Introduction
Breast cancer, which accounts for 22.9% of all
cancer cases in women, caused 458,503 deaths worldwide in 2008
(13.7% of cancer-related deaths in women) (1). As the most frequently diagnosed
life-threatening cancer in women, breast cancer is a leading cause
of cancer-related mortality among women (2), although extremely rare but highly
lethal in men (1). Breast cancer
management has become increasingly complex, requiring the data
integration of the patient’s history and specific tumor biomarkers.
Targeted and biologic therapies for breast cancer continue to
evolve rapidly with the emergence of molecular-targeted therapy for
breast cancer. In recent years, there has been an increasing and
rapid development of molecular markers as targets for
molecular-targeted therapy. A large number of molecules have
already become therapeutic targets, such as growth factors and
growth factor receptors, molecules of signal transduction,
tumor-associated antigens, molecules of intracellular protein
metabolism, factors regulating cell survival, cell cycle and cell
death, and molecules associated with invasion, metastasis and
angiogenesis (3).
Ubiquitin is a small regulatory protein that is
found in almost all tissues of eukaryotic organisms.
Ubiquitination, the covalent attachment of ubiquitin to a target
protein, is a post-translational modification that regulates the
stability, function and/or localization of the modified proteins
(4,5). It is noteworthy that many proteins
studied by clinical breast cancer researchers are involved in these
ubiquitin pathways. Such proteins include cyclins, CDK inhibitors
and the SCF in cell cycle control; the breast and ovarian cancer
suppressor BRCA1-BARD1; ErbB2/HER2/Neu and its ubiquitin ligase
c-Cbl or CHIP (6). Ubiquitination
is a strictly regulated reversible process. Deubiquitinating
enzymes (DUBs) are proteases that cleave ubiquitin or
ubiquitin-like proteins from pro-proteins or target proteins. DUBs
reverse the ubiquitination or ubiquitin-like modification of target
proteins, playing an antagonistic role against ubiquitination
(7,8).
The human genome encodes nearly 100 DUBs with
specificity for ubiquitin in 5 families: the ubiquitin C-terminal
hydrolase (UCH), ubiquitin-specific protease (USP), the ovarian
tumor (OUT), Josephin and JAB1/MPN/Mov34 metalloenzyme (JAMM)
families (9). Four families are
cysteine proteases, while the latter is a family of
metalloproteases. Although a few substrates have been identified
for a handful of DUBs, the substrates and physiological role of
most DUBs are poorly defined. USP39, also known as 65-kDa
SR-related protein of the U4/U6·U5 tri-snRNP, has been implicated
in the assembly of the mature spliceosome-complex (10). USP39 is also a factor required to
maintain the spindle checkpoint and to support successful
cytokinesis. Consistent with its previously described role in mRNA
processing, depletion of USP39 leads to specific reduction in
Aurora B mRNA levels (11). In
addition, zebrafish USP39 mutation leads to rb1 splicing
defect and pituitary lineage expansion (12).
Our study on breast cancer found that the expression
levels of USP39 in breast cancer tissues were markedly higher in
contrast to normal breast tissues, indicating that USP39 may act as
an oncogenic factor in breast cancer. To test our hypothesis, we
knocked down the expression of USP39 in breast cancer cell line
MCF-7 by RNA interference (RNAi) technology and then investigated
the proliferation, colony formation capacity, cell cycle and
apoptosis of the cells. Our data revealed that the inhibition of
USP39 significantly decreased MCF-7 proliferation and colony
formation capacity, and induced G0/G1 arrest and apoptosis,
providing us with a future target for breast cancer therapy.
Materials and methods
Immunohistochemistry
USP39 expression was evaluated by means of
immunohistochemical analysis with a rabbit monoclonal anti-USP39
antibody. Briefly, tissue sections from breast cancer specimens
were incubated with a rabbit monoclonal anti-USP39 antibody
(ab131244; Abcam, Cambridge, MA, USA) at 4°C overnight. After being
washed, the expression of USP39 in tissue sections was detected
using a polymer detection system with polymer helper and
polyperoxidase-anti-mouse/rabbit IgG (PV-9000; Zhongshan Golden
Bridge Biotechnology Co., Beijing, China), followed by
counterstaining with Mayer’s hematoxylin. The percentage of
positive cells in a total of 10 fields from each slide was examined
and graded as 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), or 4
(>75%) by 2 pathologists. The intensity of the
immunohistochemical staining was graded as follows: 0 (no
staining), 1 (bright yellow), 2 (orange), or 3 (brown). The
staining was quantified according to the sum of the positive cells
and the intensity of the immunohistochemical staining as 0, 1–2,
3–4, or 5–7, which represent negative (−), weakly positive (+),
positive (++) or hadro-positive (+++), respectively.
Lentiviral vector production
Small interfering RNAs (siRNA) targeting the USP39
sequence (AAGGTTAAGGTGAGCTCA TCG) and the non-silencing sequence
(AATTCTCCGAACG TGTCACGT) were transformed into short hairpin RNA
(shRNA) (stem-loop-stem structure) and were cloned into the
pGCSIL-GFP lentiviral vector (GeneChem Co. Ltd., Shanghai, China)
with AgeI/EcoRI sites. Then, the recombined
pGCSIL-GFP vector and two-helper vector system (GeneChem) were
transfected into the human embryonic kidney cell line 293T via
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) to generate the
lentivirus. After 3 days of incubation, the lentivirus from the
culture medium was collected and concentrated with Centricon
Plus-20 (Millipore, Billerica, MA, USA).
Cell culture and infection
The human breast cancer MCF-7 cell line and the
human embryonic kidney cell line 293T were purchased from the
American Type Culture Collection. The MCF-7 cell line was cultured
in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL) containing
4 mM L-glutamine, 0.1 mM MEM non-essential amino acid solution
(Sigma-Aldrich) and 10% fetal bovine serum (FBS; Gibco-BRL) at 37°C
in 5% CO2. The 293T cell line was cultured in DMEM
supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml
streptomycin (Sigma-Aldrich) at 37°C in 5% CO2. For
lentivirus infection, MCF-7 cells were cultured in 6-well plates,
and the USP39 shRNA-expressing lentivirus (USP39-shRNA) or
nontargeting shRNA-expressing lentivirus (control) was then added,
with a multiplicity of infection (MOI) of 10. After 5 days of
infection, cells were observed under fluorescence microscopy
(DMI4000B; Leica Microsystems, Germany).
Quantitative real-time PCR (qRT-PCR)
Total RNAs were prepared from cells using TRIzol
reagent (Invitrogen). The reverse transcription reactions were
carried out following the manufacturer’s protocol (Promega), and
each reverse transcription reaction mixture contained 2 μg total
RNA. Then, 25 μl of qRT-PCR mixtures containing 0.1 μM primers, 10
μl 2X SYBR Premix Ex Taq and 20–100 ng cDNA sample was assayed on
TP800 (both from Takara Bio, Inc.). The primers used were as
follows: USP39, 5′-TTTCCTCAACCTCCACA-3′ and
5′-ATTCAGTCCCACAATACCC-3′; GAPDH, 5′-TGACT TCAACAGCGACACCCA-3′ and
5′-CACCCTGTTGCTGTA GCCAAA-3′. The relative expression of USP39 mRNA
was calculated with the 2−ΔΔCt method, using GAPDH mRNA
expression level for normalization. All experiments were repeated
at least 3 times.
Western blot analysis
Total protein was isolated from whole cells using
ice-cold protein lysis buffer (1% Triton X-100; 50 mM Tris-HCl, pH
7.4; 150 mM NaCl; 0.1% SDS; 1 mM PMSF; 1 mM EDTA), followed by 30
min of incubation on ice and centrifugation at 10,000 × g for 10
min at 4°C. Protein concentration was determined by the BCA protein
assay (Pierce Biotechnology Inc., Rockford, IL, USA). Protein
extracts were separated on a SDS-polyacrylamide gel, blotted onto a
polyvinylidene fluoride membrane and incubated with mouse anti-FLAG
antibody (Sigma-Aldrich) or mouse anti-GAPDH antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Western blotting was
developed using horseradish peroxidase-conjugated goat anti-mouse
IgG and was detected with enhanced chemiluminescence reagent (both
from Santa Cruz Biotechnology, Inc.).
Cell colony formation assay
After 5 days of infection, cells were trypsinized,
resuspended and seeded into 6-well plates at a concentration of 200
cells/well and cultured at 37°C for 14 days. The media were
replaced every 3–4 days. At the end of incubation, the cells were
washed with phosphate-buffered saline (PBS) twice and fixed with
paraformaldehyde. Then, the cells were washed with PBS twice,
stained with Giemsa (Sigma-Aldrich) for 10 min, and washed with
ddH2O 3 times, sequentially. The plates were
photographed with a digital camera.
Monolayer growth assay
The monolayer culture growth rate was determined
using a Cellomics ArrayScan (Thermo Scientific). Briefly, after
being infected with virus for 5 days, cells at the same density
were seeded into flat-bottom 96-well plates and grown under normal
conditions. Cultured cells were observed at 1, 2, 3, 4 and 5 days
using a Cellomics ArrayScan to evaluate the cell growth with GFP
signal. Subsequently, the growth curves of the cells were measured
after each experiment. Each experiment was performed in
triplicate.
Flow cytometric analysis
Eighty-five percent confluent lentivirus-infected
cells were harvested by centrifugation at 1,200 rpm for 5 min. The
pellets were washed twice with cold PBS, fixed with cold 70%
ethanol, centrifuged at 1,500 rpm for 5 min to discard ethanol, and
resuspended with PBS, sequentially. The suspensions were filtrated
through a 400-mesh membrane and centrifuged at 1,200 rpm for 5 min.
The cells were stained with propidium iodide (PI) or Annexin V-APC
(eBioscience Inc.) and then were analyzed using a BD FACSCalibur
Flow Cytometer (BD Biosciences, San Diego, CA, USA). Each
experiment was performed in triplicate.
Statistical analysis
The data shown are presented as the mean ± standard
deviation (SD) of three independent experiments. Statistical
significance was determined with the Student’s t-test. A p-value
<0.05 was considered to indicate a statistically significant
difference.
Results
High levels of USP39 expression in breast
cancer
Since there are no reports on the USP39 expression
level in breast cancers, the levels of USP39 were detected by
immunohistochemical analysis to determine the USP39 expression in
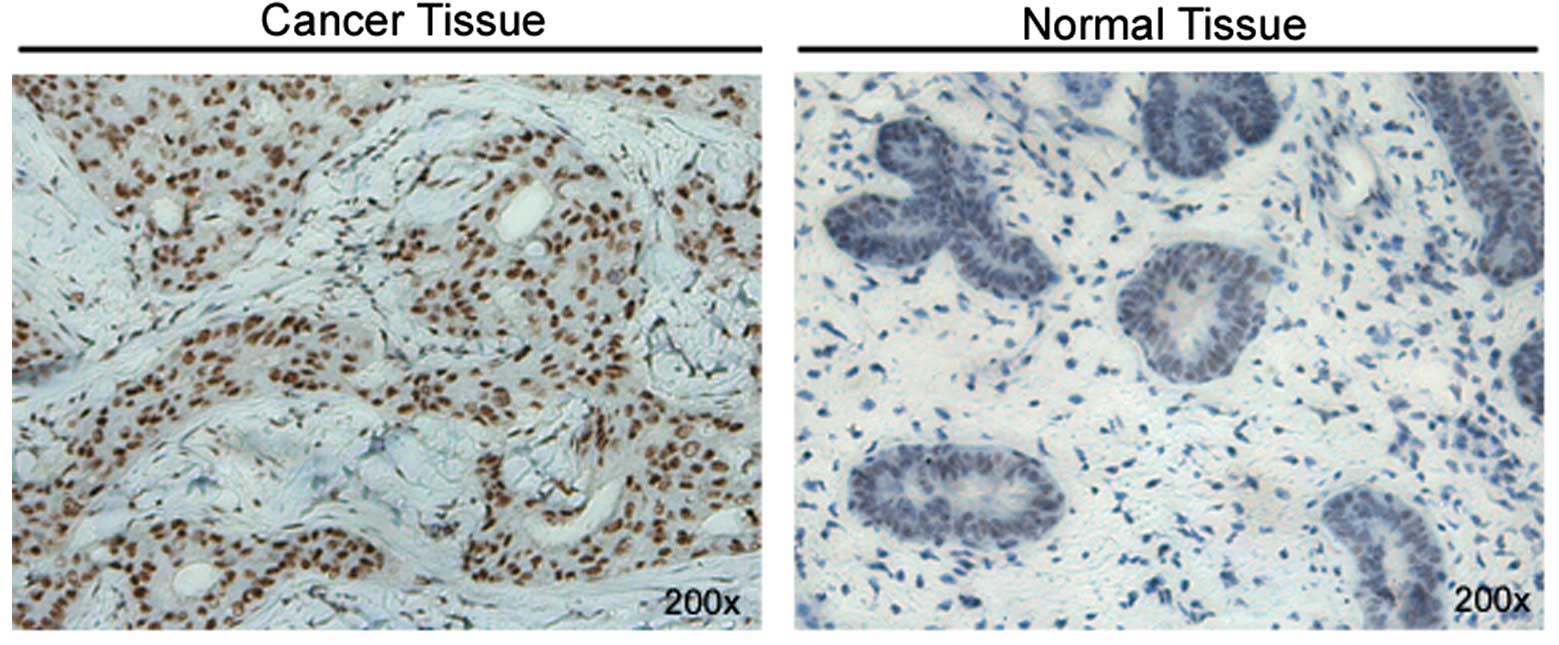
human breast cancer tissues. The overall distribution of the USP39
protein in the different histological tissue types is summarized in
Table I. Stronger staining of USP39
was observed in the breast tumor tissues than that in the normal
breast tissues (fewer normal samples were used due to the
difficulty in clinical collection) (Fig. 1). No significant correlation between
the metastatic lymph node and non-metastatic lymph node subgroups
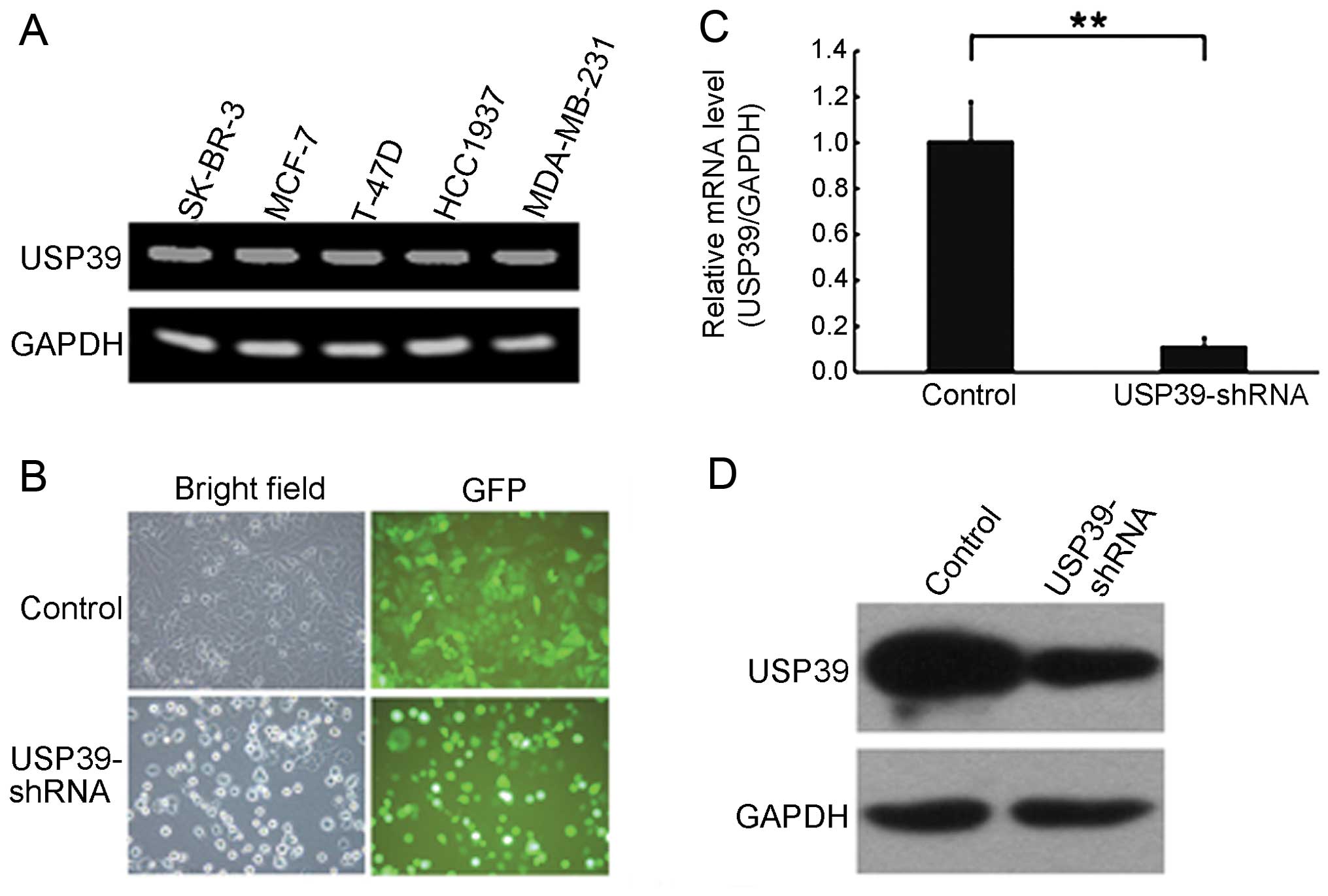
was noted in regards to USP39 expression levels (Table II). RT-PCR analysis revealed that
the expression of USP39 was positive in the SK-BR-3, MCF-7, T-47D,
HCC1937 and MDA-MB-231 breast cell lines (Fig. 2A). These data clearly indicate that
high levels of USP39 expressed selectively in human breast tumor
tissues may contribute to the development of high risk breast
cancer.
 | Table IExpression pattern of USP39 in breast
cancer and normal breast tissues. |
Table I
Expression pattern of USP39 in breast
cancer and normal breast tissues.
| | USP39 expression | |
|---|
| |
| |
|---|
| Type of tissue | No. of cases | Negative (−) | Weakly positive
(+) | Positive (++) | Hadro-positive
(+++) | P-value |
|---|
| Breast cancer | 23 | 1 | 2 | 15 | 5 | <0.05 |
| Normal breast | 6 | 3 | 2 | 1 | 0 | |
 | Table IIAssociation of USP39 expression
between metastatic and non-metastatic lymph node status. |
Table II
Association of USP39 expression
between metastatic and non-metastatic lymph node status.
| | USP39 expression | |
|---|
| |
| |
|---|
| Lymph node
status | No. of cases | Negative (−) | Weakly positive
(+) | Positive (++) | Hadro-positive
(+++) | P-value |
|---|
| Metastatic lymph
node | 10 | 3 | 0 | 6 | 1 | >0.05 |
| Non-metastatic lymph
node | 10 | 2 | 2 | 6 | 0 | |
Knockdown of USP39 by the shRNA
lentivirus system in breast cancer cells
To investigate the role of USP39 in breast cancer,
shRNA targeting USP39 or non-siliencing sequences were cloned into
the pGCSIL-GFP lentiviral vector, respectively. Then, the
USP39-shRNA lentivirus or the non-silencing shRNA lentivirus
expressing GFP were generated and infected into MCF-7 cells. The
infection efficiency of lentivirus was >90% after 5 days of
infection (Fig. 2B). The qRT-PCR
assay revealed that the USP39 mRNA level was reduced by ~88.6%
(Fig. 2C). We also determined the
level of USP39 protein in USP39-shRNA construct-transfected cells
via western blot analysis. In 293T cells co-transfected with the
USP39 plasmid and USP39-shRNA construct, the protein level of
overexpressed USP39 was significantly reduced by >60% (Fig. 2D). These results indicate that this
USP39-shRNA was specific and efficient in inhibiting the expression
of USP39.
Important role of USP39 in breast cancer
cell growth
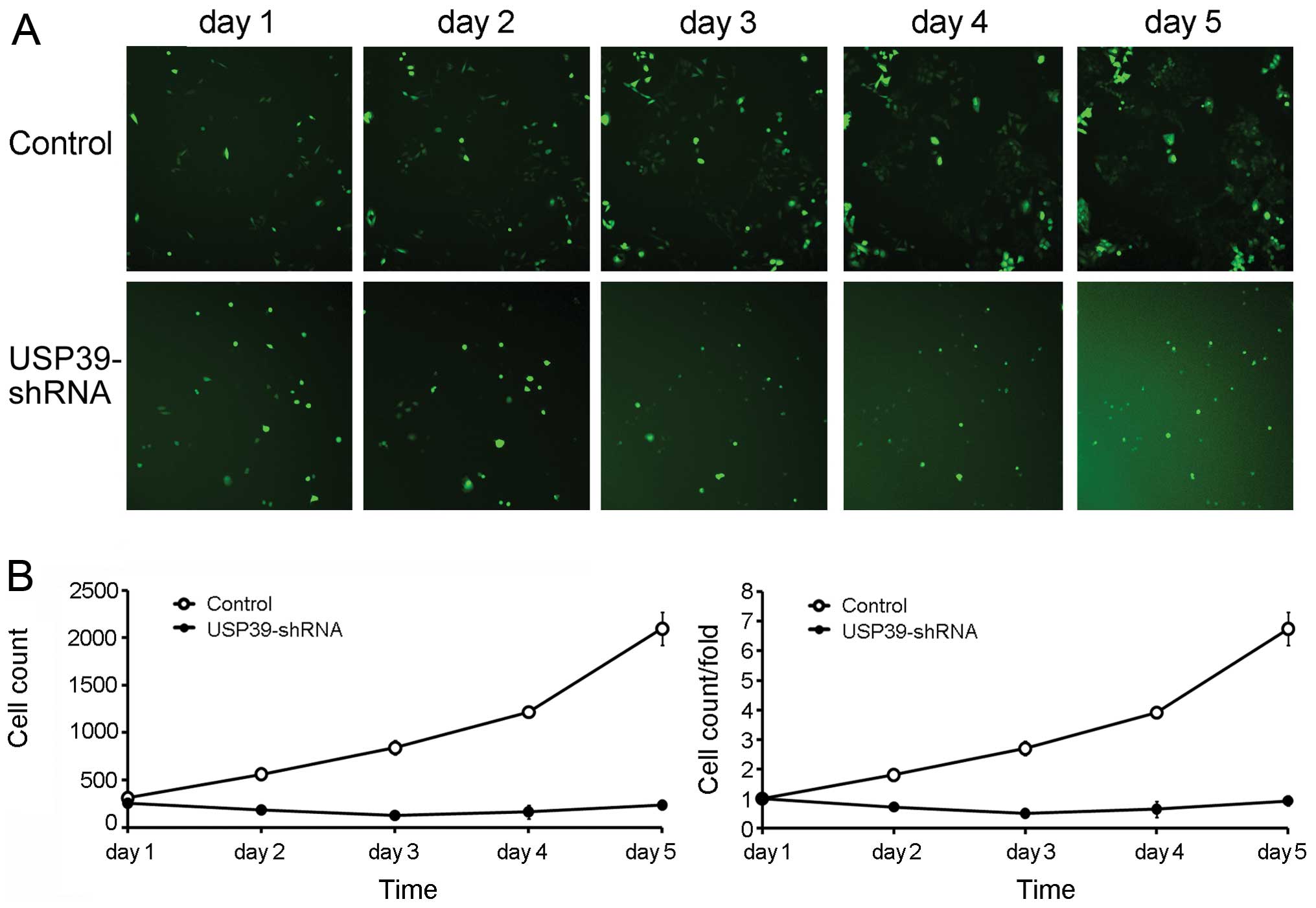
To examine the effect of USP39-shRNA-mediated
downregulation of USP39 on the growth of breast cancer cells, MCF-7
cell proliferation was assayed. Five days after infection,
USP39-shRNA and non-silencing shRNA-infected MCF-7 cells were
seeded into 96-well plates at the same density, and cell numbers
were detected through GFP signal at indicated time points using
Cellomics ArrayScan. Compared to the control, USP39-shRNA-infected
MCF-7 cells displayed significant decrease in cell proliferation
with no obvious growth observed during the entire assay period (5
days) (Fig. 3). This finding
indicates that the knockdown of USP39 markedly diminished the cell
proliferative ability in breast cancer cells.
Suppression of USP39 knockdown on cell
colony formation
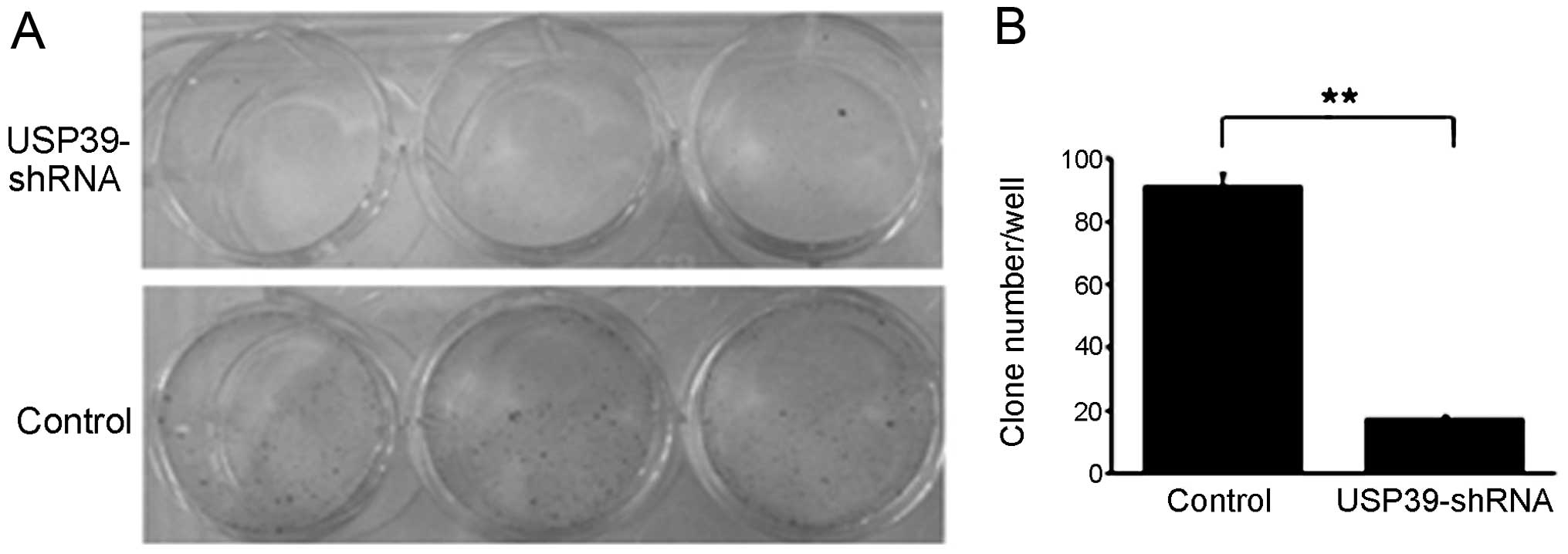
We then studied the colony formation capacity of
MCF-7 cells treated with the USP39-shRNA lentivirus. The control
group and the USP39-shRNA-infected MCF-7 cells were allowed to grow
for 14 days to form colonies. USP39 knockdown resulted in a nearly
5-fold decrease in the number of MCF-7 colonies, as compared to the
control (Fig. 4).
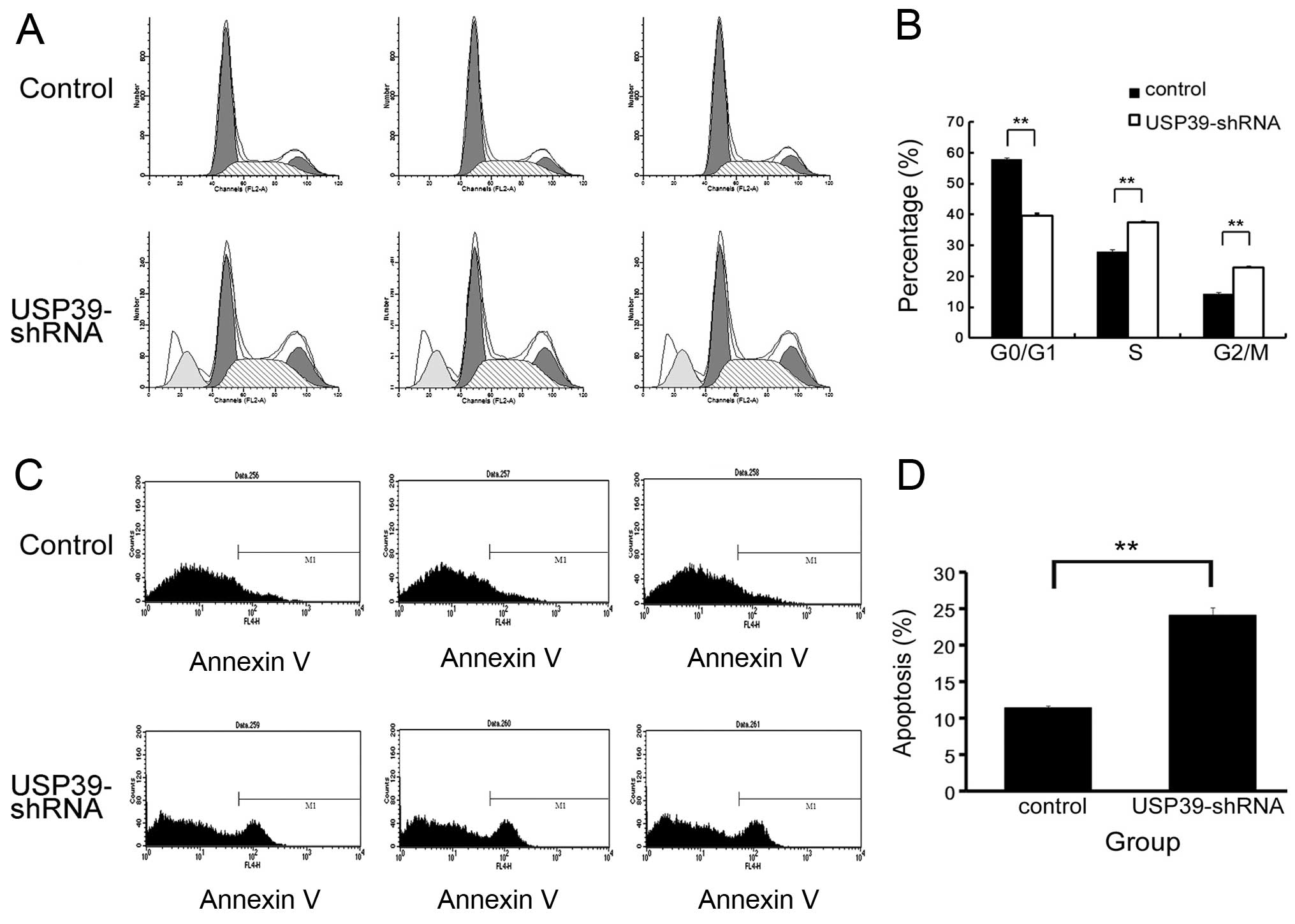
Inhibition of USP39 induces G0/G1 cell
cycle phase arrest and apoptosis
Based on the finding that inhibition of USP39 in
MCF-7 cells markedly inhibits cell proliferation and represses cell
colony formation, we further employed cell cycle analysis to
uncover the mechanism governing the inhibitory effect of
USP39-shRNA on cell proliferation and colony formation. An obvious
increase in the G0/G1-phase cell population was observed in the
USP39-shRNA-infected MCF-7 cells accompanied by a decrease in the S
and G2/M-phase cell population, when compared to these percentages
in the control (Fig. 5A and B). Our
results revealed that USP39-shRNA exerted an inhibitory effect on
breast cancer cell proliferation via G0/G1 cell cycle arrest. In
addition, we found that knockdown of USP39 led to an increase in
MCF-7 cell apoptosis (Fig. 5C and
D).
Discussion
Breast cancer affects nearly 1 out of 9 women
worldwide. There has been an intensive effort to improve treatment
for breast cancer. Novel treatment strategies have arisen from the
study of the molecular and cellular biology of breast cancer cell
lines. These studies have produced a group of agents called
targeted therapeutics for their direction at a single molecule
rather than a general process such as DNA replication or
cytoskeletal function. In our study, we found that the expression
levels of USP39 in breast cancer tissues were markedly higher,
indicating that USP39 may have an important role in breast cancer
development.
Today RNAi technology is prevalent in cancer
research and therapy (13).
Compared to chemically synthesized siRNA, shRNA encoded within an
expression vector offers advantages in silencing longevity,
delivery options and cost. To overcome limitations including
transient shRNA expression and low transfection efficiency,
lentiviral vectors have been developed (14). In this study, to confirm the role of
USP39 in breast cancer development, we used a lentivirus shRNA
system that effectively knocked down the expression of USP39 at
both the RNA and protein levels. qRT-PCR and western blot analysis
showed sufficient silencing of USP39, thus ensuring the credibility
of the subsequent assays (Fig. 2).
As expected, inhibition of USP39 in MCF-7 cells markedly decreased
breast cancer cell proliferation. We also confirmed that knockdown
of USP39 significantly inhibited the colony formation capacity of
MCF-7 cells. Intriguingly, our FACS data revealed that USP39-shRNA
had an inhibitory effect on breast cancer cell growth via
G0/G1-phase arrest and apoptosis induction.
Cell cycle regulation requires the timed degradation
of numerous checkpoint and signaling molecules to allow an orderly
progression through replication, growth and mitosis. Previous
studies have revealed roles for various DUBs in cell cycle
regulation (15,16), cell signal transduction (17,18),
regulation of the growth and development of organisms (19,20)
and DNA repair. USP1 is involved in the DNA repair processes
through its deubiquitination of the Fanconi anemia protein FANCD2
(21) and PCNA (22). USP1 is proposed to deubiquitinate
FANCD2 when cells exit the S-phase or recommence cycling after DNA
damage and may play a critical role in the FA pathway by recycling
FANCD2 (21). In a study of DNA
damage-induced apoptosis, USP28 was shown to be involved in the
checkpoint kinase 2 (Chk2)-p53-PUMA pathway, a major regulator of
DNA damage-induced apoptosis, in response to double-strand breaks
in vivo(23). USP28 also
controls the cellular levels of the transcription factor MYC
through antagonizing FBW7 (24).
USP11 has been described as a DUB that exhibits pro-survival
functions as part of the cellular response to DNA damage. In
response to mitomycin C (MMC)-induced DNA damage, USP11
participates in DNA damage repair functions within the breast
cancer 2 (BRCA2) pathways, independently of BRCA2 deubiquitination
(25). USP7, also known as
herpesvirus-associated USP (HAUSP), deubiquitinates p53 and Mdm2
and is inhibited by the Epstein-Barr nuclear antigen 1 (EBNA1)
protein of the Epstein-Barr virus (EBV) (26,27).
Recently, it was reported that USP10 is a deubiquitinase for p53.
In unstressed cells, USP10 localizes in the cytoplasm and regulates
p53 homeostasis. After DNA damage, a fraction of USP10 translocates
to the nucleus and contributes to p53 activation (28).
USP39, harboring a Dub domain, belongs to the
ubiquitin specific protease family. Previous investigations have
described the role of USP39 in mRNA processing (29). It is essential for the assembly of
mature spliceosomes (10) and
mitotic spindle checkpoint integrity (11). However no USP39-specific substrate
has been identified, and other functions of USP39 are poorly
understood. In this study, we did not discover the exact mechanism
through which USP39 influences breast cancer cell proliferation. We
infer that dysregulation of USP39 may affect the Ub pathway and
degradation of certain proteins that govern cell cycling; although
it is argued that USP39 lacks Dub activity in regulating mitotic
spindle checkpoint integrity (11).
Therefore screening USP39 downstream target proteins through high
throughput proteomics approach is the focal point of our further
research, which will facilitate the elucidation of the mechanisms
involved in the effects of USP39 on breast cancer development.
In summary, we found upregulated expression of USP39
in breast cancer tissues and proved for the first time that
RNAi-mediated knockdown of USP39 suppressed the growth and colony
formation ability of breast cancer cells. Additionally, USP39
inhibition induced G0/G1-phase arrest and apoptosis of breast
cancer cells. Our data indicate that USP39 may serve as an oncogene
in breast cancer development. Therefore, USP39 has considerable
potential to be a new therapeutic target for the treatment of
breast cancer.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Shandong Province of China (Y2008C48), the
Department of Education of Shandong Province of China (J11LF05) and
the Research Program of Qingdao South District Municipal Science
and Technology Commission (2011-5-004-YY).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Sinha D, Biswas J, Sung B, Aggarwal BB and
Bishayee A: Chemopreventive and chemotherapeutic potential of
curcumin in breast cancer. Curr Drug Targets. 13:1799–1819. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allgayer H and Fulda S: An introduction to
molecular targeted therapy of cancer. Adva Med Sci. 53:130–138.
2008.PubMed/NCBI
|
|
4
|
Pickart CM and Eddins MJ: Ubiquitin:
structures, functions, mechanisms. Biochim Biophys Acta.
1695:55–72. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pickart CM and Fushman D: Polyubiquitin
chains: polymeric protein signals. Curr Opin Chem Biol. 8:610–616.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohta T and Fukuda M: Ubiquitin and breast
cancer. Oncogene. 23:2079–2088. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilkinson KD: Regulation of
ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J.
11:1245–1256. 1997.PubMed/NCBI
|
|
8
|
Nijman SM, Luna-Vargas MP, Velds A, et al:
A genomic and functional inventory of deubiquitinating enzymes.
Cell. 123:773–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reyes-Turcu FE, Ventii KH and Wilkinson
KD: Regulation and cellular roles of ubiquitin-specific
deubiquitinating enzymes. Annu Rev Biochem. 78:363–397. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Makarova OV, Makarov EM and Luhrmann R:
The 65 and 110 kDa SR-related proteins of the U4/U6. U5 tri-snRNP
are essential for the assembly of mature spliceosomes. EMBO J.
20:2553–2563. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Leuken RJ, Luna-Vargas MP, Sixma TK,
Wolthuis RM and Medema RH: Usp39 is essential for mitotic spindle
checkpoint integrity and controls mRNA-levels of aurora B. Cell
Cycle. 7:2710–2719. 2008.PubMed/NCBI
|
|
12
|
Rios Y, Melmed S, Lin S and Liu NA:
Zebrafish usp39 mutation leads to rb1 mRNA splicing defect and
pituitary lineage expansion. PLoS Genet. 7:e10012712011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Izquierdo M: Short interfering RNAs as a
tool for cancer gene therapy. Cancer Gene Ther. 12:217–227. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qin XF, An DS, Chen IS and Baltimore D:
Inhibiting HIV-1 infection in human T cells by lentiviral-mediated
delivery of small interfering RNA against CCR5. Proc Natl Acad Sci
USA. 100:183–188. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Zhang B and Fischer JA: A specific
protein substrate for a deubiquitinating enzyme: liquid facets is
the substrate of Fat facets. Genes Dev. 16:289–294. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakurai M, Ayukawa K, Setsuie R, et al:
Ubiquitin C-terminal hydrolase L1 regulates the morphology of
neural progenitor cells and modulates their differentiation. J Cell
Sci. 119:162–171. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang JM: Emerging roles of
deubiquitinating enzymes in human cancer. Acta Pharmacologica Sin.
28:1325–1330. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paulsson K, Békassy AN, Olofsson T,
Mitelman F, Johansson B and Panagopoulos I: A novel and
cytogenetically cryptic t(7;21)(p22;q22) in acute myeloid leukemia
results in fusion of RUNX1 with the ubiquitin-specific
protease gene USP42. Leukemia. 20:224–229. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim J, Kim WJ, Liu Z, Loda M and Freeman
MR: The ubiquitin-specific protease USP2a enhances tumor
progression by targeting cyclin A1 in bladder cancer. Cell Cycle.
11:1123–1130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stevenson LF, Sparks A, Allende-Vega N,
Xirodimas DP, Lane DP and Saville MK: The deubiquitinating enzyme
USP2a regulates the p53 pathway by targeting Mdm2. EMBO J.
26:976–986. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nijman SM, Huang TT, Dirac AM, et al: The
deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway.
Mol Cell. 17:331–339. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang TT, Nijman SM, Mirchandani KD, et
al: Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat
Cell Biol. 8:339–347. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Zaugg K, Mak TW and Elledge SJ: A
role for the deubiquitinating enzyme USP28 in control of the
DNA-damage response. Cell. 126:529–542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Popov N, Herold S, Llamazares M, Schulein
C and Eilers M: Fbw7 and Usp28 regulate myc protein stability in
response to DNA damage. Cell Cycle. 6:2327–2331. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schoenfeld AR, Apgar S, Dolios G, Wang R
and Aaronson SA: BRCA2 is ubiquitinated in vivo and interacts with
USP11, a deubiquitinating enzyme that exhibits prosurvival function
in the cellular response to DNA damage. Mol Cell Biol.
24:7444–7455. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li M, Chen D, Shiloh A, et al:
Deubiquitination of p53 by HAUSP is an important pathway for p53
stabilization. Nature. 416:648–653. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cummins JM, Rago C, Kohli M, Kinzler KW,
Lengauer C and Vogelstein B: Tumour suppression: disruption of
HAUSP gene stabilizes p53. Nature. 428:1 p following 486. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuan J, Luo K, Zhang L, Cheville JC and
Lou Z: USP10 regulates p53 localization and stability by
deubiquitinating p53. Cell. 140:384–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lygerou Z, Christophides G and Seraphin B:
A novel genetic screen for snRNP assembly factors in yeast
identifies a conserved protein, Sad1p, also required for pre-mRNA
splicing. Mol Cell Biol. 19:2008–2020. 1999.PubMed/NCBI
|