Introduction
Gastric cancer is the second leading cause of
cancer-related mortality worldwide and may become one of the
leading causes of all deaths in the near future (1–3).
According to data from the National Cancer Institute (NCI), it is
estimated that more than 24,000 patients are diagnosed with gastric
cancer each year in the United States (4). The development of adjuvant
chemotherapies has improved clinical outcomes; however, advanced
gastric cancer with lymph node metastasis still has a poor
prognosis (5,6). Identification of genes responsible for
the development and progression of gastric cancer and a clear
understanding of the clinical significance of these genes are
important for the diagnosis and treatment of this disease.
The evolutionarily conserved protein COP1 has been
shown to operate as an E3 ubiquitin ligase complex, and a number of
putative substrates have been identified, including the c-Jun
oncoprotein and p53 tumor-suppressor protein (7–9). In
mammalian cells, COP1 regulates various cellular functions, such as
proliferation, cell cycle progression, apoptosis, and DNA repair
(7,10); however, the definitive role of COP1
has not yet been determined among diverse cancer types. COP1 has
been found to be overexpressed in ovarian and breast
adenocarcinomas, and its expression has been shown to correlate
with the unstable state of p53 protein in cancers that retain
wild-type p53 gene status (11).
COP1 was also found to be frequently overexpressed in human
hepatocellular carcinoma (HCC), and transfection of HCC cell lines
with COP1 siRNA in the context of wild-type p53 inhibited growth;
in contrast, p53-null Hep3B cells were resistant to the effects of
transfection with COP1 siRNA (12).
The results of these studies indicate that COP1 has significant
functions as an oncogene by degradation of p53 and could be a novel
therapeutic target in ovarian cancer and HCC. On the other hand,
several intriguing studies have also demonstrated that COP1 has a
tumor-suppressor role. Vitari et al(13) found that COP1 negatively regulates
the proto-oncogenes ETV1, ETV4 and ETV5, which
encode transcription factors in the E26 transformation-specific
(ETS) family, and that combined loss of COP1 and PTEN enhances the
invasiveness of mouse prostate adenocarcinomas. Another study
showed that COP1 specifically binds basic leucine zipper factors of
the Jun family and that expression of COP1 downregulates
c-Jun-dependent transcription as a functional consequence of
COP1-zipper factor interactions (14). Thus, it is difficult to determine
whether COP1 serves primarily as an oncogene or as a
tumor-suppressor gene among diverse malignancies and
circumstances.
In the current study, we analyzed COP1 mRNA
expression using clinical samples from 133 patients diagnosed with
primary gastric cancer. We then examined the relationships between
COP1 mRNA expression and clinicopathological factors and
determined the clinical significance of aberrant COP1
expression. Moreover, we confirmed the biological significance of
COP1 in gastric cancer cells using in vitro
assays.
Materials and methods
Clinical sample and cell lines
A total of 133 gastric cancer patients were enrolled
in the present study. All patients underwent surgery without
pre-operative treatments such as chemotherapy and radiotherapy.
Tumor and adjacent normal tissue were obtained. Total RNAs were
extracted using a QIAamp DNA Micro kit (Qiagen, Valencia, CA, USA)
following the manufacturer’s protocol. Patients were closely
observed each month after surgery, and the mean post-operative
follow-up period was 3 years. Histopathological evaluation was
assessed according to the Japanese Classification of Gastric
Cancer, 3rd English edition.
MKN-45 and NUGC4 cell lines were provided by the
American Type Culture Collection and were maintained in RPMI-1640
containing 10% fetal bovine serum (FBS) with 100 U/ml penicillin
and 100 mg/ml streptomycin. Cells were cultured in a humidified 5%
CO2 incubator at 37°C. The study protocol was reviewed
and approved by Kyushu University.
Real-time quantitative reverse
transcription (RT)-PCR
The primer sequences were as follows: COP1, forward
5′-TGCAAA GTTTGTGAGTGGTGA-3′ and reverse 5′-GAACGTAGG
CAGTATGGTTTCC-3′; MMP1, forward 5′-GCTAACCTT TGATGCTATAACTACGA-3′
and reverse 5′-TTTGTGCGC ATGTAGAATCTG-3′; MMP7, forward
5′-CTGACATCA TGATTGGCTTTG-3′ and reverse 5′-ATCTCCTCCGAG
ACCTGTCC-3′; MMP10, forward 5′-CAAAAGAGGAGG ACTCCAACA-3′ and
reverse 5′-TTCACATCCTTTTCG AGGTTG-3′; and GADPH, forward
5′-GTCAACGGATTT GGTCTGTATT-3′ and reverse 5′-AGTCTTCTGGGTGGC
AGTGAT-3′. COP1, MMP1, MMP7 and MMP10
levels were normalized to that of GAPDH. The real-time quantitative
monitoring of PCR reactions was performed using a
LightCycler® System and a LightCycler® 480
Probes Master kit (both from Roche Applied Science, Indianapolis,
IN, USA) following the manufacturer’s protocol.
COP1 RNA interference
For the siRNA knockdown experiment, double-stranded
RNA duplexes targeting human COP1
(5′-AGGAGCGUCCAGUAGAUGAACACGC-3′/5′-GCGUGU UCAUCUACUGGACGCUCCU-3′,
5′-UUCAAAUGCUUA AACUUUGGGAUUG-3′/5′-CAAUCCCAAAGUUUAAGCA UUUGAA-3′,
and 5′-UGGAAAGGAAGAUGAAGUCGCAG
GG-3′/5′-CCCUGCGACUUGAUCUUCCUUUCCA-3′) were purchased (Stealth
RNAi; Invitrogen, Carlsbad, CA, USA). Negative control siRNA was
also purchased from Invitrogen. MKN-45 and NUGC4 cells were
transfected with siRNA at a concentration of 20 μmol/l using
Lipofectamine (RNAiMAX) and were incubated in glucose-free Opti-MEM
(both from Invitrogen). Total RNA from transfected cell lines was
extracted using a QIAamp DNA Micro kit following the manufacturer’s
protocol.
Proliferation assay
MKN-45 and NUGC4 cells transfected with specific
siRNAs or negative control siRNA were seeded at 8×103
cells/well in 96-well flat-bottomed microtiter plates in a final
volume of 100 μl of culture medium/well. Cells were incubated in a
humidified atmosphere (37°C and 5% CO2) for 48 or 96 h
after initiation of transfection. The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Roche Diagnostics) was used to measure cell growth
inhibition. After incubation, 10 μl of MTT labeling reagent (final
concentration of 0.5 mg/ml) was added to each well, and the plate
was incubated for 4 h in a humidified atmosphere. Solubilization
solution (100 μl) was added to each well and the plate was
incubated overnight in a humidified atmosphere. After confirming
that the purple formazan crystals were completely solubilized, the
absorbance of each well was measured by a Model 550 series
microplate reader (Bio-Rad Laboratories, Hercules, CA, USA), at a
wavelength of 570 nm corrected to 655 nm. The assay was performed
using 6 replicates.
Statistical analysis
The significance of differences between 2 groups was
estimated with the Student’s t-test and χ2 test. Overall
survival curves were plotted according to the Kaplan-Meier method,
with the log-rank test applied for comparison. Variables with a
P-value of <0.05 by univariate analysis were used in subsequent
multivariate analysis on the basis of the Cox proportional hazards
model. All differences were considered statistically significant at
the level of P<0.05. Statistical analyses were conducted using
JMP 5 software (SAS Institute).
Results
COP1 expression in gastric cancer
tissues
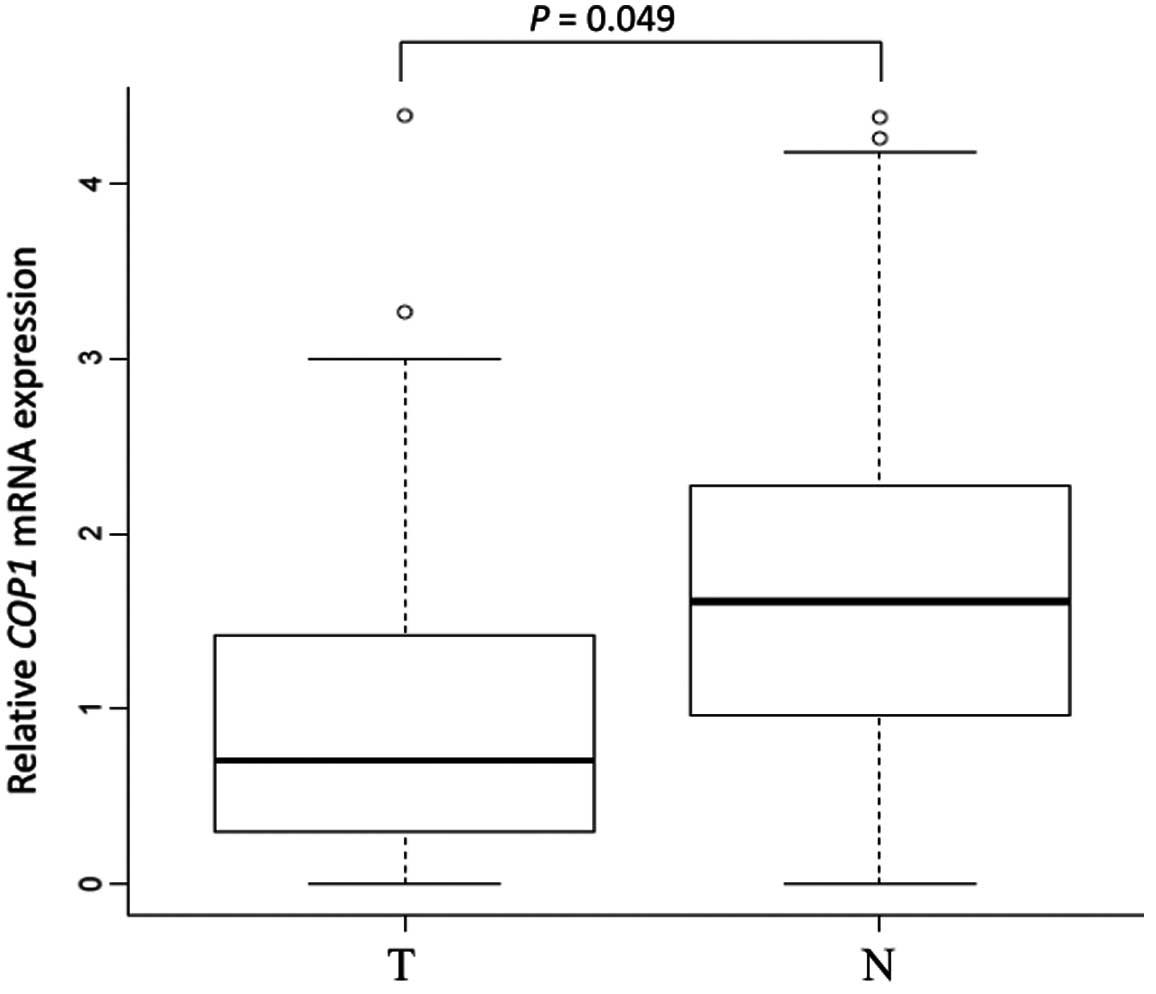
A significant difference in COP1 mRNA
expression between tumor tissue and the corresponding normal mucosa
was observed in 133 gastric cancer cases by quantitative real-time
PCR. COP1 was significantly downregulated in tumor tissues
compared to the corresponding normal mucosa (median
COP1/GAPDH ratio in tumor tissues, 1.30; median
COP1/GAPDH ratio in the corresponding normal mucosa, 1.90; P
= 0.049) (Fig. 1).
Relationship between COP1 mRNA expression
and clinicopathological factors
To evaluate the relationship between COP1
mRNA expression and clinicopathological factors, we divided the 133
patients with gastric cancer into a high COP1 expression
group (n=67) and a low COP1 expression group (n=66),
according to the COP1/GAPDH ratio of 0.80 in cancerous
tissue. There were no significant differences between the low and
high COP1 expression groups in terms of clinicopathological
factors (Table I). However, the low
COP1 expression group showed an increased tendency to be
associated with greater tumor size (P=0.099; Table I).
 | Table IRelationship between COP1 mRNA
expression and clinicopathologic factors. |
Table I
Relationship between COP1 mRNA
expression and clinicopathologic factors.
| Factors | Low expression
(n=67) | High expression
(n=66) | P-value |
|---|
| Age, mean ± SD | 65.2±11.1 | 66.4±12.5 | |
| Gender, n | | | 0.433 |
| Male | 44 | 39 | |
| Female | 23 | 27 | |
| Histological grade,
n | | | 0.187 |
| Well and mod | 34 | 26 | |
| Por and sig | 33 | 40 | |
| Tumor size (mm),
n | | | 0.099 |
| <30 | 13 | 21 | |
| >31 | 54 | 45 | |
| Depth, n | | | 0.581 |
| T1 | 18 | 15 | |
| T2–4 | 49 | 51 | |
| Lymph node
metastasis, n | | | 0.949 |
| Positive | 44 | 43 | |
| Negative | 23 | 23 | |
| Lymphatic invasion,
n | | | 0.667 |
| Positive | 46 | 43 | |
| Negative | 21 | 23 | |
| Venous invasion,
n | | | 0.8 |
| Positive | 15 | 16 | |
| Negative | 52 | 50 | |
| Liver metastasis,
n | | | 0.477 |
| Positive | 5 | 3 | |
| Negative | 62 | 63 | |
| Peritoneal
dissemination, n | | | 0.325 |
| Positive | 11 | 7 | |
| Negative | 56 | 59 | |
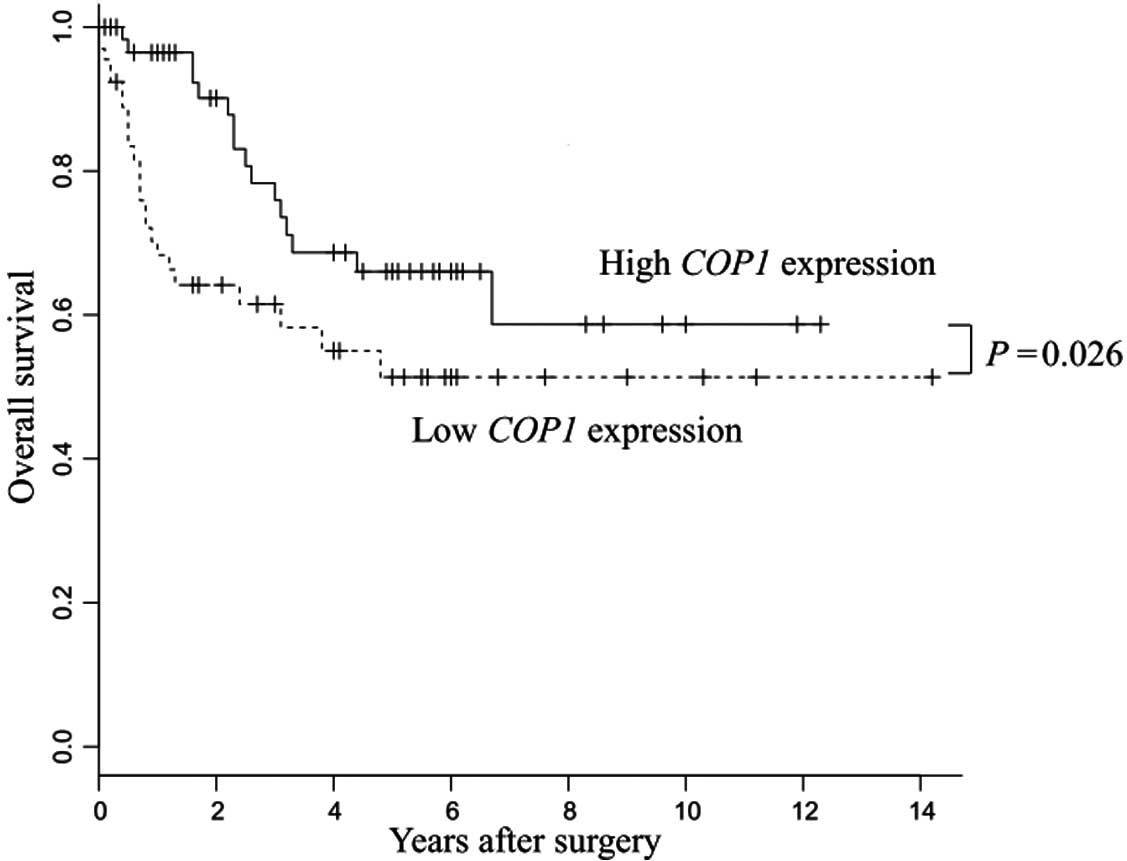
Low COP1 expression resulted in poor
prognoses in patients with gastric cancer
The low COP1 expression group showed
significantly poorer prognosis than the high COP1 expression
group (P=0.026) (Fig. 2).
Univariate analysis of overall survival revealed significant
associations with COP1 expression, depth of tumor invasion,
tumor size, lymph node metastasis, lymphatic invasion and venous
invasion in patients with gastric cancer (Table II). Thus, we applied these factors
to multivariate analysis and found that COP1 expression was
an independent prognostic indicator for overall survival in
patients with gastric cancer (P=0.0085; Table II).
 | Table IIUnivariate and multivariate analysis
for overall survival using the Cox proportional hazards regression
model. |
Table II
Univariate and multivariate analysis
for overall survival using the Cox proportional hazards regression
model.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Factor | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age | 0.7611 | 0.551–1.039 | 0.086 | - | - | - |
| Gender | 0.9837 | 0.715–1.381 | 0.9215 | - | - | - |
| Histology grade
(well and mod/por and sig) | 1.2632 | 0.919–1.772 | 0.151 | - | - | - |
| Tumor size >30
mm (negative/positive) | 1.9167 | 1.212–3.504 | 0.0035 | 1.37539 | 0.850–2.546 | 0.2093 |
| Depth
(T1/T2–4) | 4.095 | 1.908–17.252 | <0.0001 | 2.114491 | 0.857–9.353 | 0.1143 |
| Lymph node
metastasis (negative/positive) | 3.076 | 1.844–6.272 | <0.0001 | 2.177924 | 1.245–4.611 | 0.0042 |
| Lymphatic invasion
(negative/positive) | 2.221 | 1.405–4.063 | 0.0002 | 1.172472 | 0.690–2.239 | 0.577 |
| Venous invasion
(negative/positive) | 1.7543 | 1.263–2.403 | 0.0011 | 1.392089 | 0.983–1.955 | 0.0617 |
| COP1 mRNA
expression (low/high) | 0.7019 | 0.506–0.960 | 0.0269 | 0.642875 | 0.455–0.894 | 0.0085 |
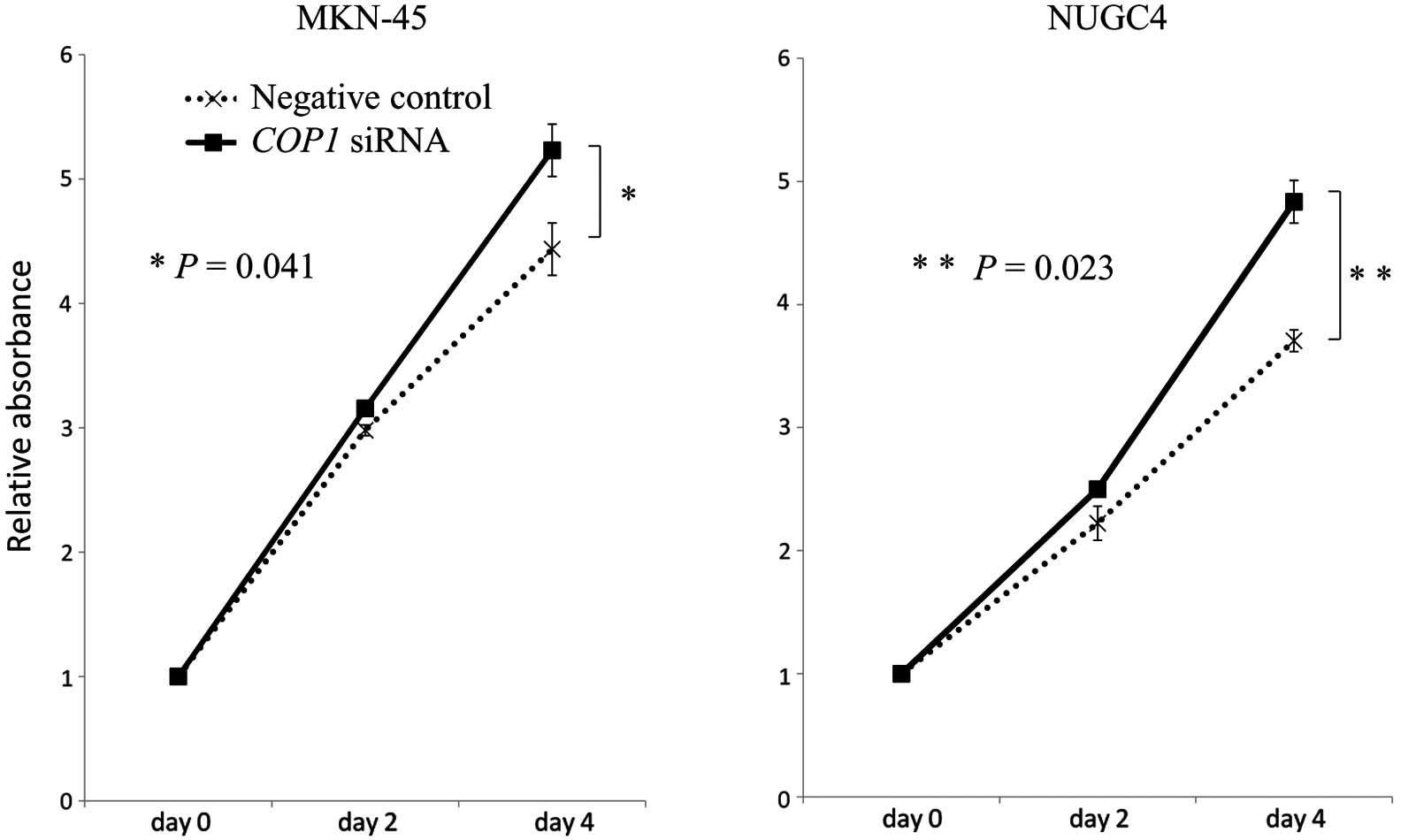
Inhibition of COP1 expression with siRNA
in gastric cancer cell lines
For proliferation assays, COP1 siRNA was
transfected into MKN-45 and NUGC4 cells (expressing wild-type p53).
A significant reduction in COP1 expression by siRNA
transfection was confirmed by quantitative real-time RT-PCR. Using
MTT assays, we found that COP1 knockdown by siRNA
significantly increased the numbers of MKN-45 and NUGC4 cancer
cells as compared with the corresponding control cells (MKN-45,
P=0.041; NUGC4, P=0.023) (Fig.
3).
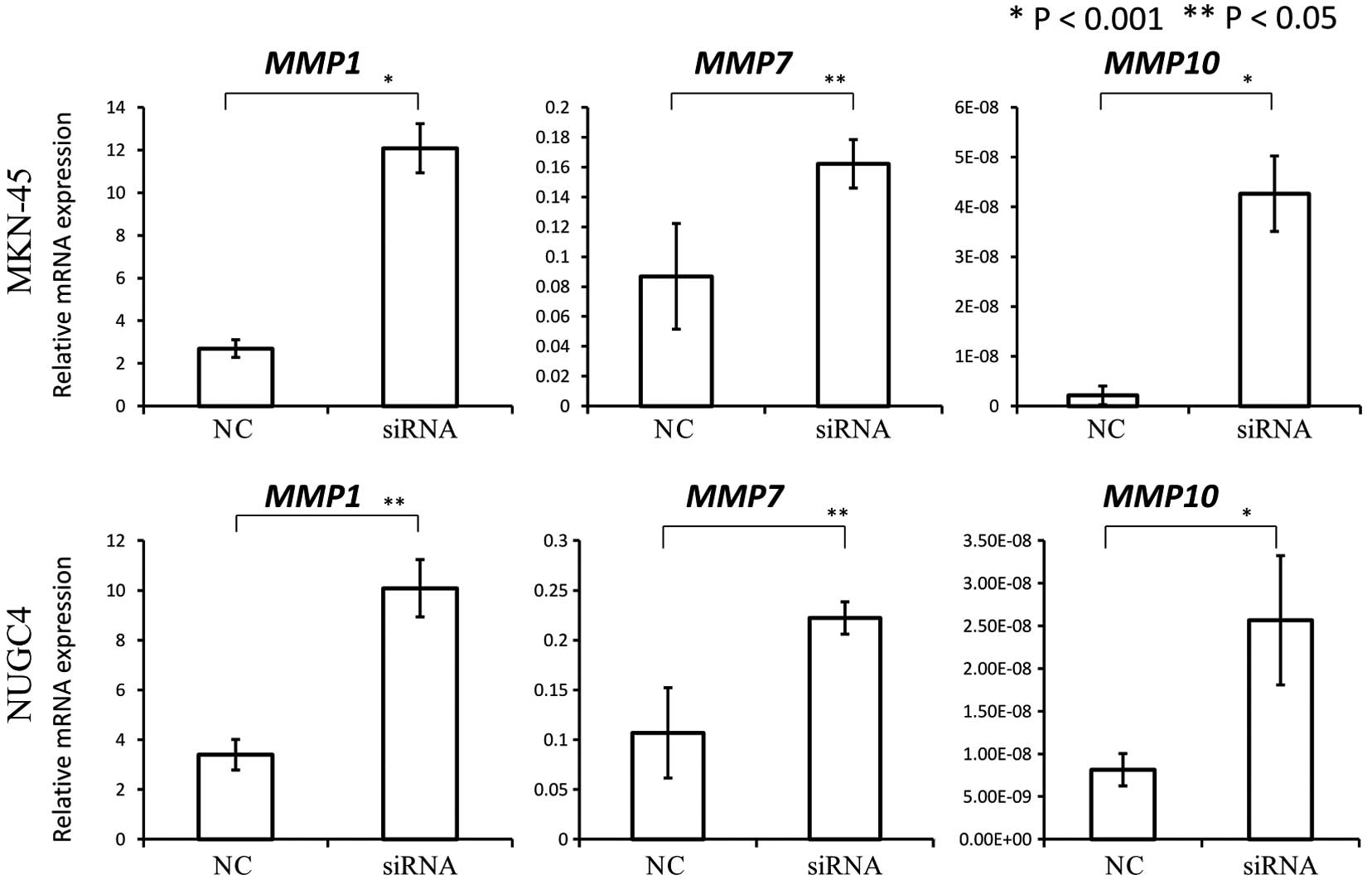
These results indicated the possibility that COP1
may function as a tumor suppressor. Therefore, we investigated the
association between COP1 expression and the expression of
MMP1, MMP7 and MMP10, transcriptional targets
of c-Jun and ETV1. We found that COP1 knockdown by siRNA
significantly increased the expression of MMP1, MMP7
and MMP10 as compared to the negative control (Fig. 4).
Discussion
The ubiqutin ligase complex, which targets proteins
for proteasome-mediated degradation, plays important roles in
maintaining cellular homeostasis (15). Accordingly, the dysregulation of
ubiquitin ligase has been implicated in a variety of diseases,
including cancer. Overexpression of COP1 was observed in ovarian
cancer, breast cancer, and HCC, and COP1 has been recognized as an
oncogene by targeting p53 for degradation in a ubiquitin-dependent
fashion (11,12). Li et al(16) also found that COP1 was overexpressed
in gastric cancer and observed a negative correlation between COP1
protein expression and p53 protein expression. In contrast, in a
mouse model of prostate cancer, COP1 was identified as a tumor
suppressor that negatively regulates ETV1, ETV4 and ETV5 (13). Moreover, identification of
well-known oncoproteins, such as c-Jun (14) and MTA1 (17), as potential COP1 substrates has
indicated the possibility that COP1 may function as a tumor
suppressor.
In the present study, we revealed that low
COP1 expression was an independent factor predicting poor
prognosis in patients with gastric cancer (Table II). These results, in contrast to
those of Li et al(16),
suggest that COP1 acted as a tumor suppressor in gastric cancer.
The major factor that may account for the observed differences
between our results and the results published by Li et al
was the p53 mutation status of clinical samples; indeed, p53
mutation status was unknown in both studies. The mutation rate of
p53 has been reported to be 41 and 73% in 2 exome sequencing
studies of gastric cancer (18,19),
suggesting that COP1-p53 interactions should not affect tumor
biology in about half of all gastric cancer cases. Thus, in order
to properly evaluate the clinical significance of COP1 expression,
it is important to investigate COP1 expression in gastric cancer
samples expressing wild-type p53.
Migliorini et al(20) utilized a genetic approach to
generate an allelic series of Cop1-mutant mice. This study
firmly established that COP1 is a ubiquitin ligase for c-Jun and
that COP1 acts as a tumor suppressor in vivo. In the present
study, we observed that COP1 expression was significantly
correlated with the expression of MMP1, MMP7 and
MMP10 and that knockdown of COP1 by siRNA increased
cell proliferation (Figs. 3 and
4). Therefore, we speculated that
the poorer prognosis in patients exhibiting low expression of
COP1 within tumor tissues may be caused by the activation of
c-Jun (AP-1) and MMP pathways.
In conclusion, while the role of COP1 in
malignancies is controversial, our current data support that COP1
acts as a tumor suppressor in gastric cancer. Further studies in
the near future will be required to clarify how the role of COP1 is
determined in various malignancies and to elucidate the mechanisms
that regulate COP1 expression.
Acknowledgements
We would like to thank T. Shimooka and M. Kasagi for
their technical assistance. The present study was funded in part by
the Funding Program for Next Generation World-Leading Researchers
(LS094).
References
|
1
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
non-tumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oh SC: Update of adjuvant chemotherapy for
resected gastric cancer. J Gastric Cancer. 12:3–6. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4
|
Jemal A, Tiwari RC, Murray T, et al:
Cancer statistics, 2004. CA Cancer J Clin. 54:8–29. 2004.
View Article : Google Scholar
|
|
5
|
Kelley JR and Duggan JM: Gastric cancer
epidemiology and risk factors. J Clin Epidemiol. 56:1–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun P, Xiang JB and Chen ZY: Meta-analysis
of adjuvant chemotherapy after radical surgery for advanced gastric
cancer. Br J Surg. 96:26–33. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dornan D, Wertz I, Shimizu H, et al: The
ubiquitin ligase COP1 is a critical negative regulator of p53.
Nature. 429:86–92. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wertz IE, O’Rourke KM, Zhang Z, et al:
Human De-etiolated-1 regulates c-Jun by assembling a CUL4A
ubiquitin ligase. Science. 303:1371–1374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Savio MG, Rotondo G, Maglie S, Rossetti G,
Bender JR and Pardi R: COP1D, an alternatively spliced constitutive
photomorphogenic-1 (COP1) product, stabilizes UV stress-induced
c-Jun through inhibition of full-length COP1. Oncogene.
27:2401–2411. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dornan D, Shimizu H, Mah A, et al: ATM
engages autodegradation of the E3 ubiquitin ligase COP1 after DNA
damage. Science. 313:1122–1126. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dornan D, Bheddah S, Newton K, et al:
COP1, the negative regulator of p53, is overexpressed in breast and
ovarian adenocarcinomas. Cancer Res. 64:7226–7230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YH, Andersen JB, Song HT, et al:
Definition of ubiquitination modulator COP1 as a novel therapeutic
target in human hepatocellular carcinoma. Cancer Res. 70:8264–8269.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vitari AC, Leong KG, Newton K, et al: COP1
is a tumour suppressor that causes degradation of ETS transcription
factors. Nature. 474:403–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bianchi E, Denti S, Catena R, et al:
Characterization of human constitutive photomorphogenesis protein
1, a RING finger ubiquitin ligase that interacts with Jun
transcription factors and modulates their transcriptional activity.
J Biol Chem. 278:19682–19690. 2003. View Article : Google Scholar
|
|
15
|
Deshaies RJ and Joazeiro CA: RING domain
E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li YF, Wang DD, Zhao BW, et al: High level
of COP1 expression is associated with poor prognosis in primary
gastric cancer. Int J Biol Sci. 8:1168–1177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li DQ, Ohshiro K, Reddy SD, et al: E3
ubiquitin ligase COP1 regulates the stability and functions of
MTA1. Proc Natl Acad Sci USA. 106:17493–17498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang K, Kan J, Yuen ST, et al: Exome
sequencing identifies frequent mutation of ARID1A in molecular
subtypes of gastric cancer. Nat Genet. 43:1219–1223. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zang ZJ, Cutcutache I, Poon SL, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Migliorini D, Bogaerts S, Defever D, et
al: Cop1 constitutively regulates c-Jun protein stability and
functions as a tumor suppressor in mice. J Clin Invest.
121:1329–1343. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marine JC: Spotlight on the role of COP1
in tumorigenesis. Nat Rev Cancer. 12:455–464. 2012. View Article : Google Scholar : PubMed/NCBI
|