Introduction
Lung cancer is the most common cause of
malignancy-related mortality in the world, and non-small cell lung
cancer (NSCLC) accounts for >85% of cases. Surgical resection,
providing the highest rate of complete recovery, is possible only
in early stages of NSCLC. However, <20% of newly diagnosed NSCLC
cases may qualify for radical resection. Thus, chemotherapy and
radiotherapy play a major role in the multidisciplinary and
systemic treatment of patients with advanced NSCLC (1,2).
Chemotherapy based on platinum compounds and third
generation drugs (such as vinorelbine, gemcitabine, pemetrexed,
docetaxel or paclitaxel) is commonly used and efficacious regimen
of first-line treatment of patients with advanced, unresectable
NSCLC without activating EGFR gene mutations. However, such
treatment is associated with considerable side-effects while it
benefits only a subset of patients (objective response to
first-line chemotherapy is achieved in only 20–40% of patients).
Moreover, median progression-free survival (MPFS) and median
overall survival (OS) in such patients do not exceed 5 and 10
months, respectively (2). Thus, it
is important in qualification to chemotherapy to find the patients
who would benefit most from the treatment and in whom the treatment
will contribute to prolongation of PFS and OS.
Most of the cytostatic drugs used in standard
chemotherapy (such as platinum compounds, gemcitabine) exert their
influence through the destruction of integrity of genetic
information contained in DNA. Due to the proved efficacy and
multiple potential mechanisms of action, platinum-containing drugs
are widely used in the treatment of several types of cancer
including NSCLC. Due to different mechanism of action and
non-overlapping toxicity, cisplatin and gemcitabine doublets are
favoured for combination therapy in NSCLC. The principal mechanism
of action of platinum compounds is formation of DNA-platinum
adducts and, subsequently, creation of intrastrand or interstrand
crosslinks which may cause alteration in the structure of DNA.
These phenomena generally lead to apoptosis of cancer cells.
However, such changes in the DNA helix can be easily identified and
fixed due to the presence of highly efficient DNA repair systems.
Nucleotide excision repair (NER) and mismatch repair (MMR) are
major repair systems that play a crucial role in the resistance of
tumour cells to platinum compounds. One of the multifunctional
enzymes that belong to NER complex, excision repair
cross-complementation group 1 (ERCC1) plays a key role in
recognition, stabilization, and incision (in cooperation with XPF
endonuclease) of cisplatin-induced DNA adducts (3).
Gemcitabine is a pyrimidine antimetabolite
(deoxycytidine analog) that has a similar antitumour activity as
platinum compounds. During DNA replication, active metabolites of
gemcitabine are incorporated into DNA (replacing cytosine
nucleotides) what results in interruption of the discussed process
and induction of tumour cell apoptosis. Furthermore, one of the
molecular targets of gemcitabine is ribonucleotide reductase
(RRM1). Intracellular phosphorylation of gemcitabine leads
indirectly to inhibition of DNA synthesis through the inhibition of
RRM1. Product of RRM1 gene (encodes the regulatory M1
subunit of ribonucleotide reductase) is the key protein involved in
the synthesis and repair of DNA by formation of
deoxyribonucleotides and transformation of ribonucleotides to
deoxyribonucletides (4). Moreover,
certain beneficial interactions were observed for platinum
compounds and gemcitabine in treatment of solid tumours. Prior data
showed that gemcitabine might have an inhibitory effect on the
expression of critical proteins involved in NER, thus inhibiting
repair of DNA lesions caused by platinum compounds (5).
In previously published data, some authors
demonstrated that single nucleotide polymorphism (SNP) of
ERCC1 gene (19007 C>T, Asn118Asn, rs11615) is associated
with patient response to platinum-based chemotherapy. Similarly,
some studies suggest that RRM1 gene promoter polymorphism
(−37A>C) may be linked to response to treatment with
gemcitabine. In a congress report, Bepler et al(6) showed that polymorphism of RRM1
(−37A/C) gene has been associated with level of RRM1 gene
expression. In the quoted study (analysis performed using real-time
quantitative PCR method, gene expression was normalized using 18S
rRNA as reference) median value of RRM1 expression was
respectively: 12.9 in patients with CC genotype, 22.8 in patients
with AC and 72.8 in patients with AA genotype. This confirms
concordance of expected shorter PFS and OS with AA or AC genotype
and longer PFS and OS in patients with CC genotype. Thus, this may
be one of the possible mechanisms of resistance to gemcitabine
treatment. The expression of these genes is described as a
predictive marker for the chemotherapy response in patients with
NSCLC, providing a personalized treatment. Earlier findings support
therapy individualization according to individual mRNA levels of
ERCC1 or RRM1 which can be modified by genetic
polymorphisms. Polymorphisms in ERCC1 or RRM1 genes
seem to influence the carcinogenesis, chemotherapy resistance and
prognosis of survival in NSCLC patients due to changes in protein
structure. However, other available data indicate that these
polymorphisms are not related to the phenotypic differences in
ERCC1 or RRM1 proteins, but, rather, may be associated with
modulation of their expression (7–9).
We performed this non-randomised, retrospective
study to investigate the relationship between polymorphisms of
ERCC1 (19007 C>T) as well as RRM1 (−37C>A)
genes and response to chemotherapy, PFS and OS in NSCLC patients
treated with platinum and gemcitabine doublets. In addition, we
assessed the utility of concerned genetic polymorphisms and
clinical factors as predictive and prognostic markers among such
treated patients.
Materials and methods
Study population
This retrospective and non-randomised study was
conducted from January 2010 to April 2012. The investigated
population consisted of 62 pathologically verified NSCLC patients
(median age, 61 years). Patients were staged as non-operative IIIA
stage, locally advanced (stage IIIB) or advanced (metastatic, stage
IV) disease using computed tomography and other available methods.
Detailed medical history of each patient was collected. Clinical
characteristics of NSCLC patients are presented in Table I. All patients received platinum and
gemcitabine doublets as a first-line chemotherapy. Response to
chemotherapy was evaluated according to RECIST criteria.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Factor | Characteristics, n
(%) |
|---|
| Gender |
| Male | 43 (69.4) |
| Female | 19 (30.6) |
| Age (years) |
| Median | 61 |
| Mean ± SD | 61.4±9.1 |
| Range | 38–76 |
| Smoking status |
| Current or former
smokers | 59 (95.2) |
| Pack-years
(median; mean ± SD) | 32.5; 31.7±17 |
| Never smoker | 3 (4.8) |
| Histopathology
diagnosis |
|
Adenocarcinoma | 27 (43.6) |
| Squamous cell
carcinoma | 10 (16.1) |
| Large cell
carcinoma | 12 (19.4) |
| NSCLC (not
otherwise specified-NOS) | 13 (20.9) |
| Disease stage |
| IIIA
(inoperable) | 6 (9.7) |
| IIIB | 16 (25.8) |
| IV | 40 (64.5) |
| No. of first-line
chemotherapy cycles |
| Mean ± SD | 3.64±1.37 |
| Median | 4 |
| First-line
radiotherapy |
| Yes | 17 (27.4) |
| No | 45 (72.6) |
| Prior surgical
treatment |
| Yes (chemotherapy
was applied after recurrence of the disease) | 14 (22.6) |
| None | 48 (77.4) |
| II/III line
treatment |
| Yes | 37 (59.7) |
| No | 25 (40.3) |
Prior to the investigation, the approval of the
Ethics Committee of the Medical University of Lublin was obtained
(KE-0254/142/2010). The retrospective study did not require
clinical trial registration.
Venous blood was collected from all patients and
genomic DNA was extracted according to the manufacturer’s protocol
using Qiagen Blood Mini kit (Qiagen, Hilden, Germany).
ERCC1 and RRM1 genotyping
For genotyping of ERCC1 (19007 C>T) and
RRM1 (−37 C>A) polymorphisms (coding and promoter
regions, respectively), PCR amplification of genomic DNA followed
by restriction enzyme digestion (PCR-RFLP) was used. The primers
used for both genes were: for ERRC1, F, 5′-AGG ACC ACA GGA
CAC GCA GA-3′ and R, 5′-CAT AGA ACA GTC CAG AAC AC-3′ and for
RRM1, F, 5′-CTG CTC AGG GGA AAG AAC TG-3′ and R, 5′-GGT CTT
GCC CAG ACT CAA CA-3′.
PCR reaction for both ERCC1 and RRM1
genes was performed in a total volume of 25 μl containing 100 ng of
template DNA, 1 μM of each primer, 0.2 mM of each dNTP, 2.4 mM
MgCl2 and 1.0 U Taq polymerase with 1X Reaction buffer
(Fermentas, Burlington, Canada). PCR amplification was carried out
in T Personal thermocycler (Biometra, Göttingen, Germany) in the
following conditions: for ERCC1: initial denaturation at
96°C for 15 min, followed by 35 cycles of 30 sec at 96°C, 30 sec at
61°C and 1.0 min at 72°C and a final elongation step of 10 min at
72°C; for RRM1: initial denaturation at 96°C for 15 min,
followed by 33 cycles of 30 sec at 96°C, 30 sec at 54°C and 30 sec
at 72°C and a final elongation step of 10 min at 72°C. PCR products
of ERCC1 and RRM1 were digested overnight with 5U of
BsrDI or BbsI enzyme (Fermentas), respectively.
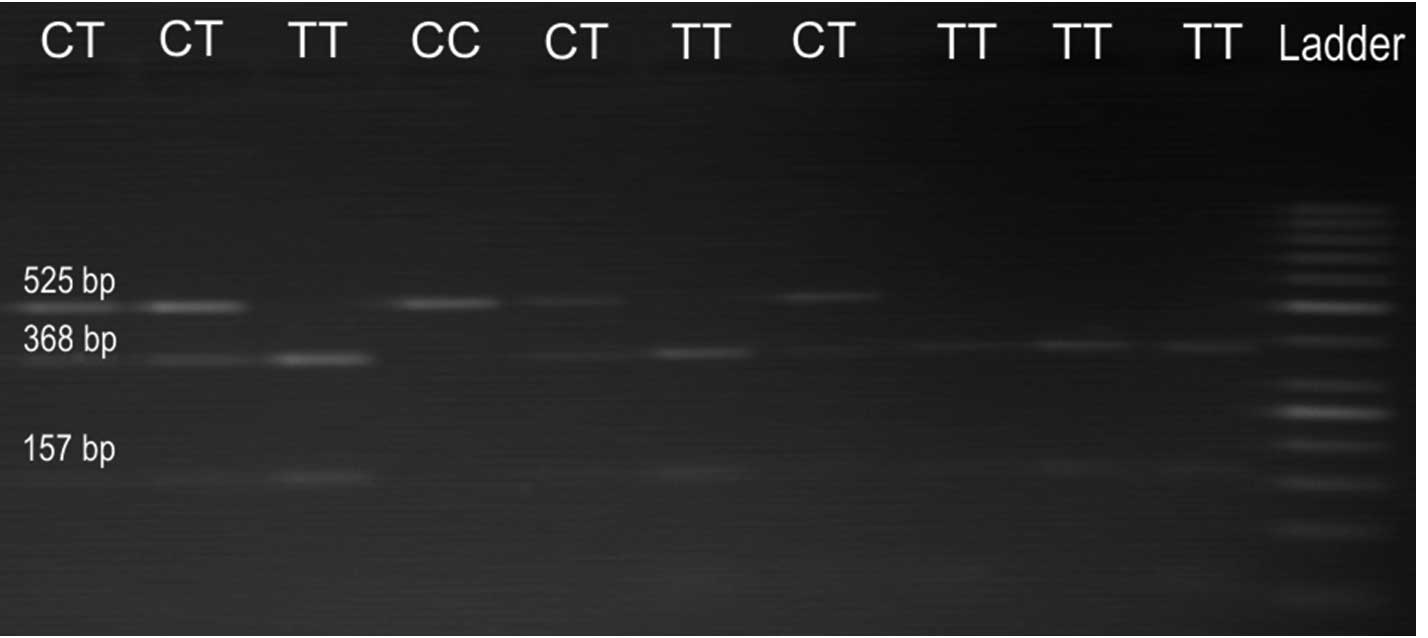
The ERRC1 19007 C>T PCR product is 525
base pairs (bp) in length, and it can be digested with BsrDI
enzyme (Fermentas) if it contains the T allele. The digestion
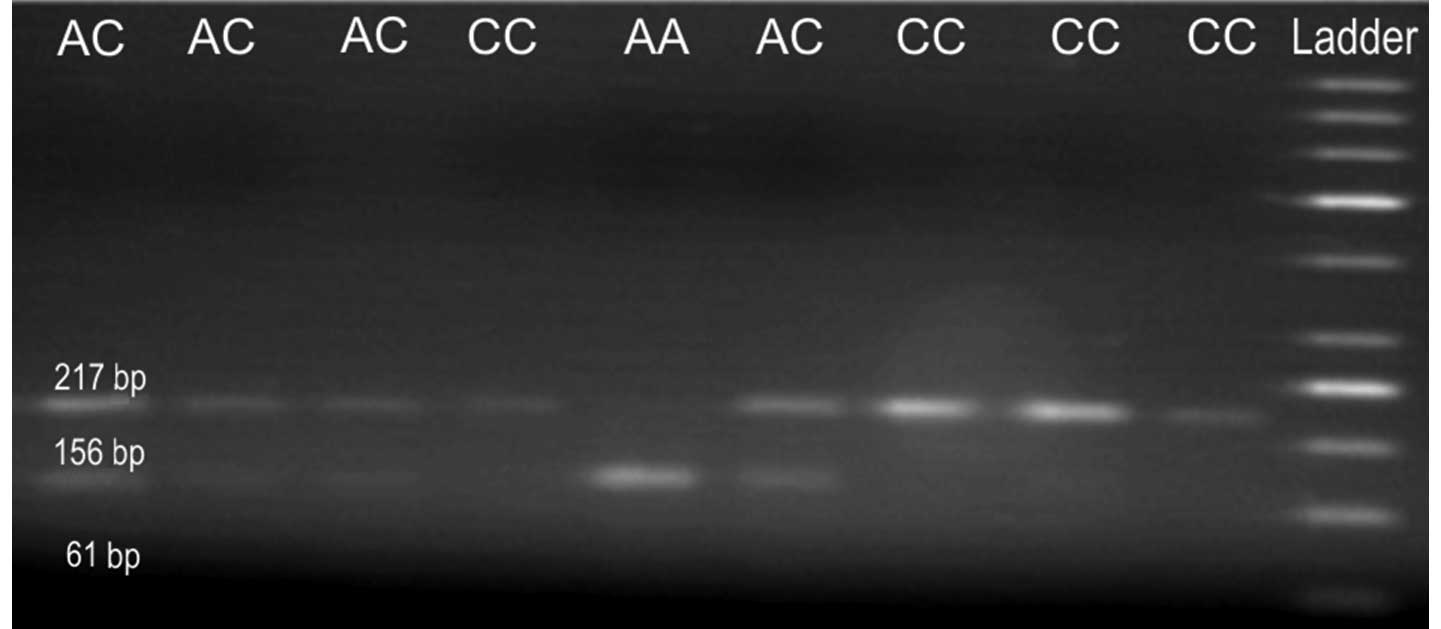
products are 368 and 157 bp respectively. The RRM1 (−37
C>A) PCR product is 217 bp in length, and it can be digested
with BbsI enzyme (Fermentas) if it contains the A allele.
The digestion products are 156 and 61 bp, respectively. The
restricted products were analysed by electrophoresis in 2% agarose
gel containing ethidium bromide. For the ERCC1, three
possible genotypes were defined by three distinct banding patterns:
homozygous for TT genotype corresponds to 368 and 157 bp fragments,
heterozygous for CT genotype corresponds to 525, 368 and 157 bp
fragments, and finally homozygous for CC genotype corresponds to
undigested band of 525 bp (Fig. 1).
For the RRM1, three possible genotypes were defined by three
distinct banding patterns: homozygous for AA genotype corresponds
to 156 and 61 bp fragments, heterozygous for AC genotype
corresponds to 217, 156 and 61 bp fragments, and finally homozygous
for CC genotype corresponds to undigested band of 217 bp (Fig. 2).
Statistical analysis
Results of ERCC1 and RRM1 genotyping
were retrospectively correlated with response to treatment, PFS and
OS of examined patients. Chi-square test was used to compare the
characteristics of the patient groups divided according to
ERCC1 19007 C>T and RRM1 −37C>A polymorphisms.
The U-Mann Whitney test was used for testing equality of population
medians among groups. The Kaplan-Meier method was used for the
comparison of survival probability between the groups of different
ERCC1 and RRM1 genotypes. Finally, the Cox regression
model with stepwise selection procedures by minimum AIC was used to
establish clinical and molecular factors affecting patient
survival. It should be noted that type 1 errors (false positive
results) could occur due to high number of factors used in
statistical analysis.
Results
Patient characteristics and frequency of
ERCC1 and RRM1 genotypes
Baseline characteristics and frequency of
ERCC1 and RRM1 genotypes in the group of 62 NSCLC
patients are shown in Table I;
69.4% of patients were male. The pack-years value was calculated as
the number of cigarette packs smoked per day multiplied by the
number of years. Median pack-years value was 32.5. Very good
performance status (ECOG PS=0) accounted for 66.1% of patients.
Squamous-cell carcinoma was diagnosed in 16.1% of patients,
adenocarcinoma in 43.5%, large-cell carcinoma in 19.3% and other
histological types in 21.1% of patients; 35.5% of patients had
locally advanced NSCLC (stage IIIB). The median number of
platinum-based chemotherapy cycles was four (range, 2–5). Platinum
(cisplatin or carboplatin) was combined with gemcitabine in all
patients. Sequential radiation therapy was administered in 17
patients (27.4%). In the study group, 14 patients (22.6%) were
previously operated due to NSCLC without adjuvant chemotherapy.
These patients were treated with first-line chemotherapy due to
NSCLC recurrence after surgical treatment.
CC homozygous variant of ERCC1 19007 C>T
polymorphism was present in 7 patients (11.3%), CT heterozygous
variant in 28 patients (45.2%) and TT homozygous variant in 27
patients (43.5%). CC homozygous variant of RRM1-37C>A
polymorphism was present in 28 patients (45.2%), AC heterozygous
variant in 32 patients (51.6%) and AA homozygous variant in only 2
patients (3.2%). The distribution of polymorphic variants of
ERCC1 gene did not depend on age, gender, histological type,
clinical stage of disease, chemotherapy regimen, smoking and
performance status of NSCLC patients (Table II). Similarly, no statistically
significant association was observed between the distribution of
polymorphic variants of RRM1 gene and demographic and
clinical factors. The only exception is smoking status due to
prevalence of AC genotype in smokers, noting that the number of
non-smokers was low in the study group (P=0.0103,
χ2=9.161) (Table
III).
 | Table IINSCLC patient characteristics
according to ERCC1 gene status. |
Table II
NSCLC patient characteristics
according to ERCC1 gene status.
| Factor | CC genotype of
ERCC1 gene | CT genotype of
ERCC1 gene | TT genotype of
ERCC1 gene | P-value | χ2 |
|---|
| Whole group | 7 (11.3) | 28 (45.2) | 27 (43.5) | | |
|
| Gender |
| Male | 4 (9.3) | 20 (46.5) | 19 (44.2) | 0.7554 | 0.561 |
| Female | 3 (15.8) | 8 (42.1) | 8 (42.1) | | |
| Age (years) |
| <70 | 5 (10.2) | 21 (42.9) | 23 (46.9) | 0.5667 | 1.136 |
| ≥70 | 2 (15.4) | 7 (53.8) | 4 (30.8) | | |
| Smoking status |
| Smoker | 7 (11.9) | 27 (45.8) | 25 (42.4) | 0.6569 | 0.840 |
| Never smoker | 0 (0) | 1 (33.3) | 2 (66.6) | | |
| Performance
status |
| PS=0/1 | 4 (9.52) | 19 (45.24) | 19 (45.24) | 0.8004 | 0.445 |
| PS=2/3 | 3 (15) | 9 (45) | 8 (40) | | |
| Disease stage |
| IIIA (inoperable),
IIIB | 0 (0) | 13 (59.09) | 9 (40.9) | 0.0682 | 5.37 |
| IV | 7 (17.5) | 15 (37.5) | 18 (45) | | |
| Chemotherapy
toxicities |
| Yes | 5 (11.9) | 21 (50) | 16 (38.1) | 0.4476 | 1.608 |
| No | 2 (10) | 7 (35) | 11 (55) | | |
| Histopathology
diagnosis |
|
Adenocarcinoma | 5 (18.5) | 10 (37) | 12 (44.4) | 0.3045 | 7.181 |
| Squamous cell
carcinoma | 1 (10) | 6 (60) | 3 (30) | | |
| Large cell
carcinoma | 0 (0) | 4 (33.3) | 8 (66.7) | | |
| NSCLC (not
otherwise specified-NOS) | 1 (7.7) | 8 (61.5) | 4 (30.8) | | |
 | Table IIINSCLC patient characteristics
according to RRM1 gene status. |
Table III
NSCLC patient characteristics
according to RRM1 gene status.
| Factor | AA genotype of
RRM1 gene n, (%) | AC genotype of
RRM1 gene n, (%) | CC genotype of
RRM1 gene n, (%) | P-value | χ2 |
|---|
| Whole group | 2 (3.22) | 32 (51.62) | 28 (45.16) | | |
|
| Gender |
| Male | 1 (2.3) | 21 (48.8) | 21 (48.8) | 0.6121 | 0.982 |
| Female | 1 (5.3) | 11 (57.9) | 7 (36.8) | | |
| Age (years) |
| <70 | 2 (4.1) | 25 (51) | 22 (44.9) | 0.7595 | 0.550 |
| ≥70 | 0 (0) | 7 (53.8) | 6 (46.2) | | |
| Smoking status |
| Smokers | 1 (1.7) | 31 (52.5) | 27 (45.8) | 0.0103 | 9.161 |
| Never smoker | 1 (33.3) | 1 (33.3) | 1 (33.3) | | |
| Performance
status |
| PS=0/1 | 2 (4.88) | 19 (46.34) | 20 (48.78) | 0.3629 | 2.027 |
| PS=2/3 | 0 (0) | 13 (61.9) | 8 (38.1) | | |
| Disease stage |
| IIIA (inoperable),
IIIB | 2 (9.1) | 11 (50) | 9 (40.9) | 0.1503 | 3.79 |
| IV | 0 (0) | 21 (52) | 19 (47.5) | | |
| Chemotherapy
complications |
| Yes | 1 (2.4) | 20 (47.6) | 21 (50) | 0.5052 | 1.365 |
| No | 1 (5) | 12 (60) | 7 (35) | | |
| Histopathology
diagnosis |
|
Adenocarcinoma | 1 (3.7) | 14 (51.9) | 12 (44.4) | 0.9306 | 1.878 |
| Squamous cell
carcinoma | 0 (0) | 6 (60) | 4 (40) | | |
| Large cell
carcinoma | 0 (0) | 6 (50) | 6 (50) | | |
| NSCLC (not
otherwise specified-NOS) | 1 (7.7) | 6 (46.2) | 6 (46.2) | | |
ERCC1 19007 C>T and RRM1-37C>A
polymorphisms, possible genotype combinations and chemotherapeutic
response
In our study group, we noted lack of complete
remission. Disease control (PR and SD) occurred in 35 patients
(56.4%), out of which: partial response and stable disease was
observed in 13 (20.9%) and 22 (35.5%) patients, respectively.
Progressive disease was observed in 27 patients (43.5%). Good
performance status (P=0.0038) and the absence of anaemia and prior
surgical treatment increased (not significantly) incidence of
disease control. The MPFS was 3 months for the whole group of
patients and 6 months for responding patients. Patients with
favourable performance status, with locally advanced NSCLC and
previously surgically treated were characterised by the longest PFS
(5.5, 6 and 8 months, respectively) (Tables IV and V).
 | Table IVThe influence of clinical and
molecular factors on early progression risk in patients with NSCLC
treated with platinum and gemcitabine-based chemotherapy. |
Table IV
The influence of clinical and
molecular factors on early progression risk in patients with NSCLC
treated with platinum and gemcitabine-based chemotherapy.
| Factor | No. | PD | SD, PR | P-value | χ2 |
|---|
| Whole group | 62 | 42 (59.2) | 29 (40.8) | | |
|
| Age (years) |
| ≤70 | 49 | 23 (46.94) | 26 (53.06) | 0.465 | 0.534 |
| >70 | 13 | 4 (30.77) | 9 (69.23) | | |
| Gender |
| Male | 43 | 22 (51.16) | 21 (48.84) | 0.1232 | 2.376 |
| Female | 19 | 5 (26.31) | 14 (73.69) | | |
| Smoking status |
| Smokers | 59 | 25 (42.37) | 34 (57.63) | 0.8173 | 0.0534 |
| Never smoker | 3 | 2 (66.66) | 1 (33.33) | | |
| Performance
status |
| PS=0/1 | 41 | 12 (29.26) | 29 (70.74) | 0.0038 | 8.399 |
| PS=2 | 21 | 15 (71.42) | 6 (28.58) | | |
| Histopathology
diagnosis |
| Squamous cell
carcinoma | 10 | 7 (70) | 3 (30) | 0.1352 | 2.232 |
| Other types of
NSCLC | 52 | 20 (38.46) | 32 (61.54) | | |
| Weight loss during
3 months |
| ≤5% | 28 | 11 (39.28) | 17 (60.72) | 0.7211 | 0.127 |
| >5% | 34 | 16 (47.05) | 18 (52.95) | | |
| Anaemia |
| Yes | 44 | 23 (52.27) | 21 (47.73) | 0.0596 | 3.550 |
| No | 18 | 4 (22.22) | 14 (77.78) | | |
| Disease stage |
| IIIA (inoperable),
IIIB | 22 | 8 (36.36) | 14 (63.64) | 0.5629 | 0.335 |
| IV | 40 | 19 (47.5) | 21 (52.5) | | |
| Prior surgical
treatment |
| Yes | 14 | 2 (14.28) | 12 (85.72) | 0.0515 | 3.793 |
| No | 48 | 23 (47.92) | 25 (52.08) | | |
| Malignant diseases
in family |
| Yes | 18 | 6 (33.33) | 12 (66.66) | 0.4500 | 0.571 |
| No | 44 | 21 (47.73) | 23 (52.27) | | |
| Genotype of
ERCC1 gene |
| CC | 7 | 4 (57.14) | 3 (42.86) | 0.5809 | 1.086 |
| CT | 28 | 13 (46.43) | 15 (53.57) | | |
| TT | 27 | 10 (37.04) | 17 (62.96) | | |
| Genotype of
ERCC1 gene |
| CC | 7 | 4 (57.14) | 3 (42.86) | 0.7147 | 0.134 |
| CT + TT | 55 | 23 (41.82) | 32 (58.18) | | |
| Genotype of
ERCC1 gene |
| CT | 28 | 13 (46.43) | 15 (53.57) | 0.8747 | 0.0249 |
| CC + TT | 34 | 14 (41.18) | 20 (58.82) | | |
| Genotype of
ERCC1 gene |
| TT | 27 | 10 (37.04) | 17 (62.96) | 0.5157 | 0.422 |
| CC + CT | 35 | 17 (48.57) | 18 (51.43) | | |
| Genotype of
RRM1 gene |
| AA | 2 | 0 (0) | 2 (100) | 0.0252 | 7.358 |
| AC | 32 | 19 (59.37) | 13 (40.63) | | |
| CC | 28 | 8 (28.57) | 20 (71.43) | | |
| Genotype of
RRM1 gene |
| AA | 2 | 0 (0) | 2 (100) | 0.5907 | 0.289 |
| AC + CC | 60 | 27 (45) | 33 (55) | | |
| Genotype of
RRM1 gene |
| AC | 32 | 19 (59.37) | 13 (40.63) | 0.0193 | 5.473 |
| AA + CC | 30 | 8 (26.7) | 22 (73.3) | | |
| Genotype of
RRM1 gene |
| CC | 28 | 8 (28.57) | 20 (71.43) | 0.0573 | 3.614 |
| AA + AC | 34 | 19 (55.9) | 15 (44.1) | | |
| Genotype of
ERCC1 and RRM1 |
| CC + AC | 4 | 2 (50) | 2 (50) | 0.8008 | 0.0636 |
| Other | 58 | 25 (43.1) | 33 (56.9) | | |
| Genotype of
ERCC1 and RRM1 |
| CC + CC | 3 | 2 (66.66) | 1 (33.33) | 0.8173 | 0.0534 |
| Other | 59 | 25 (42.4) | 34 (57.6) | | |
| Genotype of
ERCC1 and RRM1 |
| CT + AA | 2 | 0 (0) | 2 (100) | 0.5907 | 0.289 |
| Other | 60 | 27 (45) | 33 (55) | | |
| Genotype of
ERCC1 and RRM1 |
| CT + AC | 11 | 8 (72.73) | 3 (27.27) | 0.0692 | 3.301 |
| Other | 51 | 19 (37.25) | 32 (62.75) | | |
| Genotype of
ERCC1 and RRM1 |
| CT + CC | 15 | 5 (33.33) | 10 (66.66) | 0.5370 | 0.381 |
| Other | 47 | 22 (46.8) | 25 (53.2) | | |
| Genotype of
ERCC1 and RRM1 |
| TT + AC | 17 | 9 (52.94) | 8 (47.06) | 0.5289 | 0.397 |
| Other | 45 | 18 (40) | 27 (60) | | |
| Genotype of
ERCC1 and RRM1 |
| TT + CC | 10 | 1 (10) | 9 (90) | 0.0468 | 3.953 |
| Other | 52 | 26 (50) | 26 (50) | | |
 | Table VThe influence of clinical and
molecular factors on progression-free survival and overall survival
in patients treated with platinum and gemcitabine-based
chemotherapy. |
Table V
The influence of clinical and
molecular factors on progression-free survival and overall survival
in patients treated with platinum and gemcitabine-based
chemotherapy.
| Factor | Median PFS
(months) | P-value | χ2 | HR | 95% CI | Median OS
(months) | P-value | χ2 | HR | 95% CI |
|---|
| Whole group | 3 | | | | | 5.75 | | | | |
|
| Age (years) |
| >70 | 6 | 0.4627 | 0.5395 | 0.7941 | 0.4248–1.4844 | 16.5 | 0.8027 | 0.0624 | 0.895 | 0.3729–2.148 |
| ≤70 | 3 | | | | | 11 | | | | |
| Gender |
| Male | 3 | 0.8194 | 0.05214 | 1.0668 | 0.5902–1.9285 | 9.5 | 0.6864 | 0.1630 | 1.1806 | 0.5339–2.6106 |
| Female | 3.5 | | | | | 21 | | | | |
| Smoking status |
| Never smoker | 2.5 | 0.6351 | 0.2252 | 0.7674 | 0.2065–2.8523 | 9.5 | 0.5842 | 0.2996 | 1.6772 | 0.1228–3.7346 |
| Smokers | 3 | | | | | 13 | | | | |
| Performance
status |
| PS=0/1 | 5.5 | 0.0004 | 12.543 | 0.4048 | 0.1984–0.8258 | 18 | 0.0003 | 13.074 | 0.292 | 0.160–0.8043 |
| PS=2 | 2 | | | | | 5.5 | | | | |
| Histopathology
diagnosis |
| Squamous cell
carcinoma | 3.5 | 0.0902 | 2.8709 | 0.5640 | 0.2329–1.3658 | 16.5 | 0.5949 | 0.2831 | 0.7758 | 0.2704–2.226 |
| Other types of
NSCLC | 2 | | | | | 11 | | | | |
| Weight loss during
3 months |
| >5% | 3 | 0.3192 | 0.9923 | 1.2799 | 0.7417–2.2086 | 7.5 | 0.2817 | 1.1589 | 1.4393 | 0.6462–3.2058 |
| ≤5% | 4 | | | | | 16.5 | | | | |
| Anaemia |
| Yes | 2.5 | 0.0615 | 3.4955 | 1.7317 | 0.985–13.0446 | 11 | 0.6673 | 0.1847 | 0.8406 | 0.3580–1.9738 |
| No | 4 | | | | | 13 | | | | |
| Disease stage |
| IV | 3 | 0.0361 | 4.3904 | 1.7596 | 1.0175–3.0429 | 8 | 0.1586 | 1.9876 | 1.7408 | 0.8199–3.6963 |
| IIIA (inoperable),
IIIB | 6 | | | | | 16.5 | | | | |
| Radiotherapy |
| Yes | - | - | - | | | 13 | 0.2526 | 1.3089 | 0.6279 | 0.2883–1.3673 |
| No | - | - | - | | | 9.5 | | | | |
| Prior surgical
treatment |
| Yes | 8 | 0.0055 | 7.723 | 0.4291 | 0.2433–0.7567 | 33 | 0.0085 | 6.93 | 0.2503 | 0.1134–0.5524 |
| No | 2.5 | | | | | 8 | | | | |
| II/III line
treatment |
| Yes | - | - | - | | | 16.5 | 0.0041 | 8.2236 | 0.3753 | 0.1534–0.9182 |
| No | - | - | - | | | 7 | | | | |
| Chemotherapy
toxicities |
| No | 4 | 0.2176 | 1.5204 | 1.3916 | 0.7997–2.4219 | 11 | 0.7826 | 0.07617 | 1.1118 | 0.5187–2.3831 |
| Yes | 3 | | | | | 13 | | | | |
| ERCC1
genotype |
| CC | 2.5 | 0.576 | 1.1032 | | | 7.5 | 0.5143 | 1.33 | - | - |
| CT | 3 | | | | | 16.5 | | | | |
| TT | 3 | | | | | 13 | | | | |
| ERCC1
genotype |
| Other | 3 | 0.2746 | 1.1937 | 0.6634 | 0.2608–1.6871 | 13 | 0.254 | 1.3012 | 0.5545 | 0.1465–2.0988 |
| CC | 2.5 | | | | | 7.5 | | | | |
| ERCC1
genotype |
| Other | 3 | 0.5462 | 0.3642 | 1.1703 | 0.6793–2.0162 | 11 | 0.5219 | 0.4102 | 1.2672 | 0.6019–2.6678 |
| CT | 3 | | | | | 16.5 | | | | |
| ERCC1
genotype |
| Other | 3 | 0.9750 | 0.0009 | 1.0083 | 0.5818–1.7473 | 6 | 0.518 | 0.4179 | 1.1878 | 0.6888–2.0484 |
| TT | 3 | | | | | 8 | | | | |
| RRM1
genotype |
| AA | - | 0.0016 | 12.8968 | - | - | | 0.0346 | 6.7275 | - | - |
| AC | 2 | | | | | 8 | | | | |
| CC | 6 | | | | | 16.5 | | | | |
| RRM1
genotype |
| Other | 6.5 | 0.0001 | 15.1677 | 0.3927 | 0.2204–0.6992 | 16.5 | 0.0104 | 6.5631 | 0.3886 | 0.183–0.8248 |
| AC | 2 | | | | | 8 | | | | |
| RRM1
genotype |
| CC | 6 | 0.0087 | 6.8855 | 0.5134 | 0.2955–0.8919 | 16.5 | 0.0448 | 4.0263 | 0.4728 | 0.2249–0.9943 |
| Other | 2 | | | | | 8 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 3 | 0.4460 | 0.5808 | 0.6916 | 0.2088–2.2903 | 13 | 0.4272 | 0.6305 | 0.5702 | 0.09001–3.6126 |
| CC + AC | 3 | | | | | 6.75 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 3 | 0.4592 | 0.5479 | 0.6637 | 0.1631–2.6999 | 13 | 0.4407 | 0.5944 | 0.5781 | 0.09232–3.6205 |
| CC + CC | 2 | | | | | 7.5 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 3.5 | 0.0098 | 6.6753 | 0.4445 | 0.1765–1.1198 | 13 | 0.0662 | 3.3757 | 0.4489 | 0.1368–1.4730 |
| CT + AC | 2 | | | | | 5.5 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 3 | 0.2113 | 1.5624 | 1.4632 | 0.8108–2.6404 | 9.5 | 0.1501 | 2.0717 | 1.9084 | 0.8459–4.3055 |
| CT + CC | 7 | | | | | 16.5 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 4 | 0.0301 | 4.7042 | 0.5451 | 0.2691–1.1042 | 13 | 0.2345 | 1.4137 | 0.6339 | 0.2726–1.4743 |
| TT + AC | 2 | | | | | 11 | | | | |
| ERCC1 and
RRM1 genotype |
| Other | 3 | 0.0473 | 3.9339 | 2.0761 | 1.1027–3.9089 | 11 | 0.1903 | 1.7153 | 2.1411 | 0.8546–5.3647 |
| TT + CC | 6 | | | | | 13 | | | | |
There were no statistically significant
relationships between occurrences of a particular ERCC1 gene
polymorphism and the response to therapy or PFS. MPFS for CC, CT
and TT genotypes was 2.5, 3 and 3 months, respectively.
In the case of RRM1 gene polymorphism, the
carriers of AC genotype showed disease progression significantly
more frequently (P=0.0193) than carriers of AA (only two patients)
or CC genotype. Disease control occurred slightly more frequently
(P=0.0573) in patients with CC genotype compared to carriers of A
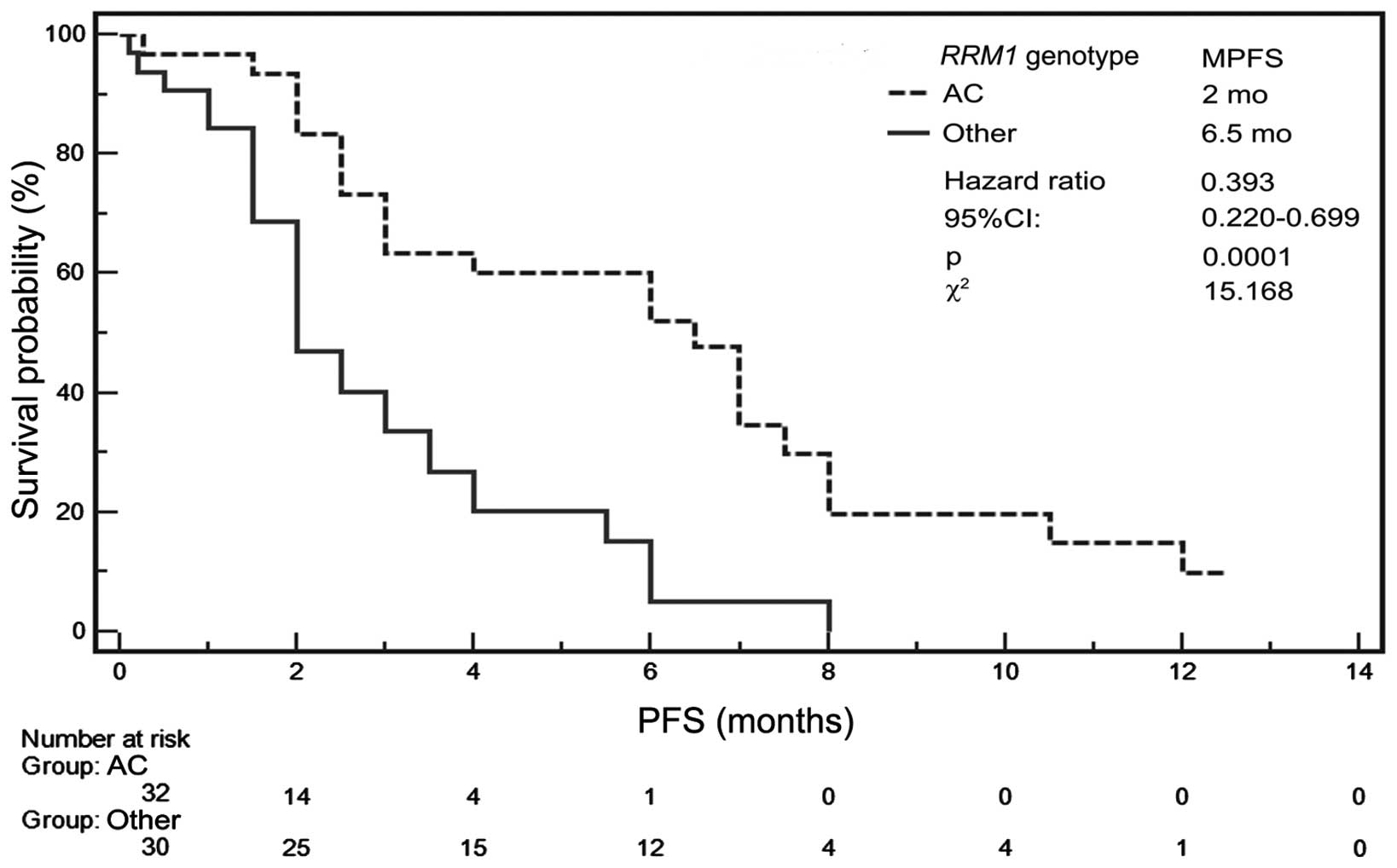
allele (patients with AA or AC genotype) (Table IV). MPFS was only 2 months for AC
heterozygous patients, but was 6.5 months for AA or CC homozygous
patients (Table V). In Kaplan-Meier
analysis, the risk of progression was significantly lower
(P=0.0001, HR=0.392, 95% CI, 0.2204–0.6992, χ2=15.167)
for patients with AA or CC genotype than for patients with AC
genotype (Fig. 3).
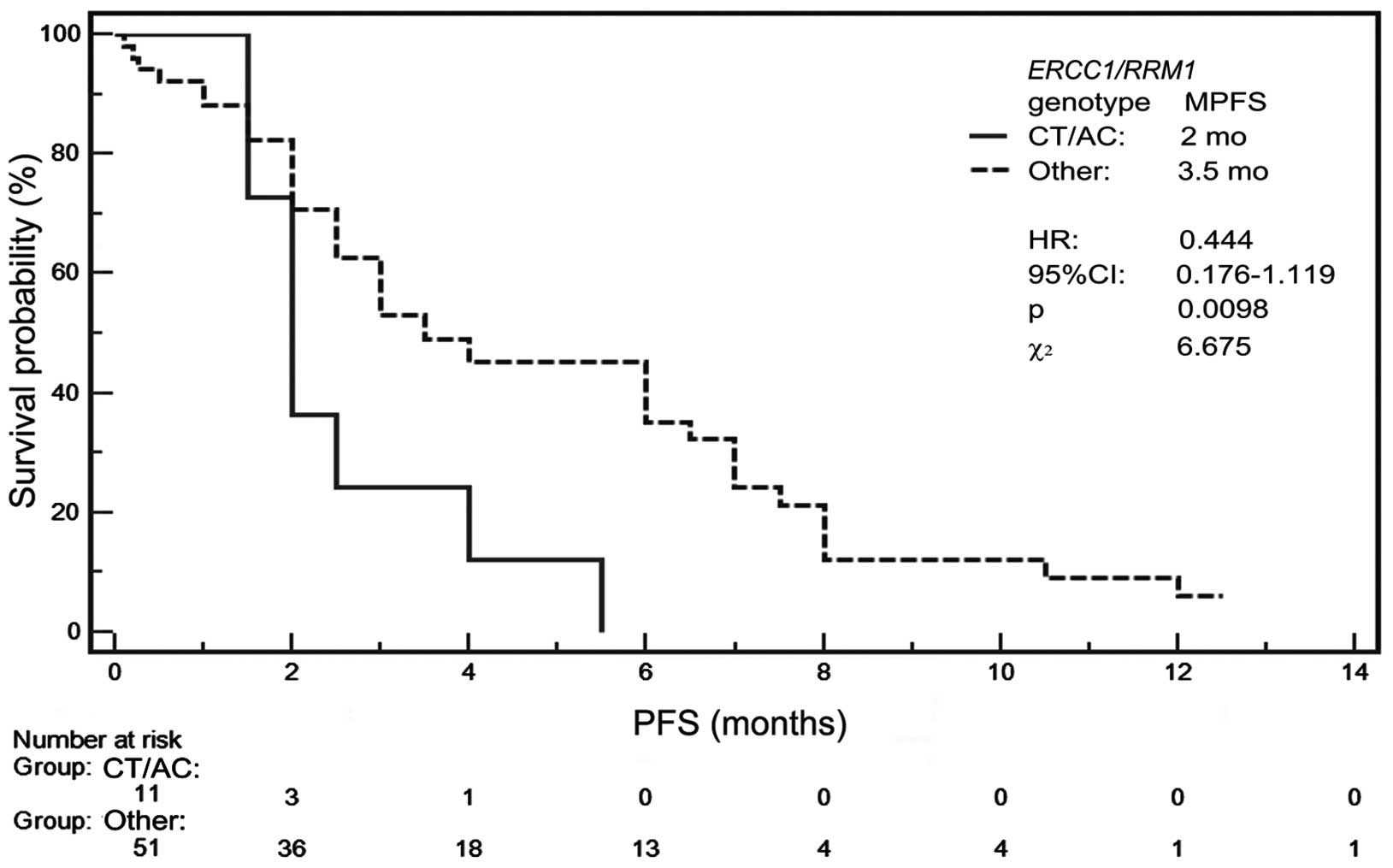
Carriers of ERCC1 and RRM1 genotype
combination TT/CC showed disease control significantly more
frequently (P=0.0468) than carriers of other genotype combinations.
Progression occurred slightly more frequently (P=0.0692) in
patients with genotype combination CT/AC compared to patients with
other genotype combinations (Table
IV). MPFS was 2 months for CT/AC genotype and 3.5 months for
other possible genotype combinations (Table V). In Kaplan-Meier analysis, the
risk of progression was the lowest (P=0.0098, HR=0.4445, 95% CI,
0.1765–1.1198, χ2=6.675) for patients with other than
CT/AC polymorphism combinations (Fig.
4).
The result of statistical analysis depends on the
effects of treatment in two patients with rare AA genotype of RRM1
gene. Thus, this result could depend on stage of disease,
performance status and molecular status different than examined in
this study. A 64-year old male patient with AA genotype with good
performance status suffered from inoperable NOS NSCLC (stage IIIA)
and showed partial response ongoing 12.5 months after only two
cycles of chemotherapy. A fifty-three-year old female patient with
adenocarcinoma remained 7.5 months in stable disease after 4 cycles
of chemotherapy. However, this patient showed EGFR gene mutation
(deletion in exon 19) and was treated with erlotinib in second-line
therapy.
ERCC1 19007 C>T and RRM1-37C>A
polymorphisms, possible genotype combinations and overall
survival
Median survival time (MST) was 5.75 months for all
NSCLC patients. OS depended on performance status and the
applicability of previous surgery as well as second-line treatment
(Table V).
ERCC1 gene polymorphism did not significantly
affect duration of survival but MST was 7.5 months for carriers of
CC genotype, 16.5 months for patients with CT genotype and 13
months for TT homozygous patients (Table V).
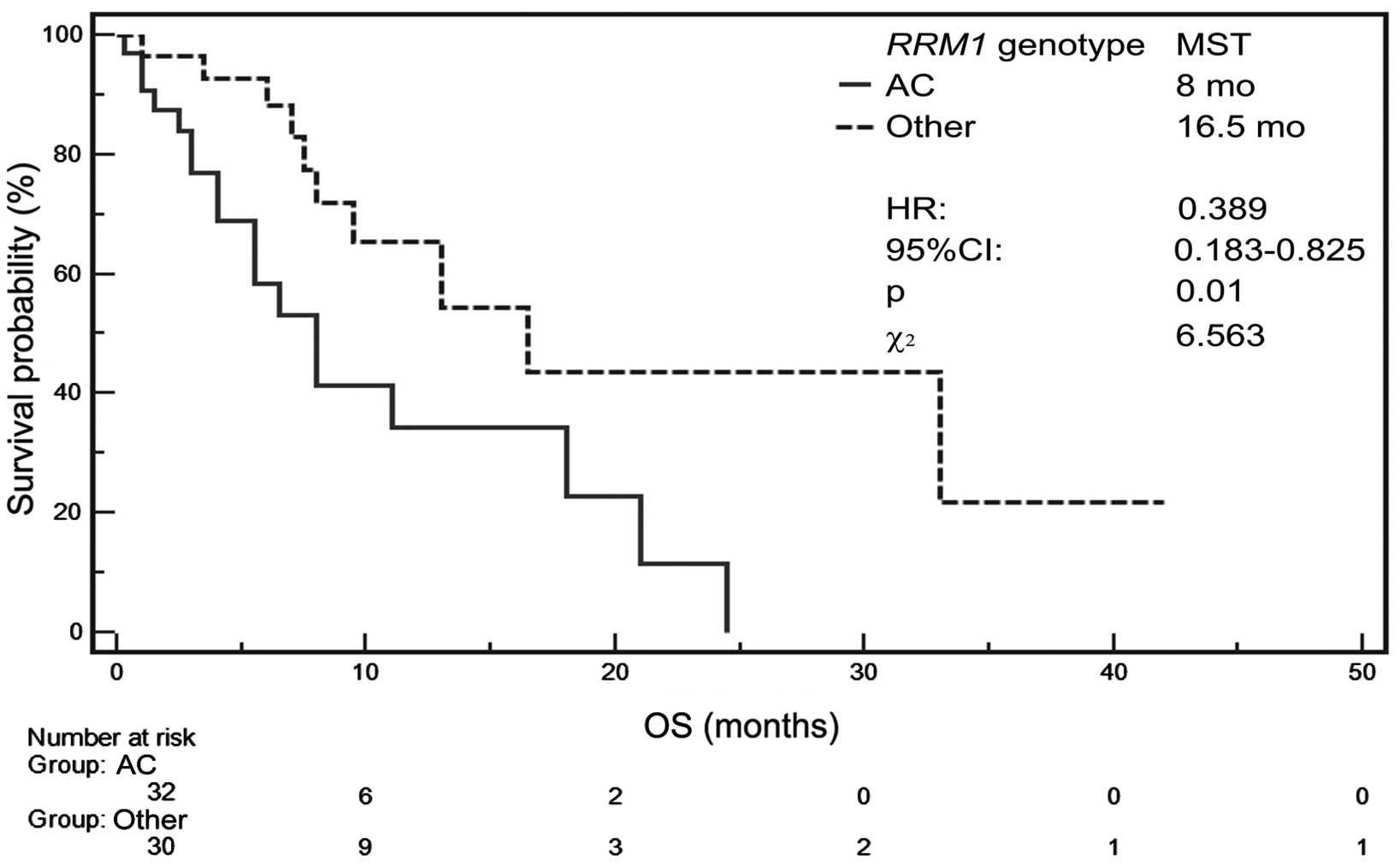
MST for patients with AA genotype of RRM1
gene was not determined but MST amounted to 8 months for carriers
of AC genotype as well as 16.5 months for patients with CC genotype
of this gene (Table V). Patients
with AA or CC genotype showed significantly higher probability of
survival (P=0.0104, HR=0.3886, 95% CI, 0.183–0.8248,
χ2=6.5631) than those with AC genotype (Fig. 5).
Patients with ERCC1 and RRM1 gene
polymorphism combinations other than CT/AC showed insignificantly
higher probability of survival (P=0.0662, HR=0.4489, 95% CI,
0.1368–1.473, χ2=3.376) than those with other genotype
combinations. However, the probability of survival was similar for
carriers of CC/AC genotype combination compared to carriers of
other possible genotypes.
In the Cox regression model, poor performance status
(HR=4.78, 95% CI, 1.82–12.56, P=0.0016), AC genotype of RRM1
gene (HR=2.47, 95% CI: 1.04–5.87, P=0.0414), lack of prior surgical
treatment (HR=4.71, 95% CI, 1.06–20.92, P=0.0425) and lack of
subsequent lines of treatment (HR=3.23, 95% CI, 1.29–8.11,
P=0.0127) were significantly associated with shortening of patient
survival (overall model fit: χ2=30.161 P<0.0001)
(Table VI).
 | Table VIFactors that significantly affect
overall survival of patients treated with platinum and gemcitabine
scheme in multiparameter analysis of Cox regression model (overall
model fit: χ2=30.161, P<0.0001). |
Table VI
Factors that significantly affect
overall survival of patients treated with platinum and gemcitabine
scheme in multiparameter analysis of Cox regression model (overall
model fit: χ2=30.161, P<0.0001).
| Factor | Coefficient β | P-value | Hazard ratio (95%
CI) |
|---|
| Poor performance
status (PS=2) | 1.5640 | 0.0016 | 4.78
(1.82–12.56) |
| AC genotype of
RRM1 gene | 0.9046 | 0.0414 | 2.47
(1.04–5.87) |
| Lack of prior
surgical treatment | 1.5504 | 0.0303 | 4.71
(1.06–20.92) |
| Lack of subsequent
line of treatment | 1.1739 | 0.0127 | 3.23
(1.29–8.11) |
Discussion
Historically, NSCLC has been classified based only
on the histological and morphological picture of the cancer tissue
as well as the anatomic site of origin. However, in NSCLC patients,
significant variation in prognosis and response to treatment was
observed regardless of histopathological diagnosis of cancer. As
demonstrated by recent studies, the eligibility of patients with
NSCLC to the ‘targeted therapies’ (such as tyrosine kinase
inhibitors, TKI) based on genetic differences (EGFR mutation
status) proved to be highly effective (10,11).
Based on recent data, genetic alterations could also
be used successfully for qualification of NSCLC patients to
appropriate standard chemotherapy regimens. Thus, the existing
approach for universal NSCLC treatment, where adjuvant chemotherapy
is provided to all patients with minor benefit and with modest
improvements in response rates and survival, is no longer suitable.
Individual approach to the selection of treatment is therefore
urgently required (3).
One of the most promising prognostic markers for
surgically treated NSCLC patients and predictive marker for
patients receiving platinum-based chemotherapy is expression of the
ERCC1 gene (7). Zheng et
al(12) showed that high
expression of both ERCC1 and RRM1 proteins in tumour cells was
associated with favourable prognosis and an excellent outcome after
surgical resection of patients in early stage of NSCLC. However,
Bepler et al(13) and
Olaussen et al(8) proved
that only ERCC1-negative NSCLC patients benefit significantly from
adjuvant chemotherapy. In their study, Bepler et al(13)demonstrated a comparable trend for
RRM1 expression, but this was statistically insignificant. In a
retrospective study, Ceppi et al(9) further validated concomitant analysis
of ERCC1 and RRM1 mRNA levels as reliable candidates
for personalized chemotherapy and showed a higher impact on the
survival of NSCLC patients treated with cisplatin and gemcitabine.
In the first prospective study, Cobo et al(14) noted the improvement in the response
rate (but not in PFS or OS) in patients qualified to cisplatin and
docetaxel or docetaxel and gemcitabine therapy based on
ERCC1 mRNA level. Another prospective research conducted by
Simon et al(15) confirmed
the benefits of determining both ERCC1 and RRM1 mRNA
gene expression levels in qualification to adequate chemotherapy
regimen in NSCLC patients.
Molecular profiles proposed above are based on the
analysis of protein or gene expression (measured by
immunohistochemistry or mRNA level in real-time PCR technique,
respectively). Initial material for such analysis is acquired from
the tumour tissue which is generally difficult to obtain.
Genetic polymorphisms may affect protein structure,
function, stability or folding. The most common form of
polymorphism in the human genome is an SNP, and some SNPs have been
shown to correlate with drug sensitivity and toxicity. SNPs,
natural genetic variation, occur in high density in the human
genome and were confirmed as predictive markers of response to
various treatment regimens. The advantage of the SNPs as predictive
markers is that genomic DNA can be examined from samples of whole
blood (peripheral blood leukocytes, PBLs), particularly when the
tumour tissue is difficult to obtain or not available (particularly
in patients with advanced NSCLC).
Tumour genotype (often heterogeneous) usually
differs from normal tissue, both in terms of copy number variations
and point mutations. Unfortunately, there is no information in the
literature that polymorphisms of ERCC1 and RRM1 genes
are constant in blood and all tumour cells. However, other
available data (including several different SNPs of genes important
in DNA repair system) indicate that concordance between blood (or
buccal) and tumour (fresh/frozen/FFPE) SNPs is 93–100%. Therefore,
the DNA isolated from PBLs seems to be sufficient material for
analysis of gene polymorphisms valid in pharmacogenetics (16).
Previous data indicate that individual ERCC1
and RRM1 mRNA levels may be related to polymorphic
difference in patient DNA. For example, 8092 C>A polymorphism
located in the 3′-untranslated region, may influence ERCC1 function
independently of the level of mRNA or protein expression (such as
by affecting mRNA stability). Bepler et al demonstrated that
RRM1 expression is controlled by the functional activity of
its promoter (due to occurrence of a particular polymorphic
variant). Polymorphisms in ERCC1 and RRM1 genes seem
to affect the cytostatic resistance, prognosis and survival in
NSCLC (17,18).
The literature presents a limited number of
scientific publications that assess the relationship between
polymorphisms of genes encoding DNA repair proteins and response to
chemotherapy based on platinum compounds and gemcitabine in
patients with locally advanced or advanced NSCLC.
When processing individual SNPs as independent
predictors, we concluded that there is no significant relationship
between different ERCC1 genotypes and response to treatment
or PFS and OS in patients treated with platinum and gemcitabine as
a first-line treatment. However, we noted that common AC genotype
in promoter region of RRM1 gene (−37 C>A) could predict
poor response, shortening of PFS and OS in such treated patients.
We observed that 59.4% of patients with AC genotype demonstrated
early progression during first or second cycle of chemotherapy in
contrast to 26.7% of patients with other possible genotypes. As a
consequence of these differences, the MPFS and OS were
significantly longer among patients with CC genotype than in
patients with AC genotype. Moreover, patients with CC genotype
showed significantly longer MPFS and insignificantly longer median
OS than carriers of A allele.
Despite the lack of statistical significance for the
risk of early progression to TT in ERCC1 and CC in
RRM1 gene polymorphism considered separately, there is a
significant relationship between TT/CC genotype (a combination of
both studied gene polymorphisms) and risk of early progression.
Moreover, MPFS was also significantly longer in carriers of
described polymorphism combinations than in other patients. The
lower MPFS in carriers of genotype combinations CT/AC and TT/AC is
probably due to the presence of an unfavourable component, the AC
genotype of RRM1 gene.
In NSCLC patients with adenocarcinoma and with
activating mutations in EGFR gene, EGFR TKI (EGFR tyrosine
kinase inhibitors) erlotinib and gefitinib are characterized by a
higher efficacy, compared to the standard chemotherapy. Therefore,
it was necessary to determine what percentage of the studied
population (in which 43.6% accounted for adenocarcinomas) received
such treatment. Based on available data (not shown in the present
study) we found that 11.3% of examined patients received erlotinib
in the second-line of treatment. Only one patient achieved stable
disease during erlotinib treatment and none of them met the
criteria of remission. However, the mutation status of EGFR
gene in these patients was unknown, since when the study was
conducted, drug registration in Poland did not require EGFR
testing in order to qualify for TKI therapy.
The limitations of our preliminary study were the
small study group and very low percentage of patients with AA
genotype of RRM1 gene. The authors are aware that due to the
small study group there may be a risk of false positive
relationship between the presence of different genotypes of
ERCC1 and RRM1 genes and the studied factors (OR, PFS
and OS). However, obtained results concerning the distribution of
genotypes are compatible (for the European population) with the
data available in the GenBank database. Available data indicates
that genotypes of ERCC1 occur with different frequency in
various groups of patients. This may be due to a small size of the
populations studied as well as ethnicity. Genotype distributions of
ERCC1 gene acquired in our study are consistent with results
of other studies conducted on Caucasian patients (19,20).
However, such conformity is not observed if we compare our findings
with research on the Asian population (21,22).
On the other hand, distribution of genotypes of RRM1 gene
achieved in this study is consistent with the distribution of
genotypes that occurs in the GenBank database and data from other
studies. Moreover, obtained results are indirectly in concordance
with Bepler et al(17)
(highest RRM1 expression is noted for AA and the lowest for
CC genotype) which confirms our results regarding concordance with
expected shorter PFS and OS in patients with AA or AC genotype and
longer in patients with CC genotype.
Moreover, the differences between the groups with
various ERCC1 and RRM1 polymorphisms according to the
PS status and advancement of NSCLC are statistically insignificant.
The genetic examination in our study was performed retrospectively
and our knowledge of patient ERCC1 and RRM1 status
was obtained after therapy termination. We speculate that AC
genotype (or, perhaps, the presence of A allele) is an unfavourable
prognostic factor and patients with AC genotype might respond worse
for chemotherapy regimens based on platinum compounds and
gemcitabine.
The different results concerning the relationship
between the presence of ERCC1 gene TT genotype and treatment
response or prolonged PFS may be due to several reasons. First, it
may be caused by a different number of groups of respondents in the
previous and the present study (n=43 and n=62, respectively) which
resulted in obtaining different distribution of genotypes in
studied populations (frequency of TT genotype was 16.3 and 43.5%
respectively), which could affect the final result of the presence
or absence of statistical significance. In addition, these
differences may be due to a significantly higher proportion of
patients with TT genotype with poor performance status, (71.4 vs.
40%, respectively), in stage IV of the disease (71.4 vs. 42.1%,
respectively) and with squamous cell carcinoma (85.7 vs. 30%,
respectively). Moreover, different schemes of first-line
chemotherapy were acceptable in our first study (23).
In a previous study, Ryu et al(21) suggested that CC genotype of
ERCC1 19007 C>T polymorphism is a marker for predicting
improved survival in NSCLC patients treated with platinum-based
chemotherapy. However, the authors did not find a correlation
between ERCC1 genotype and response to chemotherapy. Isla
et al(19) showed similar
results in advanced NSCLC patients treated with docetaxel and
cisplatin. In this study, carriers of CC genotype of ERCC1
gene demonstrated a significantly longer MPFS and median survival
than carriers of CT or TT genotype without differences in response
rate. Furthermore, they found no relationship between the
occurrence of certain RRM1 gene polymorphisms and response
to treatment, MPFS or OS.
Data presented by Ren et al(24) showed that ERCC1 118 C/T or
T/T might provide a better prognostic and predictive marker of
NSCLC patients treated with platinum-based chemotherapy, mainly in
the elderly subgroup, male, squamous carcinoma, smokers and those
treated with non-GP/GC regimen. The study of Kalikaki et
al(25) concerning the
polymorphisms of genes encoding DNA repair proteins, showed that
the joint effect of ERCC1 polymorphic variants (8092 C>A
and 19007 C>T) as well as the XRCC1 1196 A>G
polymorphism were independent prognostic factors for OS in advanced
NSCLC patients treated with platinum-based chemotherapy. The
presence of CC genotype and TT genotype of ERCC1 gene as
well as AA genotype of XRCC1 gene was associated with
shorter median survival of analysed patients. However, only
ERCC1 1907 C>T polymorphism significantly predicted
response to therapy. CR or PR was noted in 5.5% of patients with TT
genotype and in 34.7% of patients with CC or CT genotype.
However, a few studies conducted on large groups of
patients did not find the relationship between ERCC1 19007
C>T polymorphism and clinical outcome in advanced NSCLC patients
treated with chemotherapy and surgically resected tumour.
Meta-analysis performed by Yu et al(26) showed that neither ERCC1
C8092A polymorphism nor Asn118Asn variant is associated with
different response to platinum-based treatment among advanced NSCLC
patients. Additionally, these two genetic variants are not related
with treatment response in either Caucasian or Asian patients.
Moreover, Takenaka et al(27) did not observe a relationship between
ERCC1 19007 C>T polymorphism and disease-free survival or OS in
patients following tumour resection due to early stage of
NSCLC.
Recently, a large study of 192 Caucasian patients
(85.9% received cisplatin/gemcitabine regimen) showed no
significant correlations between ERCC1 19007 C>T polymorphism
and objective response to cisplatin/gemcitabine-based chemotherapy.
Moreover, the authors observed no significant differences in PFS
and OS with respect to ERCC1 genotype. Characteristics of the study
group in the publication cited, in part of demographic and clinical
factors as age, gender, smoking status, histological type and stage
of the disease is consistent with our data. However, in terms of
factors such as performance status or use of radical radiotherapy,
study groups differed significantly. Factors discussed above could
have an impact on obtaining different results (28).
Similarly, a study on a smaller group of patients
(n=62) treated with platinum/gemcitabine showed no statistically
significant relationship between the presence of polymorphisms in
ERCC1 and RRM1 genes and objective responses, PFS or OS (29).
Feng et al(30) showed that the response rates to
cisplatin-based therapy among patients with RRM1
polymorphism depended on −524 C>T polymorphism (P=0.046),
whereas it did not depend on −37 C>A polymorphism.
In contrast to our study, Song et al(31) demonstrated that patients harbouring
AC genotype of RRM1 gene −37 C>A polymorphism had a
longer PFS than patients with other possible SNPs when treated with
gemcitabine in first-line chemotherapy. In the study, researchers
showed that patients with AC genotype had MPFS of 30.7 weeks,
carriers of AA genotype, 24.7 weeks and patients with CC genotype,
23.3 weeks (P=0.043). Moreover, they demonstrated that there is no
significant correlation between sensitivity to gemcitabine and any
possible polymorphic variants of 2455 A>G or 2464 G>A of
RRM1 gene.
Bepler et al(17) not only described RRM1
promoter SNPs as a factor which may have impact on the promoter
activity, but also as a prognostic marker of outcome in patients
with resected NSCLC. The research was limited to patients with
combination of genotypes CC/TT or AC/CT in polymorphism −37 C>A
and −524 C>T of RRM1 gene. All other occurring variants
were excluded due to low patient numbers. They found that patients
with the CC/TT genotype had a better overall (P=0.06) and
disease-free (P=0.03) survival than patients with AC/CT
genotype.
In contrast to numerous studies, we have
demonstrated that RRM1 −37 C>A polymorphism analysis is
more useful than ERCC1 19007 C>T polymorphism examination
in prediction of platinum and gemcitabine effects in NSCLC
patients.
Furthermore, the results concerning lack of
significance between TT genotype in ERCC1 and CC in
RRM1 genes, and risk of early progression when considered
separately and an appearance of significance when the genotypes are
considered as a pair, allowed us to conclude that the impact of
specific genetic polymorphisms on effects of treatment should
always be viewed on a number of levels and several factors.
Thus, in patients with this genotype, platinum in
combination with gemcitabine should be considered. On the other
hand, presence of AC genotype in RRM1 gene supports the use
of non-gemcitabine-based treatment. Genetic polymorphisms could
simply be assessed using blood samples and may be easier to adopt
in the clinical setting than tumour gene expression arrays, a
prospective and randomised study should be initiated. Results of
the present study may be used as a tool in the qualification of
advanced NSCLC patients for appropriate chemotherapy regimen which
needs to be validated in prospective randomised trials. This may be
the next step towards full individualisation of chemotherapy in
patients with NSCLC.
References
|
1
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011.PubMed/NCBI
|
|
2
|
NSCLC Meta-Analysis Collaborative Group.
Chemotherapy in addition to supportive care improves survival in
advanced non-small-cell lung cancer. A systemic review and
meta-analysis of individual patient data from 16 randomized
controlled trials. J Clin Oncol. 26:4617–4625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simon GR, Ismail-Khan R and Bepler G:
Nuclear excision repair-based personalized therapy for non-small
cell lung cancer: from hypothesis to reality. Int J Biochem Cell
Biol. 39:1318–1328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jordheim LP, Sève P, Trédan O, et al: The
ribonucleotide reductase large subunit (RRM1) as a predictive
factor in patients with cancer. Lancet Oncol. 12:693–702. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart DJ: Tumor and host factors that
may limit efficacy of chemotherapy in non-small cell and small cell
lung cancer. Crit Rev Oncol Hematol. 75:173–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bepler G, Sharma S, Gautam A, et al: Tumor
genotype, RRM1 expression and outcome of patients with lung cancer.
Eur J Cancer. 38(Suppl 7): 82(Abstr 265). 2003.
|
|
7
|
Vilmar A and Sorensen JB: Excision repair
cross-complementation group 1 (ERCC1) in platinum-based treatment
of non-small cell lung cancer with special emphasis on carboplatin:
a review of current literature. Lung Cancer. 64:131–139. 2009.
View Article : Google Scholar
|
|
8
|
Olaussen K, Dunant A, Fouret P, et al: DNA
repair by ERCC1 in non-small-cell lung cancer and cisplatin-based
adjuvant chemotherapy. N Engl J Med. 335:981–983. 2006.PubMed/NCBI
|
|
9
|
Ceppi P, Volante M, Novello S, et al:
ERCC1 and RRM1 gene expression but not EGFR are predictive of
shorter survival in advanced non-small-cell lung cancer treated
with cisplatin and gemcitabine. Ann Oncol. 17:1818–1825. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paz-Ares L, Souliers D, Melezinek I, et
al: Clinical outcome in non-small cell lung cancer patients with
EGFR mutation: pooled analysis. J Cell Mol Med. 14:51–69.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomized phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
12
|
Zheng Z, Chen T, Li X, et al: DNA
synthesis and repair genes RRM1 and ERCC1 in lung
cancer. N Engl J Med. 356:800–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bepler G, Olaussen KA, Vataire AL, et al:
ERCC1 and RRM1 in the international adjuvant lung trial by
automated quantitative in situ analysis. Am J Pathol.
178:69–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cobo M, Isla D, Massuti B, et al:
Customizing cisplatin based on quantitative excision repair
cross-complementing 1 mRNA expression: a phase III trial in
non-small-cell lung cancer. J Clin Oncol. 25:2747–2754. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simon GR, Schell MJ, Begum M, et al:
Preliminary indication of survival benefit from ERCC1 and
RRM1-tailored chemotherapy in patients with advanced
non-small cell lung cancer: evidence from an individual patient
analysis. Cancer. 118:2525–2531. 2012.PubMed/NCBI
|
|
16
|
McWhinney SR and McLeod HL: Using germline
genotype in cancer pharmacogenetic studies. Pharmacogenomics.
10:489–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bepler G, Zheng Z, Gautam A, et al:
Ribonucleotide reductase M1 gene promoter activity, polymorphisms,
population frequencies, and clinical relevance. Lung Cancer.
47:183–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang PM, Tzeng CH, Chen PM, et al:
ERCC1 codon 118 C→T polymorphism associated with
ERCC1 expression and outcome of FOLFOX-4 treatment in Asian
patients with metastatic colorectal carcinoma. Cancer Sci.
100:278–283. 2009. View Article : Google Scholar
|
|
19
|
Isla D, Sarries C, Rosell R, et al: Single
nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated
advanced non-small-cell lung cancer. Ann Oncol. 15:1194–1203. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tibaldi C, Giovannetti E, Vasile E, et al:
Correlation of CDA, ERCC1, and XPD polymorphisms with response and
survival in gemcitabine/cisplatin-treated advanced non-small cell
lung cancer patients. Clin Cancer Res. 14:1797–1803. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ryu JS, Hong YC, Han HS, et al: Associated
between polymorphisms of ERCC1 and XPD and survival in
non-small-cell lung cancer patients treated with cisplatin
combination chemotherapy. Lung Cancer. 44:311–316. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su D, Ma S, Liu P, et al: Genetic
polymorphisms and treatment response in advanced non-small cell
lung cancer. Lung Cancer. 56:281–288. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krawczyk P, Wojas-Krawczyk K, Mlak R, et
al: Predictive value of ERCC1 single-nucleotide polymorphism in
patients receiving platinum-based chemotherapy for locally-advanced
and advanced non-small cell lung cancer: a pilot study. Folia
Histochem Cytobiol. 50:80–86. 2012. View Article : Google Scholar
|
|
24
|
Ren S, Zhou S, Wu F, et al: Association
between polymorphisms of DNA repair genes and survival of advanced
NSCLC patients treated with platinum-based chemotherapy. Lung
Cancer. 75:102–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kalikaki A, Kanaki M, Vassalou H, et al:
DNA repair gene polymorphisms predict favorable clinical outcome in
advanced non-small-cell lung cancer. Clin Lung Cancer. 10:118–123.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu D, Shi J, Sun T, et al: Pharmacogenetic
role of ERCC1 genetic variants in treatment response of
platinum-based chemotherapy among advanced non-small cell lung
cancer patients. Tumour Bio. 33:877–884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takenaka T, Yano T, Kiyohara C, et al:
Effects of excision repair cross-complementation group 1 (ERCC1)
single nucleotide polymorphisms on the prognosis of non-small cell
lung cancer patients. Lung Cancer. 67:101–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ludovini V, Floriani I, Pistola L, et al:
Association of cytidine deaminase and xeroderma pigmentosum group D
polymorphisms with response, toxicity, and survival in
cisplatin/gemcitabine-treated advanced non-small cell lung cancer
patients. J Thorac Oncol. 6:2018–2026. 2011. View Article : Google Scholar
|
|
29
|
Liao WY, Shih JY, Chang GC, et al: Genetic
polymorphism of XRCC1 Arg399Gln is associated with survival in
non-small-cell lung cancer patients treated with
gemcitabine/platinum. J Thorac Oncol. 7:973–981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng JF, Wu JZ, Hu SN, et al:
Polymorphisms of the ribonucleotide reductase M1 gene and
sensitivity to platin-based chemotherapy in non-small cell lung
cancer. Lung Cancer. 66:344–349. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song D, Ai-Lin G, Zhi-Hong Ch, et al: RRM1
single nucleotide polymorphism −37C>A correlates with
progression-free survival in NSCLC patients after gemcitabine-based
chemotherapy. J Hematol Oncol. 3:102010.
|