep. 25:433–441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Weber GF: The cancer biomarker
osteopontin: combination with other markers. Cancer Genomics
Proteomics. 8:263–288. 2011.PubMed/NCBI
|
|
6
|
Mirza M, Shaughnessy E, Hurley JK,
Vanpatten KA, Pestano GA, He B and Weber GF: Osteopontin-c is a
selective marker for breast cancer. Int J Cancer. 122:889–897.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sullivan J, Blair L, Alnajar A, Aziz T, Ng
CY, Chipitsyna G, Gong Q, Witkiewicz A, Weber GF, Denhardt DT, Yeo
CJ and Arafat HA: Expression of a prometastatic splice variant of
osteopontin, OPNC, in human pancreatic ductal adenocarcinoma.
Surgery. 146:232–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tilli TM, Franco VF, Robbs BK, Wanderley
JL, da Silva FR, de Mello KD, Viola JP, Weber GF and Gimba ER:
Osteopontin-c splicing isoform contributes to ovarian cancer
progression. Mol Cancer Res. 9:280–293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He B, Mirza M and Weber GF: An osteopontin
splice variant induces anchorage independence in human breast
cancer cells. Oncogene. 25:2192–2202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Senger DR, Asch BB, Smith BD, Perruzzi CA
and Dvorak HF: A secreted phosphoprotein marker for neoplastic
transformation of both epithelial and fibroblastic cells. Nature.
302:714–715. 1983. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Noda M, Yoon K, Prince CW, Butler WT and
Rodan GA: Transcriptional regulation of osteopontin production in
rat osteosarcoma cells by type β transforming growth factor.
J Biol Chem. 263:13916–13921. 1988.PubMed/NCBI
|
|
12
|
Chambers AF, Behrend EI, Wilson SM and
Denhardt DT: Induction of expression of osteopontin (OPN; secreted
phosphoprotein) in metastatic, ras-transformed NIH 3T3 cells.
Anticancer Res. 12:43–47. 1992.PubMed/NCBI
|
|
13
|
Zhang G, He B and Weber GF: Growth factor
signaling induces metastasis genes in transformed cells: molecular
connection between Akt kinase and osteopontin in breast cancer. Mol
Cell Biol. 23:6507–6519. 2003. View Article : Google Scholar
|
|
14
|
Whalen KA, Weber GF, Benjamin TL and
Schaffhausen BS: Polyomavirus middle T antigen induces the
transcription of osteopontin, a gene important for migration of
transformed cells. J Virol. 82:4946–4954. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giacopelli F, Marciano R, Pistorio A,
Catarsi P, Canini S, Karsenty G and Ravazzolo R: Polymorphisms in
the osteopontin promoter affect its transcriptional activity.
Physiol Genomics. 20:87–96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hummelshoj T, Ryder LP, Madsen HO, Odum N
and Svejgaard A: A functional polymorphism in the Eta-1
promoter is associated with allele specific binding to the
transcription factor Sp1 and elevated gene expression. Mol Immunol.
43:980–986. 2006.PubMed/NCBI
|
|
17
|
Deng QL, Ishii S and Sarai A: Binding site
analysis of c-Myb: screening of potential binding sites by using
the mutation matrix derived from systematic binding affinity
measurements. Nucleic Acids Res. 24:766–774. 1996. View Article : Google Scholar
|
|
18
|
Schultz J, Lorenz P, Ibrahim SM, Kundt G,
Gross G and Kunz M: The functional −443T/C osteopontin promoter
polymorphism influences osteopontin gene expression in melanoma
cells via binding of c-Myb transcription factor. Mol Carcinog.
48:14–23. 2009.
|
|
19
|
Zhao F, Chen X, Meng T, Hao B, Zhang Z and
Zhang G: Genetic polymorphisms in the osteopontin promoter
increases the risk of distance metastasis and death in Chinese
patients with gastric cancer. BMC Cancer. 12:4772012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashkar S, Weber GF, Panoutsakopoulou V,
Sanchirico ME, Janssen M, Zawaideh S, Rittling S, Denhardt DT,
Glimcher MJ and Cantor H: Eta-1 (osteopontin): an early component
of type-1 (cell-mediated) immunity. Science. 287:860–864. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shaker OG, Sadik NA and El-Dessouki A:
Single-nucleotide polymorphism in the promoter region of the
osteopontin gene at nucleotide −443 as a marker predicting the
efficacy of pegylated interferon/ribavirin-therapy in Egyptians
patients with chronic hepatitis C. Hum Immunol. 73:1039–1045.
2012.
|
|
22
|
Mochida S, Hashimoto M, Matsui A, Naito M,
Inao M, Nagoshi S, Nagano M, Egashira T, Mishiro S and Fujiwara K:
Genetic polymorphisms in promoter region of osteopontin gene may be
a marker reflecting hepatitis activity in chronic hepatitis C
patients. Biochem Biophys Res Commun. 313:1079–1085. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheema BS, Iyengar S, Ahluwalia TS, Kohli
HS, Sharma R, Shah VN, Bhansali A, Sakhuja V and Khullar M:
Association of an Osteopontin gene promoter polymorphism
with susceptibility to diabetic nephropathy in Asian Indians. Clin
Chim Acta. 413:1600–1604. 2012.
|
|
24
|
Trivedi T, Franek BS, Green SL, Kariuki
SN, Kumabe M, Mikolaitis RA, Jolly M, Utset TO and Niewold TB:
Osteopontin alleles are associated with clinical characteristics in
systemic lupus erythematosus. J Biomed Biotechnol. 2011:8025812011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Y, Yao M, Liu Q and Zhou C: OPN gene
polymorphisms influence the risk of knee OA and OPN levels in
synovial fluid in a Chinese population. Arthritis Res Ther.
15:R32013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Naito M, Matsui A, Inao M, Nagoshi S,
Nagano M, Ito N, Egashira T, Hashimoto M, Mishiro S, Mochida S and
Fujiwara K: SNPs in the promoter region of the osteopontin gene as
a marker predicting the efficacy of interferon-based therapies in
patients with chronic hepatitis C. J Gastroenterol. 40:381–388.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu G, Sun W, He D, Wang L, Zheng W, Nie H,
Ni L, Zhang D, Li N and Zhang J: Overexpression of osteopontin in
rheumatoid synovial mononuclear cells is associated with joint
inflammation, not with genetic polymorphism. J Rheumatol.
32:410–416. 2005.PubMed/NCBI
|
|
28
|
Gao B, Yasui T, Itoh Y, Li Z, Okada A,
Tozawa K, Hayashi Y and Kohri K: Association of osteopontin gene
haplotypes with nephrolithiasis. Kidney Int. 72:592–598. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gögebakan B, Igci YZ, Arslan A, Igci M,
Erturhan S, Oztuzcu S, Sen H, Demiryürek S, Arikoglu H, Cengiz B,
Bayraktar R, Yurtseven C, Sarıca K and Demiryürek AT: Association
between the T-593A and C6982T polymorphisms of the osteopontin gene
and risk of developing nephrolithiasis. Arch Med Res. 41:442–448.
2010.PubMed/NCBI
|
|
30
|
Schmidt-Petersen K, Brand E, Telgmann R,
Nicaud V, Hagedorn C, Labreuche J, Dördelmann C, Elbaz A,
Gautier-Bertrand M, Fischer JW, Evans A, Morrison C, Arveiler D,
Stoll M, Amarenco P, Cambien F, Paul M and Brand-Herrmann SM:
Osteopontin gene variation and cardio/cerebrovascular disease
phenotypes. Atherosclerosis. 206:209–215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hendig D, Arndt M, Szliska C, Kleesiek K
and Götting C: SPP1 promoter polymorphisms: identification
of the first modifier gene for pseudoxanthoma elasticum. Clin Chem.
53:829–836. 2007. View Article : Google Scholar
|
|
32
|
Pegoraro E, Hoffman EP, Piva L, Gavassini
BF, Cagnin S, Ermani M, Bello L, Soraru G, Pacchioni B, Bonifati
MD, Lanfranchi G, Angelini C, Kesari A, Lee I, Gordish-Dressman H,
Devaney JM and McDonald CM; Cooperative International Neuromuscular
Research Group. SPP1 genotype is a determinant of disease severity
in Duchenne muscular dystrophy. Neurology. 76:219–226. 2011.
View Article : Google Scholar
|
|
33
|
de las Fuentes L, Gu CC, Mathews SJ,
Reagan JL, Ruthmann NP, Waggoner AD, Lai CF, Towler DA and
Dávila-Román VG: Osteopontin promoter polymorphism is associated
with increased carotid intima-media thickness. J Am Soc
Echocardiogr. 21:954–960. 2008.PubMed/NCBI
|
|
34
|
Marciano R, D’Annunzio G, Minuto N,
Pasquali L, Santamaria A, Di Duca M, Ravazzolo R and Lorini R:
Association of alleles at polymorphic sites in the osteopontin
encoding gene in young type 1 diabetic patients. Clin Immunol.
131:84–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
D’Alfonso S, Barizzone N, Giordano M,
Chiocchetti A, Magnani C, Castelli L, Indelicato M, Giacopelli F,
Marchini M, Scorza R, Danieli MG, Cappelli M, Migliaresi S,
Bigliardo B, Sabbadini MG, Baldissera E, Galeazzi M, Sebastiani GD,
Minisola G, Ravazzolo R, Dianzani U and Momigliano-Richiardi P: Two
single-nucleotide polymorphisms in the 5′ and 3′ ends of the
osteopontin gene contribute to susceptibility to systemic lupus
erythematosus. Arthritis Rheum. 52:539–547. 2005.
|
|
36
|
Chen J, Wu Q, Lu Y, Xu T, Huang Y, Ribas
J, Ni X, Hu G, Huang F, Zhou L and Lu D: SPP1 promoter
polymorphisms and glioma risk in a Chinese Han population. J Hum
Genet. 55:456–461. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barizzone N, Marchini M, Cappiello F,
Chiocchetti A, Orilieri E, Ferrante D, Corrado L, Mellone S, Scorza
R, Dianzani U and D’Alfonso S: Association of osteopontin
regulatory polymorphisms with systemic sclerosis. Hum Immunol.
72:930–934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nakayama H, Nagai H, Matsumoto K, Oguro R,
Sugimoto K, Kamide K, Ohishi M, Katsuya T, Okamoto H, Maeda M,
Komamura K, Azuma J, Rakugi H and Fujio Y: Association between
osteopontin promoter variants and diastolic dysfunction in
hypertensive heart in the Japanese population. Hypertens Res.
34:1141–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|