Introduction
Particularly interesting new cysteine-histidine rich
protein (PINCH) is an adapter protein which forms a complex with
integrin-linked-kinase (ILK) and parvin and connects integrins at
the cell surface with the actin cytoskeleton of the cell. PINCH
participates in the protein-protein interaction with downstream
effectors that regulate cell shape, motility and survival (1–5), and
PINCH has been shown to be related to worse survival in colorectal
cancer, when expressed in tumor tissues, notably at the invasive
edges (6,7).
Few studies have analyzed the relationship between
PINCH and radiotherapy (RT). Eke et al(8) demonstrated, in a cell line study, that
PINCH was related to enhanced radioresistance. Our recent study
involving rectal cancer patients performed on the same material as
used in the present study showed that strong PINCH expression was
related to worse survival when compared to weak expression in
patients without RT, but not in patients with RT. A further
statistical interaction analysis between PINCH, RT and survival
showed no significant result, suggesting that RT was not directly
the reason for the differences in survival between patients with
weak and strong PINCH expression. In addition, no differences were
found when the expression of PINCH was compared between
unirradiated and radiated fibroblasts (7).
In the present study, we further investigated the
relationship between PINCH and radiation in colon cancer cells
separately cultured and in co-culture. The colon cancer cells were
co-cultured as an attempt to imitate the tumor environment inside
the body.
C-Myc is a well-known regulator in the development
of cancer, and controls key functions, such as cell proliferation,
differentiation and apoptosis (9).
Previously, it was shown in cell culture that PINCH was related to
ERK, and ERK is a well-known regulator of C-Myc (10). Since both PINCH and C-Myc have been
shown to be involved in the regulation of cell proliferation and
apoptosis, we wanted to further investigate the association between
PINCH and C-Myc (9,10).
The aim of the present study was to analyze the
expression of PINCH in relation to radiation in separately and
co-cultured colon cancer cells and to study the association between
PINCH and C-Myc. Furthermore, the relationship between PINCH and
preoperative RT was investigated in rectal cancer patients.
Materials and methods
Cell lines
The colon cancer cell lines, KM12C, KM12SM and
KM12L4a, were a kind gift from Professor I.J. Fidler (Anderson
Cancer Center, University of Texas, TX, USA). KM12C is derived from
a patient with stage II colon cancer, KM12SM is a spontaneous liver
metastasis (11) and KM12L4a is an
experimental liver metastasis (12). Western blot analysis showed no
differences in PINCH expression among the KM12C, KM12SM and KM12L4a
cell lines. The KM12C cell line was chosen for further culturing
proceedings as an attempt to imitate the tumor environment inside
the body.
The separately cultured KM12C cells, and the
co-cultured KM12C and CCD-18-Co cells were cultured in Eagle’s
minimal essential medium (MEM), and supplemented with 1%
penicillin-streptomycin and 10% FBS (ATCC; Rockville, MD, USA). The
co-cultured cells were grown in a 6-well-plate with an insert
chamber (pore size 0.4 μm).
Patients
Patients with rectal adenocarcinoma from the
Southeast Swedish Health Care region who participated in a Swedish
clinical trial of preoperative RT during 1987–1990 were included
(13). Of the 137 primary tumors,
72 patients underwent surgery alone and 65 patients underwent
preoperative RT prior to surgery. RT was delivered at 25 Gray (Gy)
in 5 fractions during a median of 6 days (range, 5–12 days).
Surgery was then performed a median of 3 days (range, 1–13 days)
after RT. None of the patients received adjuvant chemotherapy
before or after surgery. The mean age of the patients was 67 years
(range, 36–85 years) and the mean follow-up was 80 months (range,
0–193 months). Additional characteristics of the patients and
tumors are presented in Table I.
The Research Ethics Committee of Linköping University Hospital, no.
86151, approved the study.
 | Table IPatient and tumor characteristics. |
Table I
Patient and tumor characteristics.
| Characteristics | No
radiotherapy
n=72
n (%) |
Radiotherapy
n=65
n (%) | P-value |
|---|
| Gender | | | 0.702 |
| Male | 42 (58) | 40 (62) | |
| Female | 30 (42) | 25 (38) | |
| Age (years) | | | 0.842 |
| ≤67 | 30 (42) | 26 (42) | |
| >67 | 42 (58) | 39 (60) | |
| TNM | | | 0.105 |
| I | 20 (28) | 22 (34) | |
| II A | 18 (25) | 21 (32) | |
| IIIA | 8 (11) | 1 (2) | |
| IIIB | 11 (15) | 11 (17) | |
| IIIC | 11 (15) | 4 (6) | |
| IV | 4 (6) | 6 (9) | |
| Differentiation | | | 0.263 |
| Well | 2 (3) | 2 (3) | |
| Moderate | 58 (81) | 48 (74) | |
| Poor | 12 (16) | 15 (23) | |
| Local recurrence | | | 0.059 |
| No | 57 (79) | 59 (91) | |
| Yes | 15 (21) | 6 (9) | |
| Distant
recurrence | | | 0.257 |
| No | 42 (58) | 44 (68) | |
| Yes | 30 (42) | 21 (32) | |
Radiation procedure
For all experiments, cells where seeded at a density
of 60,000 cells/cm2 and irradiated with photons from a 6
MV linear accelerator (Varian Clinac 600C/D; Varian Medical
Systems, Palo Alto, CA, USA). The cells were exposed to single
doses of 0, 2, 5 or 10 Gy, respectively, at room temperature. A
dose of 2 Gy was used for further analyses since this is the most
commonly used dose in the clinic. The controls (0 Gy) were handled
under the same environmental conditions as the treated cells.
Following radiation, cells were harvested at 8 and 24 h.
Western blot analysis
After radiation, cells were washed in PBS and lysed
in RIPA buffer, containing 150 mM NaCl 2% Triton, 0.1% SDS, 50 mM
Tris pH 8.0 and a Protease Inhibitor Cocktail without chelating
reagents (Sigma-Aldrich, Stockholm, Sweden). The protein
concentration was determined using the colorimetric BCA protein
assay reagent (Pierce, Woburn, MA, USA). Samples containing 30 μg
protein were separated by electrophoresis on a Mini-PROTEAN TGX™
precast 12% gel (Bio-Rad, Hercules, CA, USA) for 55 min at 200 V.
The separated proteins were transferred to a PVDF membrane (0.45
μm; polyvinylidene fluoride transfer membrane; VWR/Life Sciences,
Pall Corp., Pensacola, FL, USA). The membranes were blocked with 5%
non-fat dried milk in Tris-buffered saline (TBS) containing 0.1%
Tween-20 (TBST) and incubated with the primary PINCH antibody (1
μg/ml). The antibodies were incubated overnight at 4°C in TBST in
1% non-fat dried milk. The membranes were washed and incubated with
an HRP-conjugated polyclonal goat anti-mouse secondary antibody
(1:5000; DakoCytomation, Glostrup, Denmark) for 1 h at room
temperature, followed by enhanced chemiluminescence (ECL) (Amersham
Biosiences/GE Healthcare). To verify equal loading of the wells,
the membranes for PINCH were re-incubated with a primary mouse
polyclonal anti-β-actin antibody (1:5000; Sigma-Aldrich, Steinheim,
Germany). All experiments were repeated three times.
RT-PCR
The relative abundances of PINCH and C-Myc mRNA were
determined by real-time PCR (RT-PCR) with duplicates of each
sample, repeated 3 times. Total RNA was extracted from KM12C and
CCD-18-Co cells by using the RNA Blood Mini kit (Qiagen), and cDNA
was transcribed using the High Capacity cDNA reverse transcription
kit according to the manufacturer’s instructions (Applied
Biosystems). PINCH and C-Myc mRNA expression was determined by
using specific primers Hs00757864_m1 for PINCH and Hs00905030_m1
for C-Myc (Applied Biosystems). The RT-PCR reactions were performed
using the 7500HT Fast RT-PCR system, and the data were displayed
graphically using the SDS 3.2 software program (Applied Biosystem).
The scores from the gene of interest and the mean scores of two
reference genes, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
and β-actin (Applied Biosystems), were used for further
calculations by using the ΔΔCt method (14).
Protein-protein interaction analysis
A protein-protein interaction analysis was carried
out to determine the possible interaction between PINCH and C-Myc.
Amino acid sequences of human LIM and senescent cell
antigen-like-containing domain (LIMS1) (Uniprot ID, P48059) (PINCH)
and human Myc proto-oncogene protein (Myc) (Uniprot ID, P01106)
were downloaded from UniProt. Subsequently, sequences were
submitted to I-TASSER server. C-score was preferably within the
range of 2 to −1.5. A further protein interaction analysis by using
the PIPS software method was used, where four different features
were considered; expression data, orthology interaction
relationships, protein features and network topology. The scores
calculated by each of these features were combined to output the
final interaction score.
Immunohistochemistry
Formalin-fixed paraffin-embedded sections were
deparaffinized in xylene and rehydrated with a graded series of
ethanol. The sections were treated by high pressure cooking for 10
min with Tris-ethylenediaminetetraacetic acid (EDTA) buffer (pH
9.0) and stored at room temperature for 30 min. Following
pre-incubation in methanol with 0.3% H2O2 for
20 min, the sections were incubated with Dako Protein Block (Dako,
Carpinteria, CA, USA) for 10 min and further incubated with rabbit
anti-PINCH antibody at 6 μg/ml in antibody diluent (Dako) for 1 h
at room temperature. After washing in phosphate-buffered saline
(PBS, pH 7.4), the sections were incubated with a mouse anti-rabbit
secondary antibody provided in the Dako ChemMate EnVision detection
kit (Dako) at room temperature for 25 min and then washed with PBS.
The sections were further subjected to 3,3′-diaminobenzidine
tetrahydrochloride for 8 min and counterstained with hematoxylin.
The positive controls were primary colorectal tumors known to stain
positive for PINCH, and the negative controls were primary rectal
tumors where PBS was used instead of the primary antibody. In all
staining procedures, the positive control showed clear staining,
and no staining was observed in the negative controls.
The results of PINCH expression in tumors consisted
of the scores determined by two independent authors (7) in a blinded fashion without any
knowledge of the clinical and biological information. The
percentage of stained cells was estimated among the total number of
cells by reading 10–20 areas at a magnification of ×400, regardless
of the staining intensity. The cases were scored as <25%,
25–49%, 50–74% or ≥75%, respectively. To avoid an artificial
effect, the cells on the margins of sections and areas with poorly
presented morphology were not counted. The results of the staining
intensity was presented in our previous study (7) and will therefore not be further
discussed in this study.
Statistical analysis
For the KM12C cell results analyzed by western
blotting and RT-PCR, an independent t-test by group was used to
evaluate whether there were any significant differences between
radiated and unirradiated cells in the expression of PINCH and
C-Myc separately cultured or in co-culture. For the patients, the
Chi-square method was used to analyze the relationship between
PINCH expression in tumors and the clinical or pathological
factors. Cox’s proportional hazard model was used to estimate the
relationship between PINCH expression and survival, including both
univariate and multivariate analyses. Survival curves were computed
according to the Kaplan-Meier method. For all statistical analysis
including the interaction analysis between PINCH, RT and survival
the statistical software program, STATISTICA, was used. Tests were
two-sided, and P<0.05 was considered to indicate a statistically
significant result.
Results
PINCH expression in KM12C and CCD-18-Co
cells cultured separately or co-cultured and treated with radiation
or without
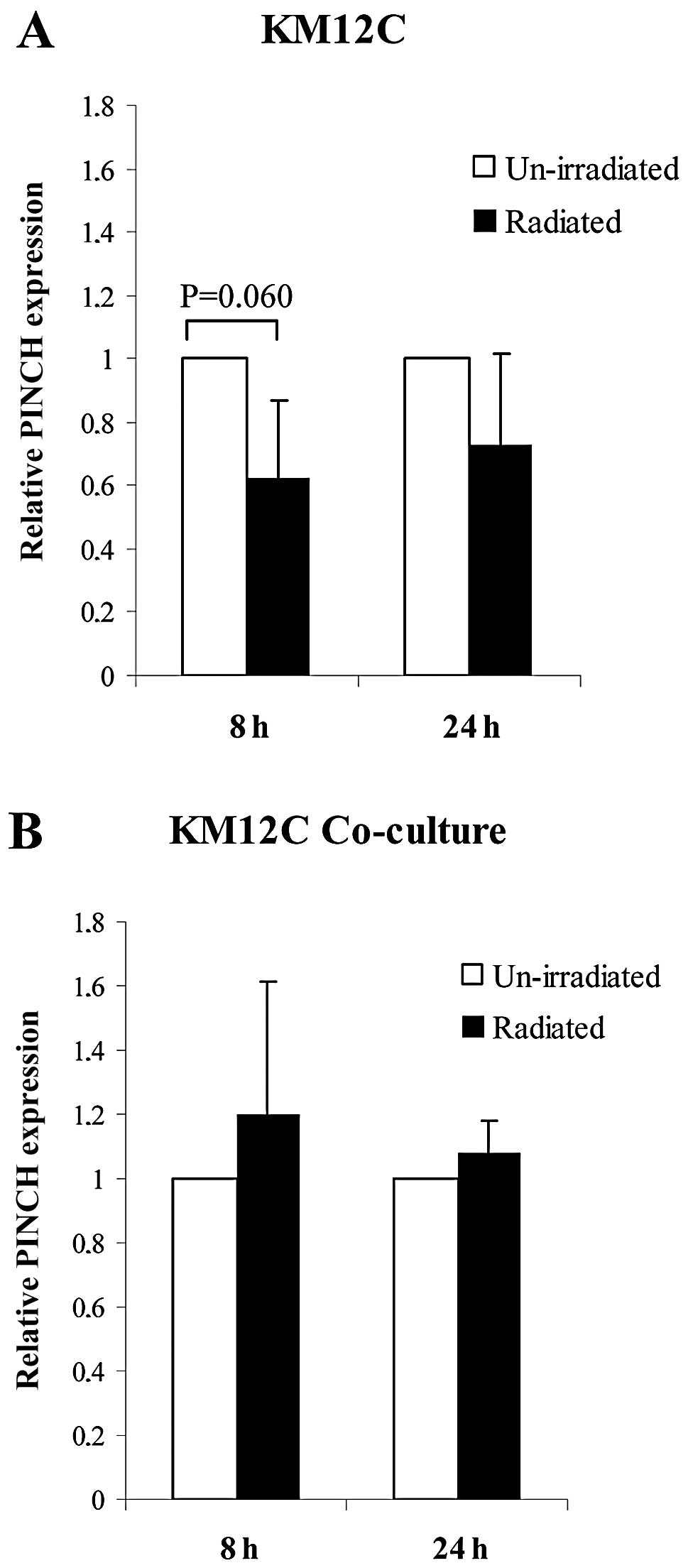
The expression of PINCH in KM12C cells cultured
separately and in co-culture by RT-PCR was compared between
unirradiated and radiated cells at 8 and 24 h. PINCH expression
tended to decrease from separately cultured KM12C cells without
radiation to cells with radiation at 8 h (P=0.060), but not at 24 h
(Fig. 1A). In co-cultured KM12C
cells, no significant differences in PINCH expression were found
between unirradiated and irradiated cells at 8 and 24 h (Fig. 1B). A similar, but weaker expression
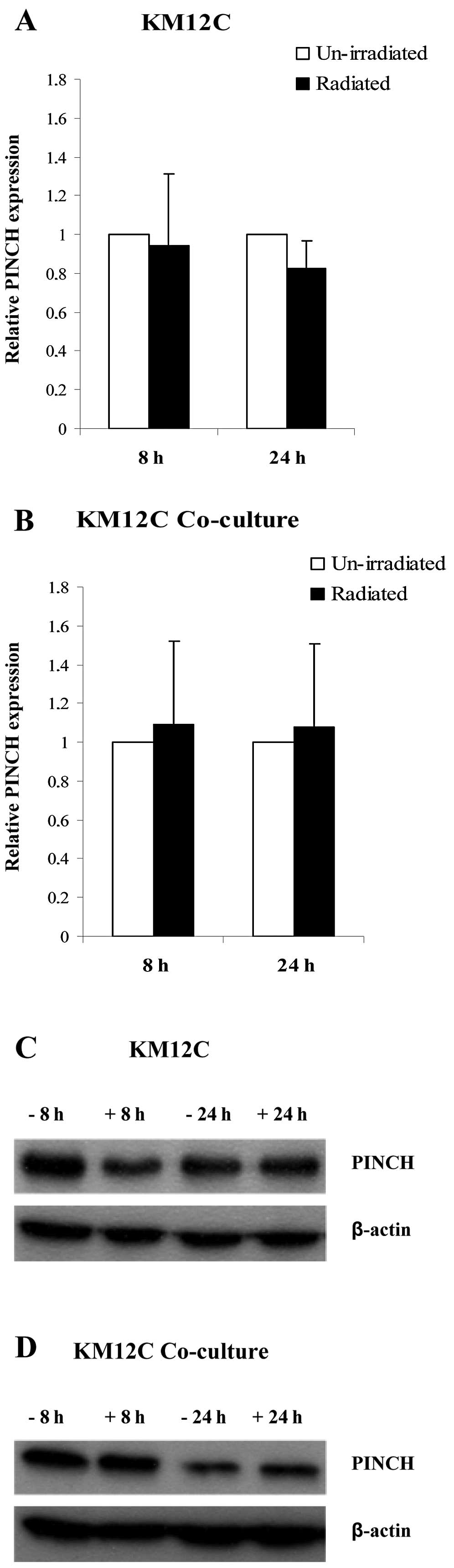
pattern, compared to RT-PCR was found when the expression of PINCH
was analyzed by western blotting in unirradiated and irradiated
KM12C cells cultured separately and in co-culture at 8 and 24 h,
but no significant difference was achieved (Fig. 2A–D).
C-Myc expression in KM12C and CCD-18-Co
cells cultured separately or in co-culture and treated with
radiation or without
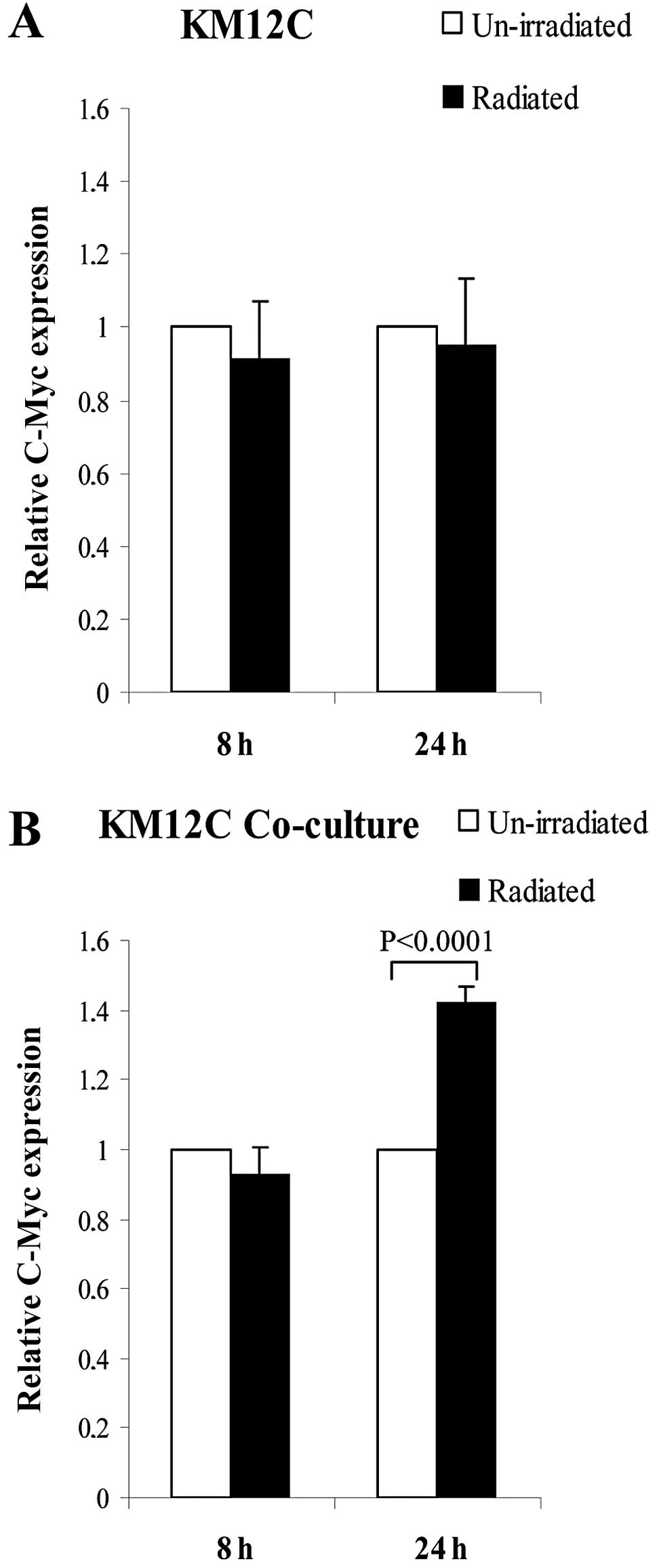
The C-Myc expression as analyzed by RT-PCR in
separately and co-cultured KM12C cells was further compared between
unirradiated and irradiated cells at 8 and 24 h. In the separately
cultured KM12C cells with radiation, compared to the cells without
radiation, C-Myc was equally expressed at 8 and 24 h (Fig. 3A), while in co-cultured KM12C cells
with radiation, C-Myc showed an equivalent expression at 8 h and a
significantly increased expression at 24 h when compared to the
cells without radiation (P<0.0001, Fig. 3B).
Interaction analysis between human PINCH,
senescent cell antigen-like-containing domain (LIMS1) and human Myc
proto-oncogene protein (C-Myc)
The interaction between human LIM and senescent cell
antigen-like-containing domain (LIMS1) (PINCH) (Uniprot ID, P48059)
and human Myc proto-oncogene protein (C-Myc) (Uniprot ID, P01106)
was further studied. To model the protein structure, the sequences
of both the proteins were PSI-BLASTed against the PDB proteins. Due
to the unavailability of appropriate template with significant
identity and query coverage, comparative modeling was not able to
be performed. Next the same fold recognition method as the I-TASSER
server was tested (Table II),
however, it also reported structures having 50% loop region and
>50% of the model lacked proper secondary structures. The
secondary structures were counter checked by predicting all
available isoforms of LIMS1 and C-Myc which confirmed the I-TASSER
results. A further protein interaction analysis using the PIPS
software showed an interaction score of 0.052, and no green domains
were found when the Chi square scores for co-occurrence of domains
were studied.
 | Table IIProtein interaction analysis between
LIMS1/PINCH and C-Myc. |
Table II
Protein interaction analysis between
LIMS1/PINCH and C-Myc.
| Best model from
I-TASSER | C-Scorea | TM-Scoreb | RMSDc (Å) |
|---|
| LIMS1/PINCHd | −3.77 | 0.31±0.10 | 15.7±3.3 |
| C-Myc | −1.18 | 0.57±0.15 | 9.7±4.6 |
PINCH expression in primary rectal tumors
with or without RT
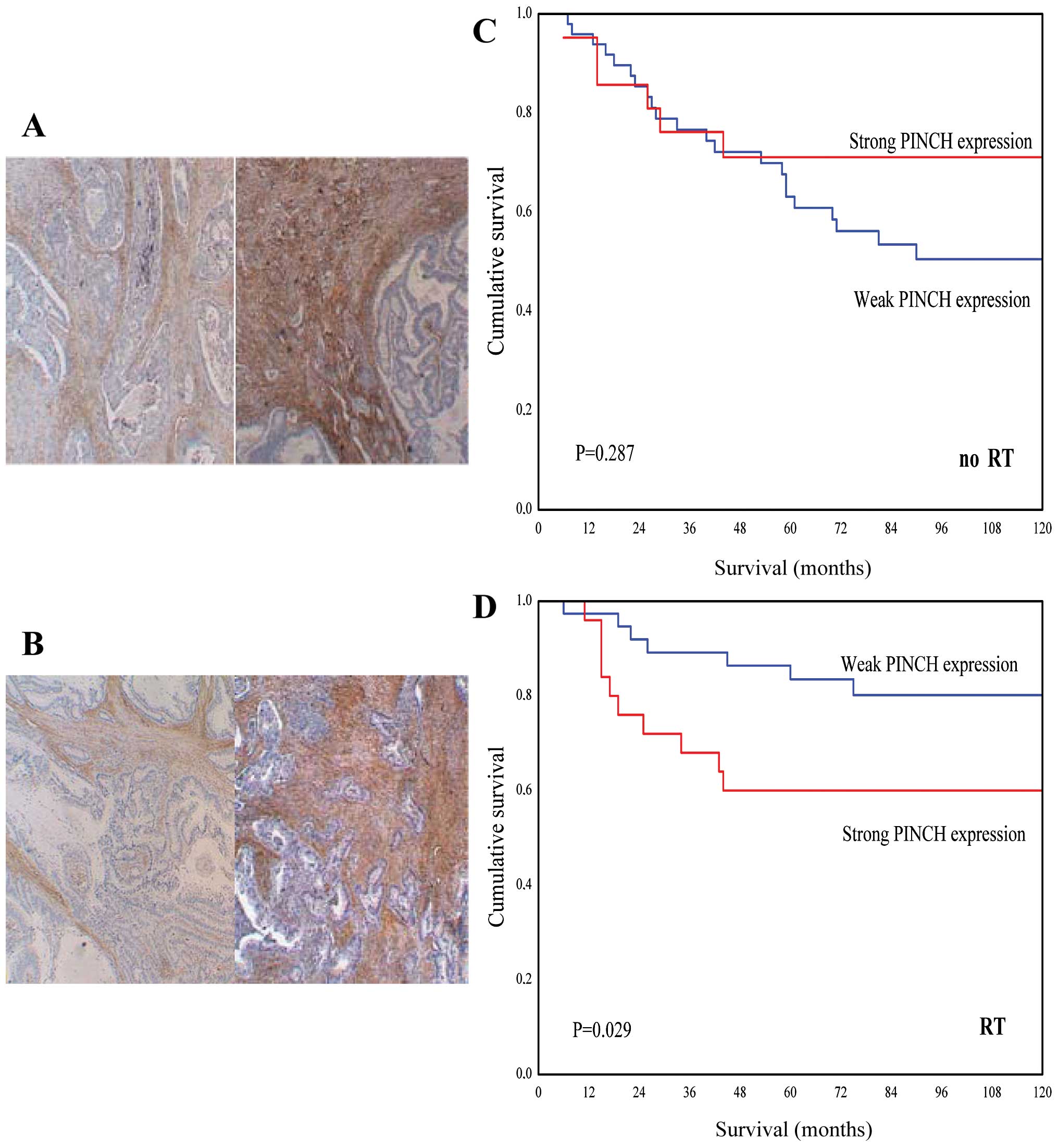
By analyzing the entire tumor area, the staining
percentage of PINCH in the primary tumors without (Fig. 4A) or with RT (Fig. 4B) was evaluated. Of the 137 tumors,
9 cases (7%) had <25% staining, 17 (12%) cases had 25–49%
staining, 62 (45%) cases had 50–74% staining and 47 (34%) cases had
≥75% PINCH staining. For further analysis, the cut-off point for
the staining percentage of PINCH expression was set to 75%. The
cases with negative and <74% stained cells were classified as
the weak expression subgroup, and the cases with ≥75% stained cells
were classified as the strongly PINCH expression subgroup.
We further analyzed the relationship between the
staining percentage of PINCH and survival in patients without or
with RT. In the patients with RT, strong PINCH expression tended to
be related to worse survival when compared to the patients with
weak PINCH expression (P=0.056). After adjusting for TNM stage,
degree of differentiation grade, age and p53 status, the
relationship reached statistically significance (P=0.029; RR, 4.03;
95% CI, 1.34–12.1) (Fig. 4D,
Table III). No statistically
significant difference was found in the patients without RT
(P=0.287, Fig. 4C). A further
statistical interaction analysis between PINCH, RT and survival
showed a trend towards significance (P=0.057). No significant
difference in PINCH expression was found in the subgroups with no
RT and with RT for disease-free survival, local recurrence-free
survival and distant recurrence-free survival (P>0.5).
 | Table IIIMultivariate analysis of PINCH
expression and clinicopathological features in rectal cancer
patients with radiotherapy. |
Table III
Multivariate analysis of PINCH
expression and clinicopathological features in rectal cancer
patients with radiotherapy.
| Features | Patients,
n=65
n (%) | Cancer death rate
ratio (95% CI) | P-value |
|---|
| PINCH expression | | | 0.029 |
| Weak | 39 (60) | 1.0 | |
| Strong | 26 (40) | 4.03 (1.34–12.1) | |
| Age (years) | | | 0.521 |
| <67 | 26 (40) | 1.0 | |
| ≥67 | 39 (60) | 0.73 (0.21–2.56) | |
| TNM stage | | | 0.0004 |
| I | 22 (34) | 1.0 | |
| IIA | 21 (32) | 3.47
(0.37–32.8) | |
| IIIA | 1 (2) | 0a (0–32.8) | |
| IIIB | 11 (17) | 7.14
(0.71–72.1) | |
| IIIC | 4 (6) | 32.3
(2.74–378.9) | |
| IV | 6 (9) | 138.2
(11.2–1701.2) | |
| Differentiation
degree | | | 0.503 |
| Well | 5 (8) | 1.0 | |
| Moderate | 39 (60) | 1.02
(0.08–12.5) | |
| Poor | 21 (32) | 1.21
(0.10–14.5) | |
| p53 status | | | 0.204 |
| Negative | 51 (76) | 1.0 | |
| Positive | 14 (24) | 2.08
(0.67–6.44) | |
Discussion
In the present study, we demonstrated that the
expression of PINCH tended to change when the separately cultured
cancer cells were compared with the co-cultured cells. In
separately cultured cancer cells at 8 h, PINCH tended to decrease
following RT when compared to cells without RT, while in
co-cultured cancer cells there was no significant difference. It
has been shown in cell culture that PINCH is regulated by EGF and
PDGF via Nck-2 (15). The different
expression pattern for PINCH in co-culture cells, compared to
separately culture cells, may be caused by growth factors that
regulate the expression of PINCH.
Even though we noted slight changes in the
expression of PINCH between separately cultured and co-cultured
cancer cells, we did not find in the present study, nor in our
previous study (7) a high enough
difference in PINCH expression between radiated and unirradiated
cells to be able to conclude that the expression of PINCH was
regulated by RT. Therefore, we re-analyzed the expression of PINCH
using the same patient material that we used in our previous study
(7). In the previous study, no
significant results were found for the staining percentage of PINCH
when the cut-off point for PINCH percentage was set to 50%. In the
present study, the cut-off point was set to 75%. Moreover, in the
present study, the PINCH staining of the entire tumor section was
analyzed compared to the previous study where only the staining
intensity at the invasive margin showed significant relationships
(7). Noteworthy, when the entire
tumor area using the same material as described was re-investigated
for the staining percentage of PINCH, we found that in the patients
with RT, the survival of patients with strong PINCH expression was
significantly decreased when compared with the patients with weak
PINCH expression after adjustment for TNM stage, differentiation
degree, age and p53 status. In the patients without RT, no
statistically significant difference was found. An interaction
analysis was further performed in order to investigate whether RT
was the reason for the change in survival between patients with
weak and strong PINCH expression. The interaction analysis showed a
trend towards significance, which may indicate that RT was the
reason for the survival differences in the patients with weak and
strong PINCH expression. In our previous study, it was shown that
strong PINCH expression was related to worse survival when compared
to weak expression in the patients without RT, while in the
patients with RT, the significant difference was lost (7). When the invasive margin of the tumor
was investigated, we found a relationship between strong PINCH
expression and reduced survival in the patients without RT
(7) while in the present study,
when the entire tumor area was investigated, the same relationship
was found but in the patients with RT, suggesting that the
relationship between PINCH, RT and survival may depend on where
PINCH is located in the tumor and how the immunostaining for PINCH
is evaluated (percentage or intensity).
Few studies have analyzed the relationship between
PINCH and RT. A recent cell line study of mouse embryonic
fibroblasts and human colon, lung, cervix, skin and pancreas tumors
showed that PINCH enhances radioresistance by activating Akt
(8) while others found PINCH
radiosensitivity to be similar even when the cells were grown in a
suspension or under adherent conditions (16). The significant differences in
survival of the patients with weak and strong PINCH expression
treated with RT, and the positive interaction analysis between
PINCH, RT and survival support our theory that the expression of
PINCH may be regulated by RT. An in-depth cell line study and a
study using a larger sample of rectal cancer patient material are
ongoing to further confirm this relationship.
We found significantly increased expression of C-Myc
at 24 h in co-cultured cancer cells treated with radiation compared
to cells without radiation, which was not found in the separately
cultured cancer cells. Previously, it was shown in a cell culture
of colonic adenoma cells that growth factors play an important role
in the regulation of the production of C-Myc (17). Other studies have shown that RT
upregulates the number of growth factor receptors in cancer cells
(18,19). The significant increase in C-Myc
expression at 24 h in co-cultured cancer cells treated with
radiation compared to cancer cells without radiation may be due to
growth factors produced in co-culture but not in separately
cultured cells that together with radiation, not at 8 h but at 24 h
after radiation, activate receptors on the cell surface, which
further increases the expression of C-Myc. Since C-Myc has been
shown to reduce apoptosis (20,21),
we suggest that in co-culture compared to separately cultured
cells, cancer cells may become resistant to radiation by
upregulation of C-Myc.
We found almost the same but a weaker expression
pattern for PINCH compared to C-Myc in separately and co-cultured
cancer cells with or without radiation, suggesting that there may
be some relationship between PINCH and C-Myc. To further study the
interaction between PINCH and C-Myc, a DNA sequence analysis was
performed. Due to the unavailability of appropriate secondary
structures, we were unable to further study the interaction between
PINCH and C-Myc by bioinformatics approach. However, a
bioinformatics analysis depends on previously published studies
concerning the protein structure. Thus, it is evident that studies
concerning these proteins have not been previously reported. We
conclude that the two proteins do not directly interact, but may
influence the expression of each other depending on other proteins
that are yet to be identified.
PINCH is an independent prognostic factor in rectal
cancer patients with RT, but not in patients without RT. The
expression of PINCH in radiated colon cancer cells changed when
analyzing expression in separately cultured to that in co-cultured
cells, suggesting that the expression of PINCH may be regulated by
radiation and by environmental factors surrounding the cancer
cells. C-Myc significantly increased in co-cultured colon cancer
cells with radiation compared to cell without radiation. In
co-culture, C-Myc may be upregulated to protect cells from
apoptosis induced by radiation. An in-depth cell line study and a
study using a larger sample of rectal cancer patient material are
ongoing to further confirm this relationship.
Acknowledgements
The authors are grateful to Dr Dianne Langford
(Department of Neuroscience, Temple University School of Medicine,
Philadelphia, PA, USA) who kindly provided us with the PINCH
antibody. This study was supported by grants from the Foundation of
Oncological Clinical Research in Linköping, the Swedish Cancer
Foundation, Swedish Research Council and the Health Research
Council in Southeast Sweden.
References
|
1
|
Rearden A: A new LIM protein containing an
autoepitope homologous to ‘senescent cell antigen’. Biochem Biophys
Res Commun. 201:1124–1131. 1994.PubMed/NCBI
|
|
2
|
Cabodi S, del Pilar Camacho-Leal M, Di
Stefano P and Defilippi P: Integrin signalling adaptors: not only
figurants in the cancer story. Nat Rev Cancer. 10:858–870. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukuda T, Chen K, Shi X and Wu C: PINCH-1
is an obligate partner of integrin-linked kinase (ILK) functioning
in cell shape modulation, motility, and survival. J Biol Chem.
278:51324–51333. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Chen K, Tu Y, et al: Assembly of
the PINCH-ILK-CH-ILKBP complex precedes and is essential for
localization of each component to cell-matrix adhesion sites. J
Cell Sci. 115:4777–4786. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu C: The PINCH-ILK-parvin complexes:
assembly, functions and regulation. Biochim Biophys Acta.
1692:55–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao J, Arbman G, Rearden A and Sun XF:
Stromal staining for PINCH is an independent prognostic indicator
in colorectal cancer. Neoplasia. 6:796–801. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holmqvist A, Gao J, Holmlund B, et al:
PINCH is an independent prognostic factor in rectal cancer patients
without preoperative radiotherapy - a study in a Swedish rectal
cancer trial of preoperative radiotherapy. BMC Cancer. 12:652012.
View Article : Google Scholar
|
|
8
|
Eke I, Koch U, Hehlgans S, et al: PINCH1
regulates Akt1 activation and enhances radioresistance by
inhibiting PP1alpha. J Clin Invest. 120:2516–2527. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Henriksson M and Luscher B: Proteins of
the Myc network: essential regulators of cell growth and
differentiation. Adv Cancer Res. 68:109–182. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen K, Tu Y, Zhang Y, Blair HC, Zhang L
and Wu C: PINCH-1 regulates the ERK-Bim pathway and contributes to
apoptosis resistance in cancer cells. J Biol Chem. 283:2508–2517.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morikawa K, Walker SM, Nakajima M, Pathak
S, Jessup JM and Fidler IJ: Influence of organ environment on the
growth, selection, and metastasis of human colon carcinoma cells in
nude mice. Cancer Res. 48:6863–6871. 1988.PubMed/NCBI
|
|
12
|
Morikawa K, Walker SM, Jessup JM and
Fidler IJ: In vivo selection of highly metastatic cells from
surgical specimens of different primary human colon carcinomas
implanted into nude mice. Cancer Res. 48:1943–1948. 1988.PubMed/NCBI
|
|
13
|
No authors listed. Improved survival with
preoperative radiotherapy in resectable rectal cancer. Swedish
Rectal Cancer Trial. N Engl J Med. 336:980–987. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tu Y, Li F and Wu C: Nck-2, a novel Src
homology2/3-containing adaptor protein that interacts with the
LIM-only protein PINCH and components of growth factor receptor
kinase-signaling pathways. Mol Biol Cell. 9:3367–3382. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sandfort V, Eke I and Cordes N: The role
of the focal adhesion protein PINCH1 for the radiosensitivity of
adhesion and suspension cell cultures. PLoS One. 5:e13056. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hague A, Hicks DJ, Bracey TS and Paraskeva
C: Cell-cell contact and specific cytokines inhibit apoptosis of
colonic epithelial cells: growth factors protect against
c-myc-independent apoptosis. Br J Cancer. 75:960–968. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruifrok AC, Mason KA, Lozano G and Thames
HD: Spatial and temporal patterns of expression of epidermal growth
factor, transforming growth factor alpha and transforming growth
factor beta 1–3 and their receptors in mouse jejunum after
radiation treatment. Radiat Res. 147:1–12. 1997.
|
|
19
|
Schmidt-Ullrich RK, Valerie KC, Chan W and
McWilliams D: Altered expression of epidermal growth factor
receptor and estrogen receptor in MCF-7 cells after single and
repeated radiation exposures. Int J Radiat Oncol Biol Phys.
29:813–819. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greco C, Alvino S, Buglioni S, et al:
Activation of c-MYC and c-MYB proto-oncogenes is associated with
decreased apoptosis in tumor colon progression. Anticancer Res.
21:3185–3192. 2001.PubMed/NCBI
|
|
21
|
Park EJ, Kiselev E, Conda-Sheridan M,
Cushman M and Pezzuto JM: Induction of apoptosis by
3-amino-6-(3-aminopropyl)-5,6-dihydro-5,11-dioxo-11H-indeno[1,2-c]isoquinoline
via modulation of MAPKs (p38 and c-Jun N-terminal kinase) and c-Myc
in HL-60 human leukemia cells. J Nat Prod. 75:378–384.
2012.PubMed/NCBI
|