Introduction
Human mesenchymal stem cells (hMSCs) are an
important type of adult stem cell and have multiple differentiation
activities and high self-renewal potential. The ease of isolation
and availability of hMSCs make them attractive tools for potential
use in cell transplantation therapy and tissue repair (1,2).
However, the mechanism of the self-renewal mechanisms and multiple
differentiation capabilities of hMSCs have not been fully
elucidated, thus limiting the use of these powerful tools.
Recently, many research groups have investigated the
regulation of hMSC proliferation and differentiation, and several
important regulatory pathways have been revealed. For instance, the
role of the Wnt (wingless) signaling pathway on the fate of hMSCs
has been established. However, the effects of Wnt signaling can
induce different or even opposite biological functions (1,3). In
the differentiation process of hMSCs, the role of Wnt signaling is
dependent on the level of the signals, and high levels of Wnts
promote osteogenesis of hMSCs, whereas low levels inhibit
osteogenesis (4). Thus, the complex
effect of Wnt signaling is context-dependent and is closely related
to its target genes (3).
Furthermore, Shtutman et al(5) reported that the promyelocytic leukemia
(PML) gene can function as a target of the Wnt signaling pathway,
and its gene product is involved in the regulation of various cell
functions. Thus, the role of the PML protein in hMSCs aroused our
interest.
PML, as an important tumor-suppressor, was initially
discovered in patients with acute promyelocytic leukemia (APL)
(6). In normal cells, PML is an
important regulator of numerous fundamental processes, such as
apoptosis, cellular proliferation, senescence and cell cycle
regulation. Recently, PML was found to regulate neural progenitor
cells (NPCs), leukemia-initiating cells (LICs) and hematopoietic
stem cells (HSCs) (7–9). Loss of PML increases the number of
NPCs and can impair their differentiation. PML is highly expressed
in HSCs and is responsible for normal cellular function and
maintenance, while its deficiency results in progressive impairment
of cell long-term repopulating capacity (9). However, the role of PML in regulating
hMSCs remains unclear.
In the present study, we investigated the regulation
of PML in hMSC proliferation and the progression of osteoblast
differentiation. The possible underlying mechanisms involved in the
regulation of proliferation and differentiation of hMSCs were also
explored.
Materials and methods
Cell isolation and culture
The present study was approved by the Ethics
Committee of our hospital, and informed consent was obtained from
each patient prior to obtaining bone marrow samples. hMSCs were
obtained from the bone marrow of patients undergoing total hip
replacement surgeries. The mononuclear cells were then isolated by
density gradient centrifugation. Cells were maintained in
low-glucose Dulbecco’s modified Eagle’s medium (L-DMEM)
supplemented with 10% fetal bovine serum (FBS) (both from
Gibco-BRL, Carlsbad, CA, USA), which was replaced every 3 days.
When cells reached 70–80% confluence, they were passaged by
trypsinization. All cells were grown in a monolayer culture in a
humidified incubator containing 5% CO2 at 37°C. Flow
cytometry was used to identify the phenotype of the hMSCs, and
those in the third passage from the same batch were used in the
following experiments. Every experiment was repeated using 3
different hMSCs strains isolated from 3 different randomly chosen
samples.
Cell proliferation analysis
Cell viability was determined using the Cell
Counting Kit-8 assay (CCK-8; Dojin, Tokyo, Japan). Briefly,
cultured cells were plated at a density of 3,000 cells/well in
96-well plates and incubated at 37°C in a humidified incubator
containing 5% CO2 for 0, 1, 3, 5, or 7 days. At the
indicated time points, cells were incubated with 20 μl of CCK-8
solution for 2 h at 37°C, and the absorbance at 490 nm was measured
using a microplate reader (Bio-Rad Laboratories, Tokyo, Japan).
Three contiguous wells were used for each testing group.
Flow cytometric analysis of the cell
cycle and apoptosis
After the designated treatments, the adherent and
supernatant cells were harvested. For cell cycle analysis, the
cells were harvested, washed with phosphate-buffered saline (PBS)
containing 0.1% bovine serum albumin (BSA), fixed with 70% ethanol
for 2 h, and washed twice with PBS. Then cells were stained with
0.5 ml PI/RNase staining buffer for 15 min at room temperature (BD
Biosciences, San Diego, CA, USA). Apoptosis analysis was performed
using the BD Pharmingen™ PE Annexin V Apoptosis Detection kit (BD
Biosciences). Briefly, the cells were washed and re-suspended in
500 μl of binding buffer, then incubated with 5 μl of Annexin V-PE
and 5 μl of 7-amino-actinomycin D (7-AAD) solution for 15 min in
the dark at room temperature. The DNA content and cell apoptosis
were then analyzed by flow cytometry (FC500; Beckman Coulter, Inc.,
Brea, CA, USA). Three contiguous wells were used for each testing
group.
Induction of osteogenic
differentiation
For osteogenic differentiation, cells were plated in
6-well plates at a density of 50,000 cells/well. When the cells
reached 70% confluence, they were treated with or without
osteogenic supplements (10 nM dexamethasone, 10 mM
β-glycerophosphate and 50 μg/ml L-ascorbic acid-2-phosphate; all
from Sigma-Aldrich, Poole, UK) for 7 or 14 days. The medium was
replaced every 3 days. Cells were verified by von Kossa staining
and alkaline phosphatase (ALP) activity analysis. ALP activities
were normalized to total protein content using the BCA protein
assay kit (Beyotime, Jiangsu, China) and were expressed as units/mg
protein. Three contiguous wells were used for each testing
group.
Immunofluorescence staining
hMSCs at passage 3 were seeded on glass coverslips
in 6-well plates at a density of 3×104/well. For
immunofluorescence staining, the coverslips were removed and washed
twice with PBS, fixed with 4% paraformaldehyde for 15 min, and then
permeabilized with 0.2% Triton X-100 for 10 min at room
temperature. Next, the cells were washed 3 times with PBS, blocked
with 10% BSA at room temperature for 60 min, and then incubated at
4°C overnight with the rabbit anti-PML polyclonal antibody (1:500
dilution; Abcam, Hong Kong). The cells were then washed 3 times
with PBS and incubated for 30 min at room temperature with
fluorescein isothiocyanate-conjugated anti-rabbit antibodies
(1:1,000 dilution; Boster Biological Technology, Wuhan, China). The
cells were washed 3 times, stained with
4′6′-diamidino-2-phenylindole (DAPI) (10 μg/ml) for 3 min and then
photographed via fluorescence microscopy.
Lentiviral vectors and hMSC
transduction
Recombinant lentiviral vector plenti6.3-PML-EGFP
which encodes the full-length human PML-cDNA (NM_002675) and the
enhanced green fluorescent protein (EGFP) was synthesized by
Invitrogen Life Technologies (Shanghai, China). The empty vector,
plenti6.3-EGFP, was used as the control. The lentiviruses were
produced as described previously. Briefly, human embryonic kidney
293T cells were used as packaging cells. One day before
transfection, the 293T cells were seeded at a density of
5×106 cells/10-cm dish and cultured to 90–95%
confluence. Plenti6.3-PML-EGFP or control vectors (3 μg), together
with packaging plasmids (9 μg; pLP1, pLP2 and pLP/VSVG), were
cotransfected into 293T cells using Lipofectamine™ 2000 (both from
Invitrogen Life Technologies) according to the manufacturer’s
instructions. After 48 h, the cell culture supernatant was
collected and passed through a 0.45-μm filter. When the hMSCs
reached 50–60% confluence, they were transfected with either the
PML or control lentivirus at a variety of multiplicity of
infections (MOIs) ranging from 0–100. After incubation for 72 h,
the transfection efficiency was estimated by evaluation of EGFP
expression via fluorescence microscopy, and PML gene expression was
verified by quantitative real-time PCR (qRT-PCR) and western blot
analysis.
Short hairpin RNA (shRNA) preparation and
gene transfection
Four different shRNAs against PML in a lentiviral
vector with green fluorescent protein and the control vector were
designed and synthesized by GenePharma Inc. (Shanghai, China).
Prior to lentivirus-mediated knockdown of PML in hMSCs, the
inhibitory activity of the shRNAs was tested in 293T cells, and the
most effective sequence was used for transfection into the hMSCs.
As described earlier, high-titer lentivirus was produced in 293T
cells by transfection of the lentiviral expression vector and
packaging vectors. The lentivirus was harvested after 48 h,
filtrated and was used to infect hMSCs which had reached 50–60%
confluence. After transfection for 72 h, the efficiency was
estimated by evaluation of EGFP expression via fluorescence
microscopy, and PML gene expression was verified by qRT-PCR and
western blot analysis. The DNA sequences of the PML shRNA was
5′-TGGAGGAG GGTTCCAGTTTCTTTCAAGAGAAGAAACTGGAACTC CTCCTCCTTTTTTC-3′
and 5′-TCGAGAAAAAAGGAGG AGGAGTTCCAGTTTCTTCTCTTGAAAGAAACTGGAA
CTCCTCCTCCA-3′. The control shRNA was 5′-TGTTCTCC
GAACGTGTCACGTTTCAAGAGAACATGACACGTTCG GAGAACTTTTTTC-3′ and
5′-TCGAGAAAAAAGTTCTC CGAACGTGTCACGTTCTCTTGAAACGTGACACGTTC
GGAGAACA-3′.
Reverse transcription and real-time PCR
analysis
Total cellular RNA was extracted using TRIzol
reagent (Invitrogen Life Technologies), and 1 μg RNA was
reverse-transcribed to cDNA using Moloney murine leukemia virus
(MMLV) reverse transcriptase (Fermentas-Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Qualitative analysis of RNA expression was
carried out by reverse transcription-polymerase chain reaction
(RT-PCR) using β-actin mRNA as the internal control. cDNA was
amplified in a final reaction volume of 20 μl using a Takara Taq™
kit. The amplification consisted of 35 cycles for 30 sec at 95°C,
45 sec at 59°C and 1 min at 72°C. Then, the PCR products were
electrophoresed on a 1.5% agarose gel containing ethidium bromide
and visualized by UV absorption.
Quantitative analysis of RNA expression was carried
out by qRT-PCR. The cDNA amplification was performed in a total
volume of 20 μl in 40 cycles of 95°C for 5 sec, and 59°C for 20 sec
using the LightCycler RT-PCR system (Roche, Mannheim, Germany)
according to the manufacturer’s instructions. Data were analyzed
using the relative standard curve method, and each sample was
normalized to β-actin to correct differences in RNA quality and
quantity. In each experiment, every sample was assayed in
triplicate, and each experiment was performed 3 times. The forward
and reverse primers for amplification of PML, ALP, IBSP, and
β-actin are as follows: PML forward, 5′-TGTACCG GCAGATTGTGGAT-3′
and reverse, 5′-AGATGTTGTTGGT CTTGCGG-3′; ALP forward,
5′-GACCATTCCCACGTCTT CACATT-3′ and reverse, 5′-CAGACTGCGCCTGGTAGTT
GT-3′; IBSP forward, 5′-CCAGAGGAAGCAATCACCA AA-3′ and reverse,
5′-TTGAGAAAGCACAGGCCATTC-3′; β-actin forward,
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse,
5′-GGGCACGAAGGCTCATCATT-3′.
Western blot analysis
hMSCs proteins were lysed with
radioimmunoprecipitation assay (RIPA) buffer and proteinase
inhibitors (Beyotime). Protein concentrations were determined using
the bicinchoninic acid protein assay with BSA as a standard
(Beyotime). Equal amounts of cell proteins (30 μg) were subjected
to 10% SDS-polyacrylamide gel electrophoresis and
electrotransferred onto polyvinylidene difluoride (PVDF) membranes,
which were blocked with 5% phosphatase-free powdered milk in
Tris-buffered saline with 0.1% Tween (TBST) for 2 h. Then, the
membranes were incubated overnight at 4°C with the primary
antibodies consisting of PML (Abcam), caspase-3, poly(ADP-ribose)
polymerase (PARP) and β-actin antibody (Cell Signaling Technology,
Inc.). The membranes were then rinsed with TBST and incubated with
horseradish peroxidase-conjugated goat anti-mouse or goat
anti-rabbit immunoglobulin G antibody (Abcam) for 1.5 h. The
membranes were exposed to X-ray film, and the density of each band
was quantified using ImageJ software (National Institutes of
Health). Expression of the protein of interest was normalized to
β-actin expression in the same lane. All experiments were repeated
at least 3 times and each yielded similar results.
Microarray and data analyses
The microarray analyses were performed by Kangchen
Biotech Co., Ltd. (Shanghai, China) using standard protocols.
Briefly, when PML was overexpressed for 3 days, total RNA from
normal, mock-, or PML-transfected hMSCs was isolated using TRIzol
reagent. Double-stranded cDNA was synthesized from total RNA using
the SupersScript® Double-Stranded cDNA Synthesis kit
(Invitrogen Life Technologies) and labeled in accordance with the
NimbleGen Gene Expression Analysis protocol (Roche NimbleGen
Systems, Inc., Madison, WI, USA).
An Axon GenePix 4000B scanner (Molecular Devices,
Inc., Downingtown, PA, USA) and NimbleScan software (version 2.5)
was used to analyze the expression data, which were normalized
through quantile normalization using the Robust Multichip average
algorithm included in the NimbleScan software. All gene level files
were imported into GeneSpring GX software (version 11.5.1; Agilent
Technologies, Inc., Santa Clara, CA, USA) for further analysis.
Differentially expressed genes with statistical significance were
identified through volcano plot filtering and hierarchical
clustering using the Agilent GeneSpring GX software.
Statistical analysis
All statistical analyses were performed using SPSS
software (SPSS, Inc., Chicago, IL, USA), and all data are expressed
as means ± standard deviation. Differences between groups were
evaluated using one-way analyses of variance (ANOVA), and a P-value
<0.05 was considered to indicate a statistically significant
result.
Results
Phenotypes of the purified bone marrow
cells
The hMSCs exhibited typical spindle shapes, and
significant colonies were observed on day 7 of the primary culture
(Fig. 1A). After reaching
confluence, the cells were arranged in various orientations and
exhibited vortex-like growth (Fig.
1B). The hMSCs phenotypes were identified by flow cytometry
which demonstrated that they were uniformly positive for cluster of
differentiation CD29, CD44 and CD166, but negative for CD34, CD45
and human leukocyte antigen DR (data not shown), consistent with
the cytobiological characteristics reported in our previous studies
and others (10–12). Isolated hMSCs were successfully
transformed into osteoblasts, as indicated by positive von Kossa
staining after 14 days of induction (Fig. 1C).
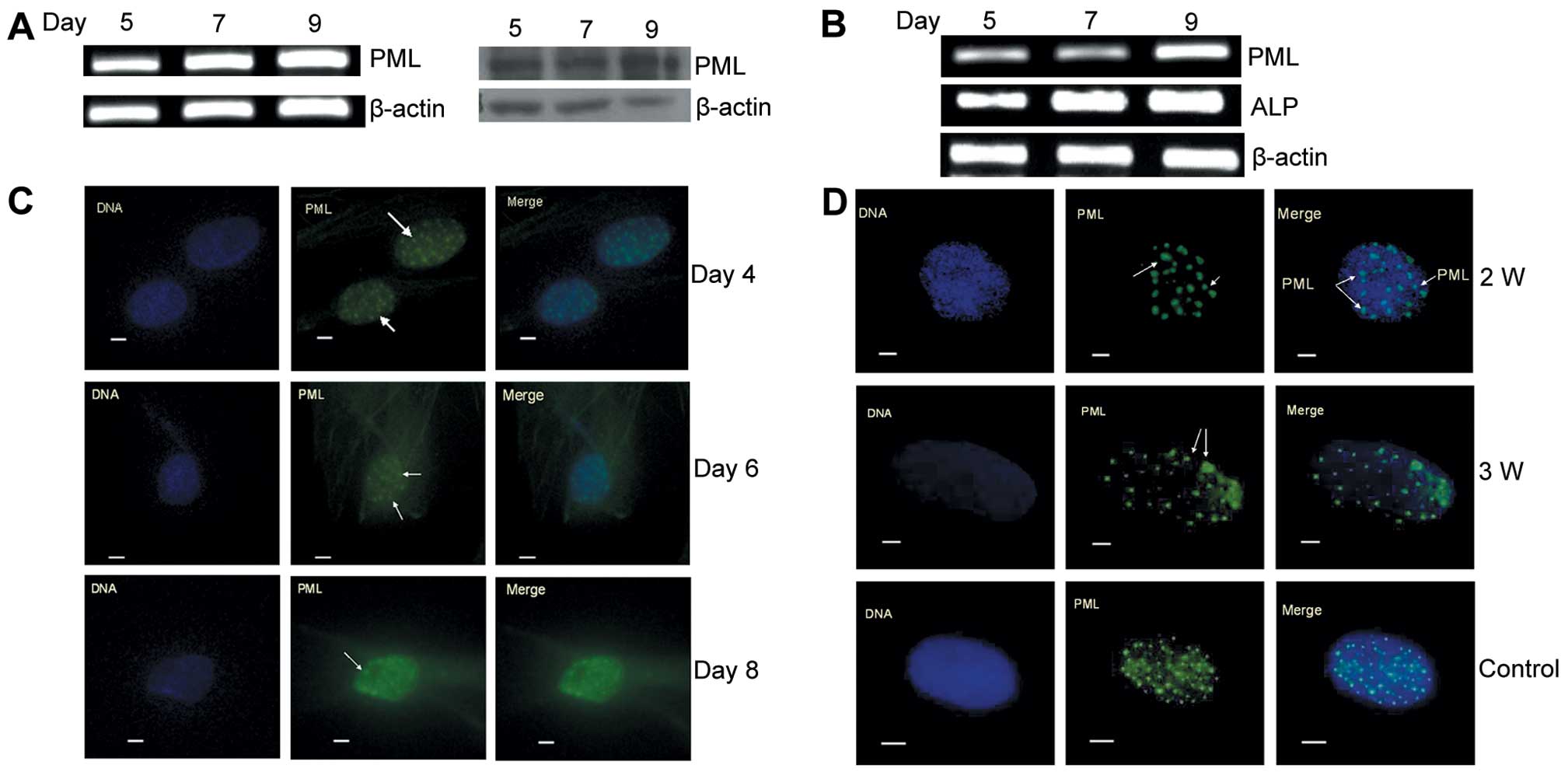
PML is stably expressed in hMSCs and
expression is increased time-dependently along with cell osteogenic
differentiation
PML was positively expressed in the hMSCs, and both
mRNA and protein concentrations were detectable over 9 days along
with cell proliferation (Fig. 2A).
The induction of hMSCs to differentiate into osteoblasts increased
the expression of PML, which was consistent with the upregulation
of ALP mRNA in a time-dependent manner (Fig. 2B). The PML protein epitomizes the
PML-nuclear body (PML-NB), which is involved in the regulation of
cell function, and the relative size of PML-NBs can change
depending on cell status. PML-NBs in hMSCs were not significantly
altered along with cell proliferation as detected by
immunofluorescence staining on day 4, 6 and 8 (Fig. 2C). However, when cells were induced
to differentiate into osteoblasts for 2 and 3 weeks, the PML-NBs
became larger and unequal in size when compared to those of normal
cells (Fig. 2D). Our preliminary
results showed that PML was stably expressed in hMSCs and was
associated with osteogenic differentiation of hMSCs.
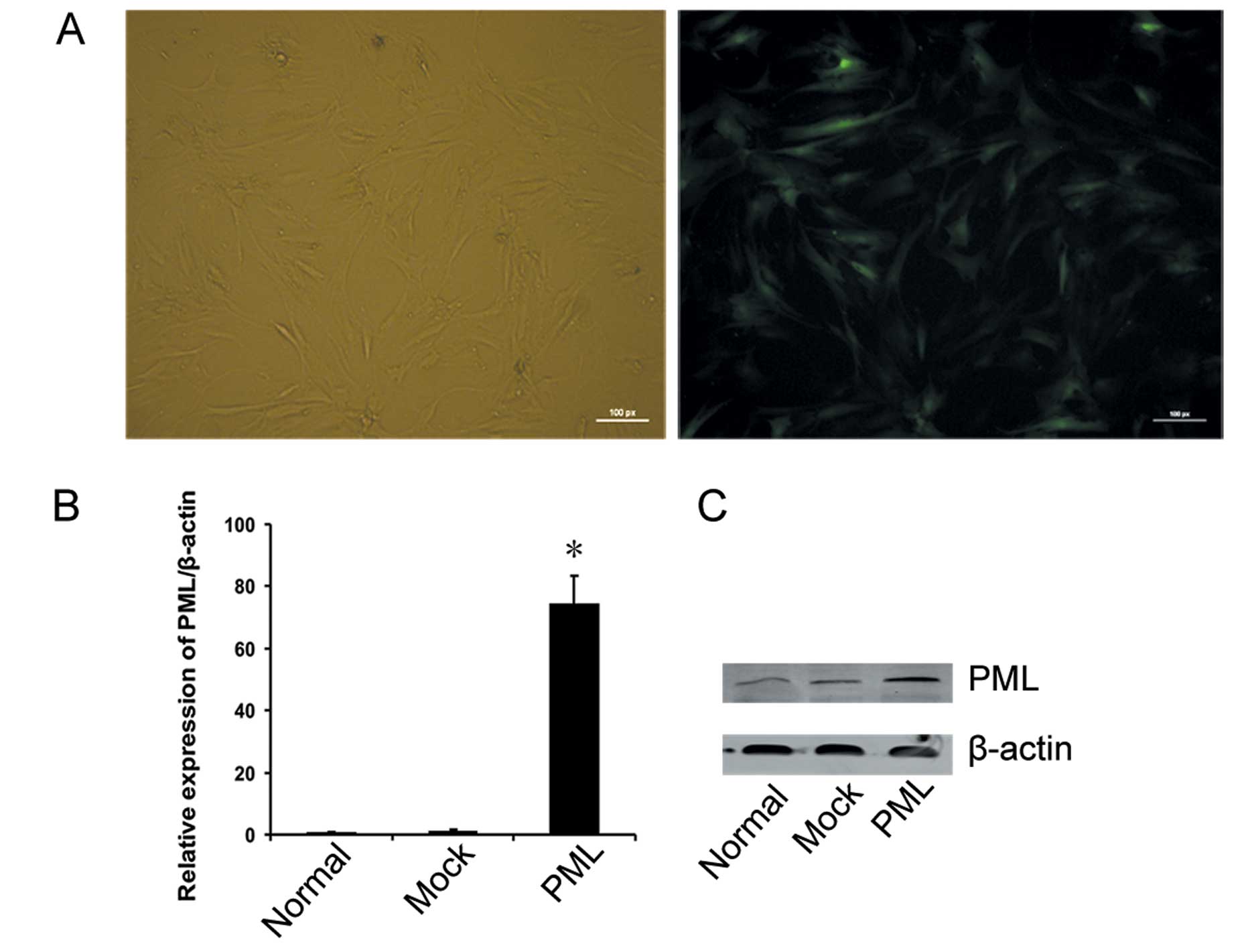
PML overexpression in hMSCs by stable
transduction
In order to determine the impact of PML on hMSCs
proliferation and differentiation, a lentiviral vector encoding
full-length human PML cDNA was transfected into hMSCs. A vacant
vector was transfected as a negative control, and non-transfected
hMSCs served as normal controls. At an MOI of 40, >80% of the
hMSCs were EGFP-positive on day 3, as assessed by fluorescence
microscopy (Fig. 3A), and flow
cytometry. At the same time, PML expression in transfected hMSCs
was detected by qRT-PCR and western blotting. As shown in Fig. 3B, PML RNA expression was
significantly increased in PML-transfected cells when compared with
that in the untreated and mock-transfected cells, and the PML
protein was readily detected in the PML-transfected cells, but was
very low in the untreated and mock-transfected cells, respectively
(Fig. 3C).
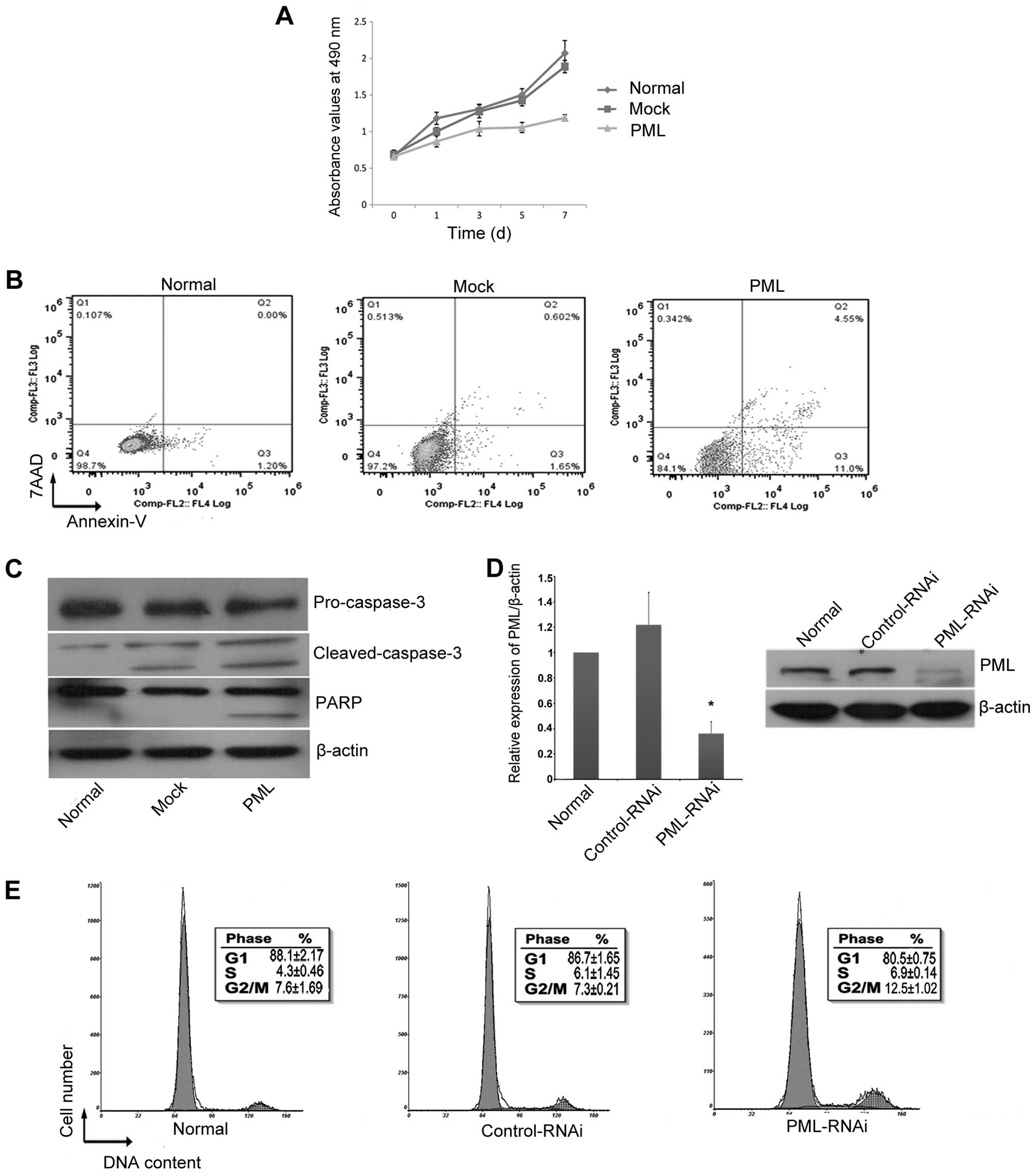
PML overexpression inhibits hMSCs
proliferation
PML overexpression in hMSCs induced a significant
decrease in cell proliferation as indicated by the change in
absorbance from day 0 to day 7 (Fig.
4A). Cell proliferation was inhibited by 20.57±4.18% in hMSCs
overexpressing PML as compared to that of the normal controls
(P<0.05) and by 18.46±3.53% when compared to the
mock-transfected cells on day 3 (P<0.05). PML overexpression
also significantly increased cell apoptosis. After 3 days of PML
overexpression, the percentage of early apoptotic cells in the
PML-transfected cells was 10.3±4.23%, which was significantly
higher than that in the mock-transfected cells (2.53±0.77%)
(P<0.05) and normal control cells (2.18±1.88%) (P<0.05)
(Fig. 4B). Western blot analysis
showed that this process occurred concomitantly with a decrease in
production of the apoptosis marker proteins pro-caspase-3 and PARP,
and an increase in production of cleaved caspase-3 (Fig. 4C). There were no obvious changes to
the cell cycle (data not shown) when PML expression was enhanced.
However, when the expression of PML was knocked down for 3 days
(Fig. 4D), the proportion of hMSCs
in the S/G2+M phases was clearly increased (Fig. 4E).
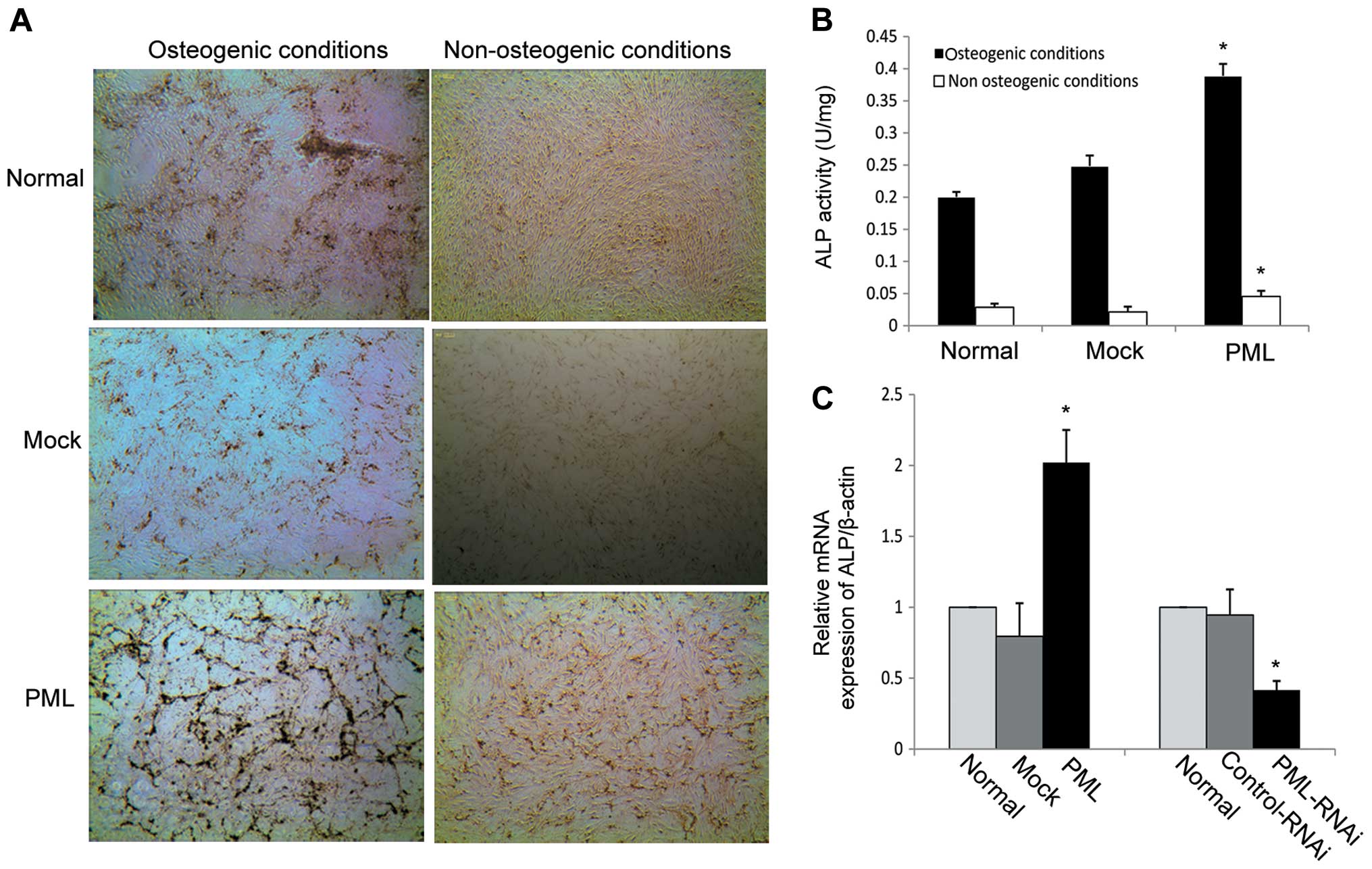
PML overexpression promotes osteogenic
differentiation of hMSCs
To evaluate the effects of PML overexpression on the
osteogenic differentiation of hMSCs, PML-transfected cells were
induced to osteogenic differentiation, while normal and
mock-transfected cells served as controls. von Kossa staining and
ALP activity were used to evaluate the degree of osteogenic
differentiation. As shown in Fig.
5A, mineralized matrix production was strongly enhanced on day
7 in the PML-overexpressing hMSCs under osteogenic differentiation
conditions and the ALP activity was obviously increased in
comparison to the mock-transfected and normal MSCs (0.39±0.019 vs.
0.24±0.016 and 0.2±0.008 U/mg, respectively) (P<0.05 for both)
(Fig. 5B). Next, calcium deposition
and ALP activity were detected in PML-transfected cells under
non-osteogenic differentiation conditions (DMEM-LG containing 10%
FBS). We found that PML-overexpressing hMSCs exhibited an obvious
increase in mineralized matrix production on day 7 when compared
with that in the normal and mock-transfected cells (Fig. 5A). Under normal culture conditions
for 7 days, ALP activity was also obviously increased in comparison
to the mock-transfected and normal MSCs (0.049±0.006 vs.
0.024±0.005 and 0.028±0.005 U/mg, respectively) (P<0.05 for
both) (Fig. 5B). Next, alterations
in ALP mRNA expression were investigated by RT-PCR. mRNA expression
of ALP as an early marker of osteoblastic differentiation, was
dramatically enhanced on day 7 in PML-overexpressing hMSCs.
Knockdown of PML expression decreased ALP mRNA expression when the
cells were cultured under normal conditions for 7 days (Fig. 5C). Together, these findings suggest
that PML promotes the osteogenic differentiation potential of hMSCs
even under non-osteogenic differentiation conditions.
Typical gene expression from the
microarray and confirmation
To explore the potential mechanism of PML in the
regulation of hMSC proliferation and differentiation, we
investigated genome-wide expression profiles of the normal, mock-
and PML-transfected hMSCs. The microarray employed for the present
analysis consisted of 45,033 genes that were collected from genetic
databases, including that from the National Center for
Biotechnology Information. After performing t-test analyses, a
total of 357 genes were found to have significantly altered
expression levels of >1.5-fold (P<0.05), of which 222 were
upregulated and 135 were downregulated in the PML-transfected hMSCs
when compared to the normal and mock-transfected cells. In the
PML-transfected cells, the 3 most upregulated genes were
interleukin (IL)-1β, IL-8 and IBSP, while the most downregulated
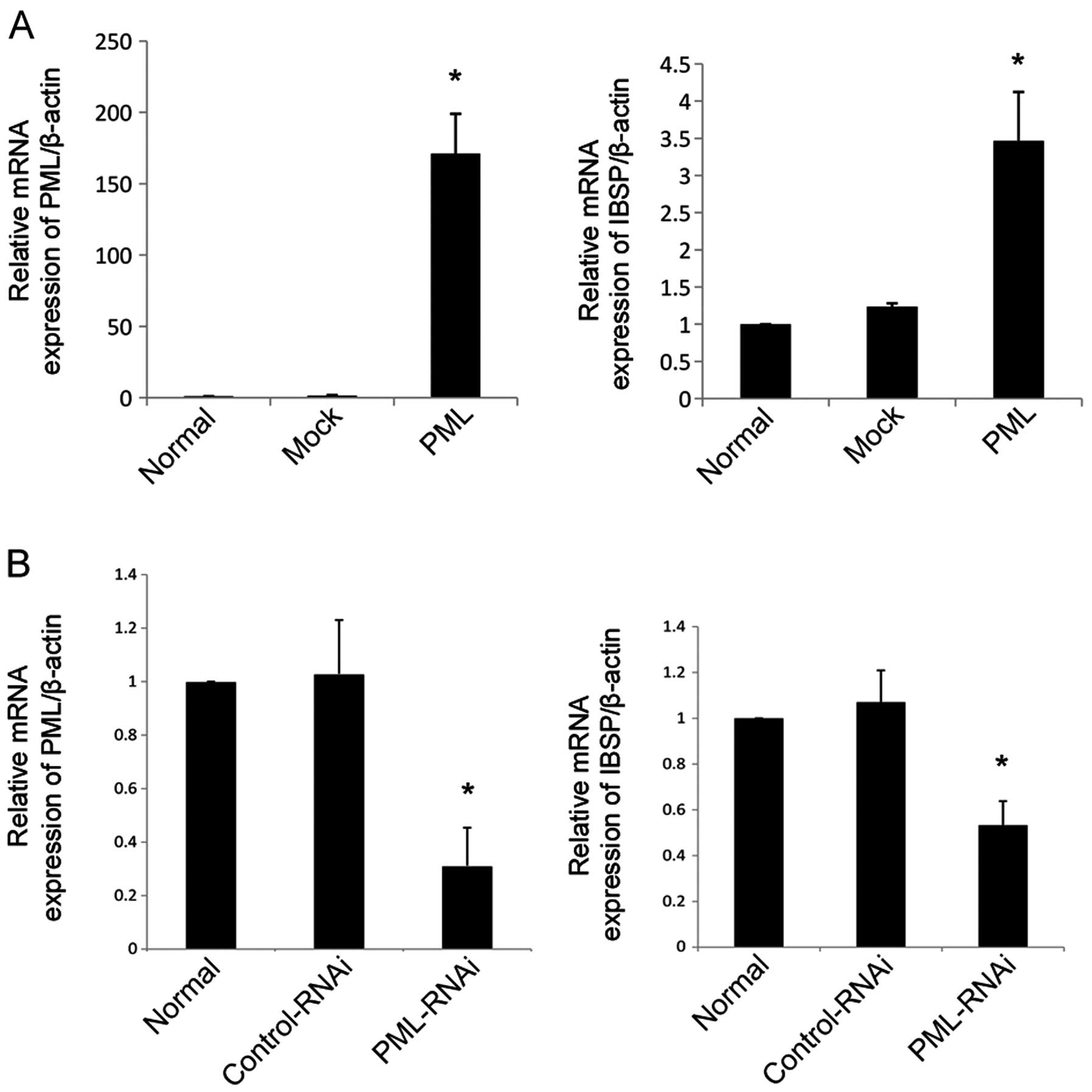
gene was fibroblast growth factor receptor 2 (FGFR2) (Table I). IBSP, as an important marker of
osteogenic differentiation, plays a key role in the regulation of
osteogenic differentiation. Next, we validated the IBSP expression
levels by qRT-PCR. Previous studies have shown that IBSP is
detectable in the early stage of osteogenesis (13). With the increased expression of PML
for 3 days, hMSCs cultured in normal conditions (DMEM-LG containing
10% FBS) exhibited strong upregulation of IBSP. Moreover, IBSP
expression was reduced when the PML gene was knocked down for 3
days as compared to the mock-transfected and normal cells
(P<0.05 for all) (Fig. 6).
 | Table IThe most differentially expressed
genes between the mock and PML-transfected hMSCs as detected by
genome-wide microarray. |
Table I
The most differentially expressed
genes between the mock and PML-transfected hMSCs as detected by
genome-wide microarray.
| | | Normalized intensity
of each group | |
|---|
| | |
| |
|---|
| Accession no. | Gene name | Regulation | Mock | PML | P-value |
|---|
| NM-022971 | FGFR2 | Downregulation | 280.16 | 97.79 | 0.014 |
| NM-022975 | FGFR2 | Downregulation | 117.37 | 51.71 | 0.017 |
| NM-022976 | FGFR2 | Downregulation | 596.47 | 307.03 | 0.045 |
| NM-000576 | IL-1B | Upregulation | 47.05 | 139.58 | 0.046 |
| NM-000584 | IL-8 | Upregulation | 67.36 | 211.69 | 0.005 |
| J05213 | IBSP | Upregulation | 90.37 | 267.03 | 0.012 |
Discussion
In the present study, we investigated the effect of
PML in hMSCs and found that PML inhibits cell proliferation, yet
promotes the osteogenic differentiation of hMSCs.
To date, the biochemical and molecular functions of
PML have been studied extensively, and the roles of PML as a
tumor-suppressor in cell proliferation, differentiation, and
apoptosis have been shown in many types of tumor cells (14,15).
In cancer cells, enhanced PML expression can arrest the cell cycle
in the G1 phase (15), and PML
overexpression has been shown to induce tumor cell apoptosis via
the caspase-dependent pathway (16). Recently, several reports have
demonstrated an unexpected and critical role of PML in stem cell
and progenitor cell biology. PML is required for HSCs maintenance
and NPCs differentiation (7,9). Yet,
the role of PML in hMSCs remains unknown.
In the present study, we found that PML was
expressed in hMSCs, and the expression was increased
time-dependently along with cell osteogenic differentiation. To
further investigate the role of PML in hMSCs proliferation and
osteogenic differentiation, we established an hMSCs cell line that
stably overexpressed PML. PML, as a key tumor-suppressor, can
suppress cell growth in many types of cancer cells via a
caspase-associated pathway (17).
In hMSCs, enhanced PML expression also decreased cell proliferation
by inducing caspase-associated apoptosis, which is in accordance
with previous studies in tumor cells (18,19).
The PML-induced hMSCs apoptosis was verified by the decreased
expression of apoptosis-associated proteins, such as pro-caspase-3
and PARP, and by an increased production of cleaved caspase-3 as
detected by western blot analysis. Cell apoptosis is strongly
dependent on cell cycle arrest; however, PML overexpression induced
no significant changes to the cell cycle. One possible explanation
for this phenomenon is that hMSCs have different cell cycle
condition as compared to cancer cells. Cell cycle analysis using
hMSCs in normal culture showed that >90% of cells were in the
G0/G1 phase (20); thus it is
difficult to detect cell cycle arrest in hMSCs. Nevertheless, when
PML expression was inhibited by RNAi, the proportion of hMSCs in
the S/G2 phase of the cell cycle had clearly increased. In other
words, loss of PML expression was in favor of cell proliferation by
promoting cell cycle progression. These results imply that a stable
and a certain level of expression of PML plays an important role in
maintaining the normal proliferation capacity of hMSCs. When the
expression of PML is altered, proliferation of hMSCs is
affected.
Calcium deposition is a major marker of osteogenic
differentiated hMSCs and is positively correlated with the degree
of differentiation. Under osteogenic differentiation conditions,
hMSC calcium deposition becomes evident after 7 days (21), as was indicated by von Kossa
staining, which is an important tool to examine mineralization
in vitro(22). In our study,
mineralization and matrix production of hMSCs cultured in
osteogenic differentiation medium were increased in comparison to
those cultured under non-osteogenic differentiation conditions by
day 7. When PML was overexpressed, hMSCs under an osteogenic
differentiation condition caused a dramatic increase in calcium
deposition when compared to the normal and mock-transfected cells.
To determine whether the increased PML level causes osteogenic
differentiation, hMSCs were cultured under non-osteogenic
differentiation condition for 7 days. We also found a significant
calcium deposition in the PML-overexpressing cells, when compared
with the normal and mock-transfected cells. ALP protein activity is
another important marker of early osteogenetic progression
(23). We found that ALP protein
activity was enhanced by PML overexpression even when the cells
were cultured under a non-osteogenic growth condition and ALP
expression was coincidental with ALP activity. These results
indicated that the osteogenic differentiation potential of hMSCs
was enhanced by PML overexpression.
When evaluating the microarray expression data, we
found that IBSP was significantly increased in the PML-transfected
hMSCs, but no other osteogenic-related gene was noted. IBSP, as an
important osteogenic-related gene, is a member of the small
integrin-binding ligand N-linked glycoprotein (SIBLING) family and
is responsible for mineralization and matrix production (24,25).
Upregulation of IBSP gene expression is associated with osteoblast
differentiation. cAMP response element (CRE) is an important
promoter of the IBSP gene, which is required for gene
transcriptional activation (26).
The phosphorylation of CRE-binding protein (p-CREB) stimulates IBSP
gene expression. However activation of the IBSP gene by p-CREB
partly requires co-activation of the CREB-bind protein (CBP)
(27). PML, as the major
constituent of PML-NBs, has been proven to interact with CBP in
vitro and in vivo. CBP was co-localized with PML in
PML-NBs, and upregulation of PML protein has been shown to promote
the function of CBP as a potent coactivator (28). When PML was overexpressed in hMSCs,
the enhanced activity of CBP stimulated IBSP gene transcription. In
the present study, we proved that IBSP expression displayed the
same tendency with PML.
In addition, FGFR2 was the most highly downregulated
gene in the PML-transfected cells. The fibroblast growth factor
(FGF) family comprises 22 proteins and plays a key role in bone
formation, homeostasis, and repair through activation of the FGF
receptor (FGFR) (29), and the
expression of FGFR in hMSCs has been well acknowledged. Thus, in
hMSCs, FGFR signaling activation can increase cell proliferation
and is essential for cell differentiation (30,31).
However, the mechanisms of FGFR2 downregulation caused by PML
warrant further investigation.
In conclusion, the present study revealed a
previously unknown role of PML in the regulation of cell
proliferation and osteogenic differentiation of hMSCs. Briefly, PML
regulates hMSCs as an inhibitor of cell proliferation, but promotes
osteogenic differentiation, which is associated with the
upregulation of IBSP. Thus, our findings offer new insights into
the role of PML in hMSC proliferation and differentiation. Future
studies will enable us to better discern the functions of hMSCs in
order to broaden the spectrum of their applications.
Acknowledgements
We thank Shanghai KangChen Bio-Tech for their
excellent technical assistance with the microarray analysis. This
study was supported by the National Natural Science Foundation of
China (nos. 81170526, 81100364, 2009C14011), the Major Science, and
the Technology Program of Zhejiang Province (no. Z2100097).
References
|
1
|
Chen BY, Wang X, Chen LW and Luo ZJ:
Molecular targeting regulation of proliferation and differentiation
of the bone marrow-derived mesenchymal stem cells or mesenchymal
stromal cells. Curr Drug Targets. 13:561–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Satija NK, Gurudutta GU, Sharma S, et al:
Mesenchymal stem cells: molecular targets for tissue engineering.
Stem Cells Dev. 16:7–23. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ling L, Nurcombe V and Cool SM: Wnt
signaling controls the fate of mesenchymal stem cells. Gene.
433:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Boer J, Wang HJ and Van Blitterswijk C:
Effects of Wnt signaling on proliferation and differentiation of
human mesenchymal stem cells. Tissue Eng. 10:393–401.
2004.PubMed/NCBI
|
|
5
|
Shtutman M, Zhurinsky J, Oren M, Levina E
and Ben-Ze’ev A: PML is a target gene of β-catenin and
plakoglobin, and coactivates β-catenin-mediated transcription.
Cancer Res. 62:5947–5954. 2002.
|
|
6
|
Scaglioni PP, Yung TM, Cai LF, et al: A
CK2-dependent mechanism for degradation of the PML tumor
suppressor. Cell. 126:269–283. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Regad T, Bellodi C, Nicotera P and
Salomoni P: The tumor-suppressor Pml regulates cell fate in the
developing neocortex. Nat Neurosci. 12:132–140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salomoni P, Dvorkina M and Michod D: Role
of the promyelocytic leukaemia protein in cell death regulation.
Cell Death Dis. 3:e2472012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ito K, Bernardi R, Morotti A, et al: PML
targeting eradicates quiescent leukaemia-initiating cells. Nature.
453:1072–1078. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meuleman N, Tondreau T, Delforge A, et al:
Human marrow mesenchymal stem cell culture: serum-free medium
allows better expansion than classical α-MEM medium. Eur J
Haematol. 76:309–316. 2006.PubMed/NCBI
|
|
11
|
Liu L, Yu Q, Lin J, et al:
Hypoxia-inducible factor-1α is essential for hypoxia-induced
mesenchymal stem cell mobilization into the peripheral blood. Stem
Cells Dev. 20:1961–1971. 2011.
|
|
12
|
Zhang L, Zheng W, Wang Y and Huang H:
Human bone marrow mesenchymal stem cells support the derivation and
propagation of human induced pluripotent stem cells in culture.
Cell Reprogram. 15:216–223. 2013.PubMed/NCBI
|
|
13
|
Gay IC, Chen S and MacDougall M: Isolation
and characterization of multipotent human periodontal ligament stem
cells. Orthod Craniofac Res. 10:149–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferbeyre G, de Stanchina E, Querido E,
Baptiste N, Prives C and Lowe SW: PML is induced by oncogenic
ras and promotes premature senescence. Genes Dev.
14:2015–2027. 2000.
|
|
15
|
Salomoni P, Ferguson BJ, Wyllie AH and
Rich T: New insights into the role of PML in tumour suppression.
Cell Res. 18:622–640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang ZG, Ruggero D, Ronchetti S, et al:
PML is essential for multiple apoptotic pathways. Nat Genet.
20:266–272. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Lu Y, Huang W, et al: In vitro
effect of adenovirus-mediated human Gamma Interferon gene transfer
into human mesenchymal stem cells for chronic myelogenous leukemia.
Hematol Oncol. 24:151–158. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lallemand-Breitenbach V and de Thé H: PML
nuclear bodies. Cold Spring Harb Perspect Biol. 2:a0006612010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takahashi Y, Lallemand-Breitenbach V, Zhu
J and de Thé H: PML nuclear bodies and apoptosis. Oncogene.
23:2819–2824. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Conget PA and Minguell JJ: Phenotypical
and functional properties of human bone marrow mesenchymal
progenitor cells. J Cell Physiol. 181:67–73. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pittenger MF, Mackay AM, Beck SC, et al:
Multilineage potential of adult human mesenchymal stem cells.
Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
George J, Kuboki Y and Miyata T:
Differentiation of mesenchymal stem cells into osteoblasts on
honeycomb collagen scaffolds. Biotechnol Bioeng. 95:404–411. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ogawa R, Mizuno H, Watanabe A, Migita M,
Shimada T and Hyakusoku H: Osteogenic and chondrogenic
differentiation by adipose-derived stem cells harvested from GFP
transgenic mice. Biochem Biophys Res Commun. 313:871–877. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keselowsky BG, Collard DM and García AJ:
Integrin binding specificity regulates biomaterial surface
chemistry effects on cell differentiation. Proc Natl Acad Sci USA.
102:5953–5957. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ilmer M, Karow M, Geissler C, Jochum M and
Neth P: Human osteoblast-derived factors induce early osteogenic
markers in human mesenchymal stem cells. Tissue Eng Part A.
15:2397–2409. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shimizu E, Matsuda-Honjyo Y, Samoto H, et
al: Static magnetic fields-induced bone sialoprotein (BSP)
expression is mediated through FGF2 response element and
pituitary-specific transcription factor-1 motif. J Cell Biochem.
91:1183–1196. 2004. View Article : Google Scholar
|
|
27
|
Zhang X, Odom DT, Koo SH, et al:
Genome-wide analysis of cAMP-response element binding protein
occupancy, phosphorylation, and target gene activation in human
tissues. Proc Natl Acad Sci USA. 102:4459–4464. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Doucas V: The promyelocytic (PML) nuclear
compartment and transcription control. Biochem Pharmacol.
60:1197–1201. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marie PJ: Fibroblast growth factor
signaling controlling bone formation: an update. Gene. 498:1–4.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ryu E, Hong S, Kang J, et al:
Identification of senescence-associated genes in human bone marrow
mesenchymal stem cells. Biochem Biophys Res Commun. 371:431–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Coutu DL, François M and Galipeau J:
Inhibition of cellular senescence by developmentally regulated FGF
receptors in mesenchymal stem cells. Blood. 117:6801–6812. 2011.
View Article : Google Scholar : PubMed/NCBI
|