Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
eighth leading cause of cancer-related mortality worldwide, and
laryngeal squamous cell carcinoma (LSCC) is a major type of HNSCC
(1). In China, the incidence of
LSCC has been gradually increasing, particularly in the Northeast.
LSCC is a heterogeneous disease involving deregulation of multiple
pathways involved in cellular differentiation, the cell cycle,
apoptosis, angiogenesis and metastasis (2). Despite advancement in local control
and overall improvement in the quality of life achieved with the
use of combined modality therapies, the survival rates for LSCC
patients have not improved significantly over the past 2 decades.
Therefore, the current research is often focused on the
identification of useful biologic and molecular markers for the
diagnosis and therapy of LSCC (3,4).
As one of the epigenetic types, DNA methylation can
cause the silencing of gene transcription, genetic imprinting,
genomic stability and × chromosome inactivation (5–7).
Aberrant methylation of genes controlling the cell cycle,
proliferation, apoptosis, metastasis, drug resistance and
intracellular signaling has been identified in multiple types of
cancers (8–10). Tumor-suppressor genes are often
inactivated in cancer tissues by hypermethylation within CpG
islands (11,12). Hypomethylation usually leads to the
activation of oncogenes in cancer tissues (13,14).
In our previous study, we found that the S100A4 gene is
overexpressed in laryngeal cancer tissues and that RNAi
S100A4 decreased the invasiveness of HEp2 cells, which
implies that S100A4 is involved in the progression of LSCC.
We also found significant hypomethylation within the promoter
region of S100A4 in LSCC tissues when compared to that in
paired adjacent tissues. Statistical analysis showed the
association of DNA hypomethylation and the overexpression of the
S100A4 gene in laryngeal cancer tissues (15). However, the exact mechanism remains
unclear. We speculated that the methylated sites of the
S100A4 promoter affects the binding of the corresponding
transcription factors to S100A4. In the present study, we
identified the putative transcription factors that bind to
S100A4 and explored the effect of hypomethylation on the
binding.
Materials and methods
Cell culture and reagents
The human laryngeal carcinoma cell line HEp2 (Cell
Biology Institute of Shanghai, Chinese Academy of Science) was
maintained in RPMI-1640 (Gibco-BRL, Los Angeles, CA, USA) with 10%
fetal bovine serum, 100 units/ml penicillin and 100 μg/ml
streptomycin in a humidified atmosphere at 37ºC with 5%
CO2. 5′-Aza-2′-deoxycytidine (5-Aza) was purchased from
Sigma-Aldrich (St. Louis, MO, USA). Cells were treated with 4 μM
5-Aza for 72 h. PGL3-basic, pRL-TK and Dual-Luciferase reporter
assay system were obtained from Promega Corporation (Madison, WI,
USA). The Plasmid Miniprep kit and Lipofectamine™ 2000 transfection
reagents were purchased from Invitrogen Life Technologies
(Carlsbad, CA, USA). The primary antibodies, protein A/G-agarose
and the anti-IgG antibody used in the present study were purchased
from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Plasmid construction
Human genomic DNA was isolated from HEp2 cells using
a genomic DNA isolation kit (Beyotime, Jiangsu, China) according to
the manufacturer’s instructions. Over 1.2 kb fragment of the
S100A4 5′-flanking sequence was amplified using Probest
Taq DNA polymerase (Takara, Japan). KpnI and
XhoI restriction sites were integrated into the fragment
above using primers P0 and P1 (Table
I). The fragment was then cloned into the plasmid PGL3-Basic,
named P1283 (−1283/+73). The deletion constructs P970 (−970/+73),
P912 (−912/+73), P842 (−842/+73), P530 (−530/+73) and P485
(−485/+73) were created by PCR using P1283 as a template and the
oligonucleotides P2 to P6 as primers. The reporter constructs
including point mutations and deletions at potential c-Myb and
C/EBPα binding sites were generated using PCR-based Megaprimer
technique as previously described (16). Primer P7 was used for the
construction of the mutant c-Myb vector (pcMyb-M), P8 and P9 for
the deletion cMyb vector (pcMyb-D), P10 for the C/EBPα wild-type
vector, P11 for the mutant C/EBPα vector (pC/EBPα-M), P12 and P13
for the deletion C/EBPα vector (pC/EBPα-D). All constructs were
sequenced in a capillary automatic sequencer (ABI PRISM®
3100 Genetic Analyzer; Applied Biosystems). All primer sequences
are listed in Table I.
 | Table IPrimer sequences used in the
generation of the related constructs. |
Table I
Primer sequences used in the
generation of the related constructs.
| Primer name | Sequence: 5′ to
3′ |
|---|
| P0 |
TTTAAGCTTTGCTCTGGGCAGTGAACAT |
| P1 |
TTTGGTACCGCTGGGACTACAGGCTAC |
| P2 |
TTTGGTACCGTGCCCATCTCATCCAG |
| P3 |
TTTGGTACCGTGCCCACCTGGGAACA |
| P4 |
TTTGGTACCTCAGCCCACAGCAGGAAG |
| P5 |
TTTGGTACCCACACACACATGCACGTAAG |
| P6 |
TTTGGTACCTGAGCAAGTGACTGAA |
| P7 |
TGGGCTTGCACATTCTGTTGCTATAGTACG |
| P8 |
TGAGATGTGGGCTTGCACATGTTGCTATAGTACGTGTTGGT |
| P9 |
ACCAACACGTACTATAGCAACATGTGCAAGCCCACATCTCA |
| P10 |
TTTGGTACCGGCTCATGTTTGCTGGGTT |
| P11 |
GGCTCATGTTTGCTGGGTTGTACACTAAGGAGCAGGAAGC |
| P12 |
ACTGGCTCATGTTTGCTGGGGAGCAGGAAGCAAAGGAAAGGCA |
| P13 |
TGCCTTTCCTTTGCTTCCTGCTCCCCAGCAAACATGAGCCAGT |
Transient transfection and luciferase
assays
HEp2 cells were seeded in 24-well plates, grown to
60–80% confluence, and transfected by the constructs using
Lipofectamine™ 2000 according to the manufacturer’s instructions
(Invitrogen Life Technologies). pRL-TK plasmid containing the
Renilla luciferase gene was used as the internal control in
each experiment. After 24 h of transfection, the cells were
incubated with 4 μM 5-Aza for an additional 3 days. Firefly and
Renilla luciferase activities were determined using a Lumat
LB 9507 luminometer (Bethold Technologies, Bad Wildbad,
Germany).
Semi-quantitative RT-PCR
Total RNA was isolated by TRIzol reagent according
to the manufacturer’s instructions, and cDNA was reversibly
transcribed from the isolated RNA using an AMV RNA PCR kit (Takara,
Japan) in line with the standard operation protocol. The primer
sequences were as follows: S100A4 forward,
5′-CCCTGGATGTGATGGTGTC-3′ and reverse, 5′-CTCGTTGTCCCTGTTGCTG-3′,
which were expected to produce a 185-bp DNA fragment; β-actin
forward, 5′-CCAGATCATGTTTGAGACCT-3′ and reverse,
5′-TTGAAGGTAGTTTCGTGGAT-3′, which were expected to produce a 480-bp
DNA fragment. The PCR reaction was performed in a 25-μl reaction
system, starting with denaturation at 94ºC for 4 min, then 30
cycles of denaturation at 94ºC for 30 sec, annealing at 55ºC for 30
sec, extension at 72ºC for 45 sec, followed by an extra extension
at 72ºC for 5 min. The PCR products were run on a 1.5% agarose gel,
and the band intensity was measured and normalized to the internal
control β-actin.
Western blotting
Total protein was extracted from HEp2 cells. In
brief, cells were lysed using 250 μl of RIPA lysis buffer and then
underwent a process of homogenization for 10 min, ice-bath for 1 h
and centrifugation at 12,000 × g for 30 min at 4ºC. The supernatant
was finally collected, and the protein concentration was determined
by the BCA protein assay system (Pierce Biotechnology, Inc.,
Rockford, IL, USA). Extracts equivalent to 50 μg of total protein
were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel
electrophoresis and transferred onto nitrocellulose membranes.
After being blocked overnight at 4ºC with 1× PBS, 0.1% Tween-20 and
5% non-fat milk, the membranes were incubated with the primary
antibodies against S100A4 (1:2,000) or β-actin (1:1,000) for 3 h at
room temperature, washed twice and then incubated with the
secondary horseradish peroxidase-conjugated goat anti-rabbit
antibody (1:5,000; Zhongshan, China) for 2 h at room temperature.
Immunodetection was performed with chemiluminescence (ECL reagent;
Beyotime, China), and the membranes were exposed to film. The
densities of the bands on the filters were quantified by
densitometric analysis using the UVP GelWorks ID advanced version
2.5 software (Bio-Rad, USA).
Bisulfite modification and
bisulfite-specific PCR
Genomic DNA isolated from the HEp2 cells was used to
detect the methylation status of the S100A4 promoter. In
brief, ~1 μg of genomic DNA was bisulfite-modified using the EZ DNA
Methylation-Gold™ kit (Zymo Research, Orange, CA, USA) according to
the manufacturer’s recommendation. Based on the functional promoter
sequence of the S100A4 gene, the primers (forward,
5′-TAGAAAAGTGAGTAAGTGATTGA-3′ and reverse, 5′-TC
TACCTTTCCTTTACTTCCTAC-3′) were used in bisulfite-specific PCR
detection, and the amplified fragment was 170 bp. The PCR reaction
was performed in a 25-μl reaction system, starting with
denaturation at 94ºC for 4 min, then 42 cycles of denaturation at
94ºC for 30 sec, annealing at 53ºC for 30 sec, extension at 72ºC
for 45 sec, followed by an extra extension at 72ºC for 5 min. The
BSP products were then cloned into a T-vector (Takara, Japan), and
JM109 E. coli competent cells (Takara, Japan) were used for
transformation according to the manufacturer’s instructions.
Bioinformatics
The human S100A4 promoter sequence was
obtained from e-Ensembl with the accession no. ENST00000368715. The
binding sites were predicted by using the online P-Match™ program
(http://www.gene-regulation.com/pub/programs.html#pmatch).
Electromobility shift assay (EMSA)
Non-denatured cellular nuclear proteins were
prepared using a nuclear protein extraction kit according to the
manufacturer’s instructions (Active Motif, Carlsbad, CA, USA). EMSA
was carried out using a commercially available LightShift
Chemiluminescent EMSA kit (Pierce Biotechnology, Inc.). The probes
used in EMSA were synthesized by Sangene (Beijing, China), and the
sequences were as follows: c-Myb wild-type,
5′-tgcacacgctgttgctatagta-3′ and mutant,
5′-tgcacacgctgaatctatagta-3′; cEBpα wild-type,
5′-gggttgcgcactcgggagcagg-3′ and mutant,
5′-gggaatcgcactcgggagcagg-3′. Two micrograms of nuclear protein
extracts was incubated with 3′-end-biotin-labeled S100A4 DNA
probes in binding buffer for 30 min on ice, separated on a 6%
non-denaturing polyacrylamide gel in 0.5× TBE, and then transferred
onto a nylon membrane and fixed by ultraviolet cross-linking. The
biotin-labeled probes were detected with streptavidin-horseradish
peroxidase (Pierce Biotechnology, Inc.). For the competition
experiments, a 100-fold excess of unlabeled doubled-stranded cold
probes was added to the binding reaction. To determine the effect
of the antibodies on protein-DNA binding, 1 μg primary antibodies
against c-Myb, C/EBPα, Ap2 and Msx-1 (Santa Cruz Biotechnology,
Inc.) was incubated with the nuclear extracts for 30 min on ice
prior to the addition of the biotin-labeled DNA probes,
respectively.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP-IT kit was purchased from Active Motif, and
the ChIP assay was carried out according to the manufacturer’s
protocol. One microgram of antibodies against c-Myb or C/EBPα was
utilized for immunoprecipitation. After proteinase K digestion and
DNA precipitation, 3 μg/l of diluted DNA was used for PCR. The
primer sequences for amplifying the S100A4 promoter and
GAPDH were as follows: S100A4 forward, 5′-CAAACAG
AAAAGTGAGCAAGTGAC-3′ and reverse, 5′-AGAGGCAA GGGTTGGAAGAG-3′ to
generate a 289-bp fragment; GAPDH forward,
5′-CGACCACTTTGTCAAGCTCA-3′ and reverse, 5′-AGGGGTCTACATGGCAACTG-3′
to generate a 332-bp fragment.
Statistical analysis
Unless otherwise stated, each experiment was
performed for a minimum of 3 times. Data were subjected to
statistical analysis using SPSS 13.0 software and are shown as
means ± standard error of the mean (SEM). Differences in mean
values were analyzed using one-way analysis of variance (ANOVA) or
t-test as appropriate. Statistical significance was assumed for a
two-tailed P<0.05.
Results
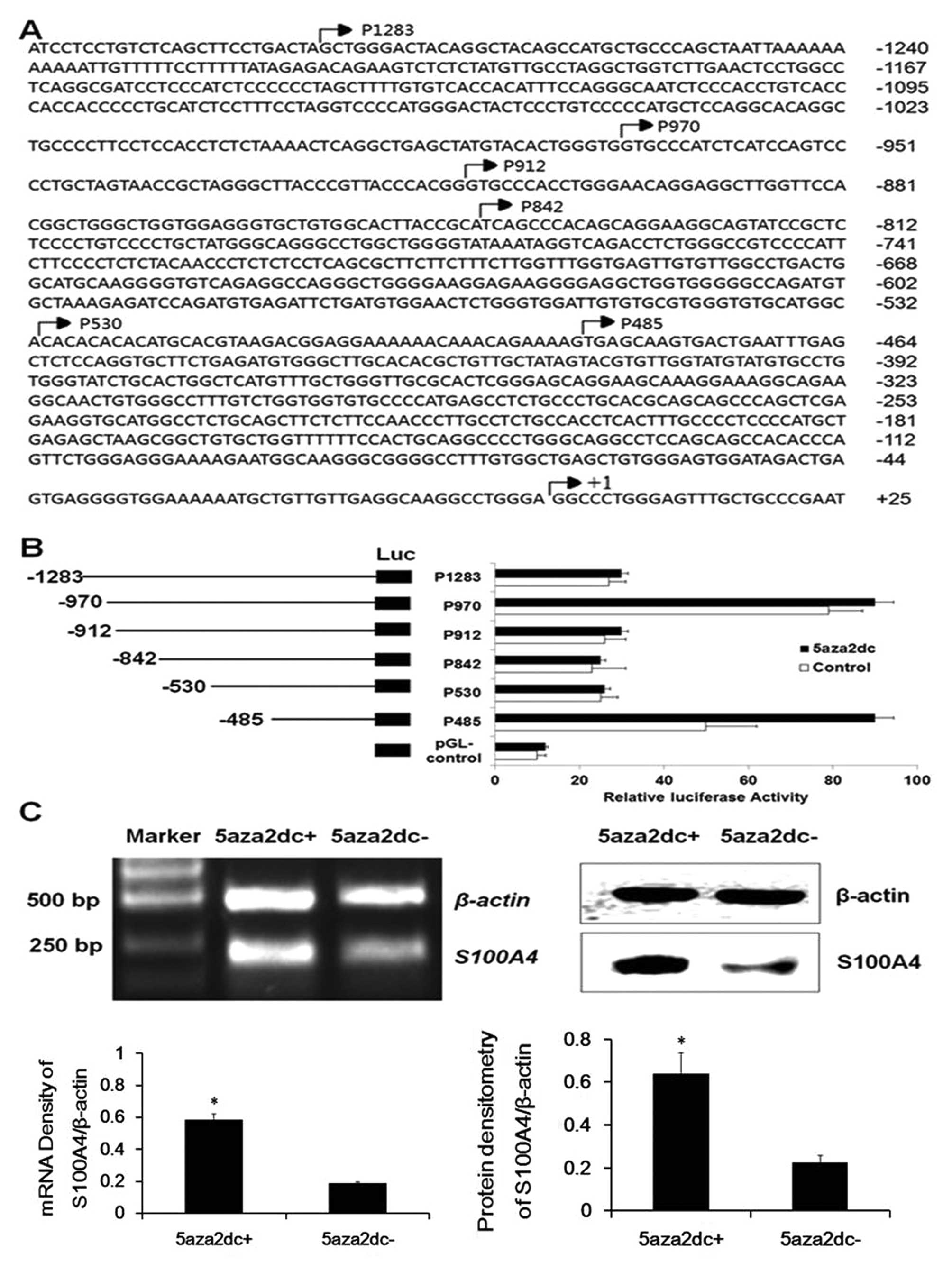
S100A4 promoter region −485 – +73 harbors
major transcription factor binding sites related to the methylation
status in laryngeal cancer cells
In search of the potential cis-acting
elements of the S100A4 promoter, several constructs
including the region −485 – +73 of S100A4 were generated
(Fig. 1A), and transient
transfection was then performed to assess their relative reporter
gene activities. As shown in Fig.
1B, under basal conditions, the constructs p970-luc and
p485-luc in HEp2 cells produced much higher luciferase activities
than those of the other truncated vectors (P<0.05), indicating
that the region −485 – +73 harbors the positive cis-acting
elements, and the fragment −486 – −530 the negative
cis-acting elements. Following a 72-h exposure to 4 μM
5-Aza, the HEp2 cells transfected by p485-luc resulting in a
significantly high level of luciferase activity when compared to
the other groups (P<0.05). RT-PCR and western blot results also
indicated that 5-Aza induced significant overexpression of
S100A4 both at the mRNA and protein levels (Fig. 1C, P<0.05). The MSP-based
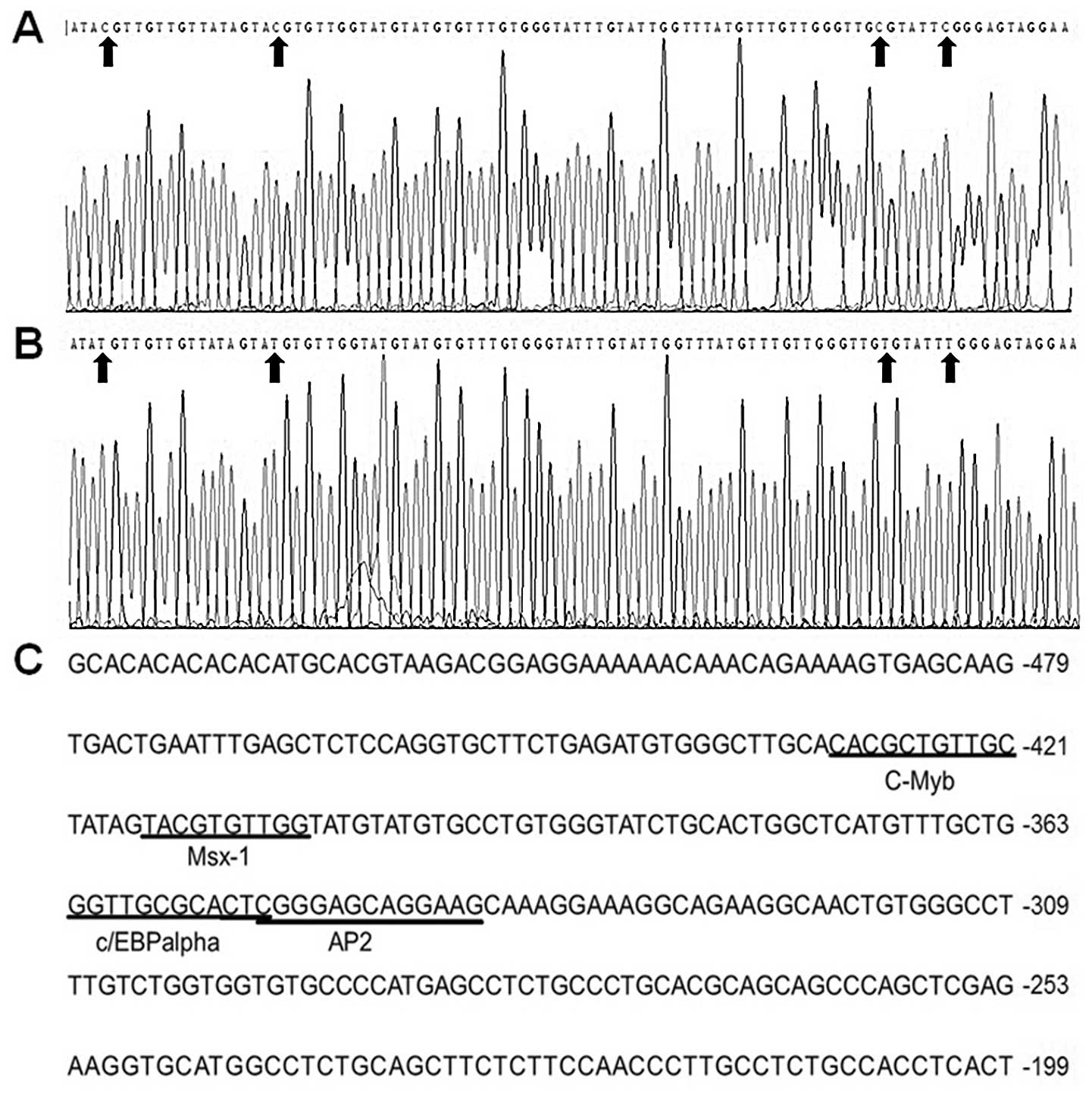
sequencing result showed that the cytosines of the dinucleotide CG
at position −485 – +73 were methylated in the untreated HEp2 cells
and the methyl groups of the cytosines were removed in HEp2 cells
treated with 5-Aza (Fig. 2A and B).
These findings suggest that demethylation increases the expression
of endogenous transcription factor binding to the area and results
in the methylation-free status in the region of S100A4 as
well. Therefore, there could exist important cis-acting
elements within the promoter region −485 – +73, which is affected
by the methylation status of either the gene itself or the upstream
genes regulating the transcription of S100A4.
Putative cis-acting elements in the
S100A4 promoter region −485 – +73 are predicted
The P-Match software was used to predict the
cis-acting elements in the S100A4 promoter region
−485 – +73. The result indicted that this region contained the
binding motifs of certain transcription factors including c-Myb,
C/EBPα, Ap2 and Msx-1 (Fig. 2C).
The cytosines in these binding sites were methylated in HEp2 cells
(Fig. 2A and B).
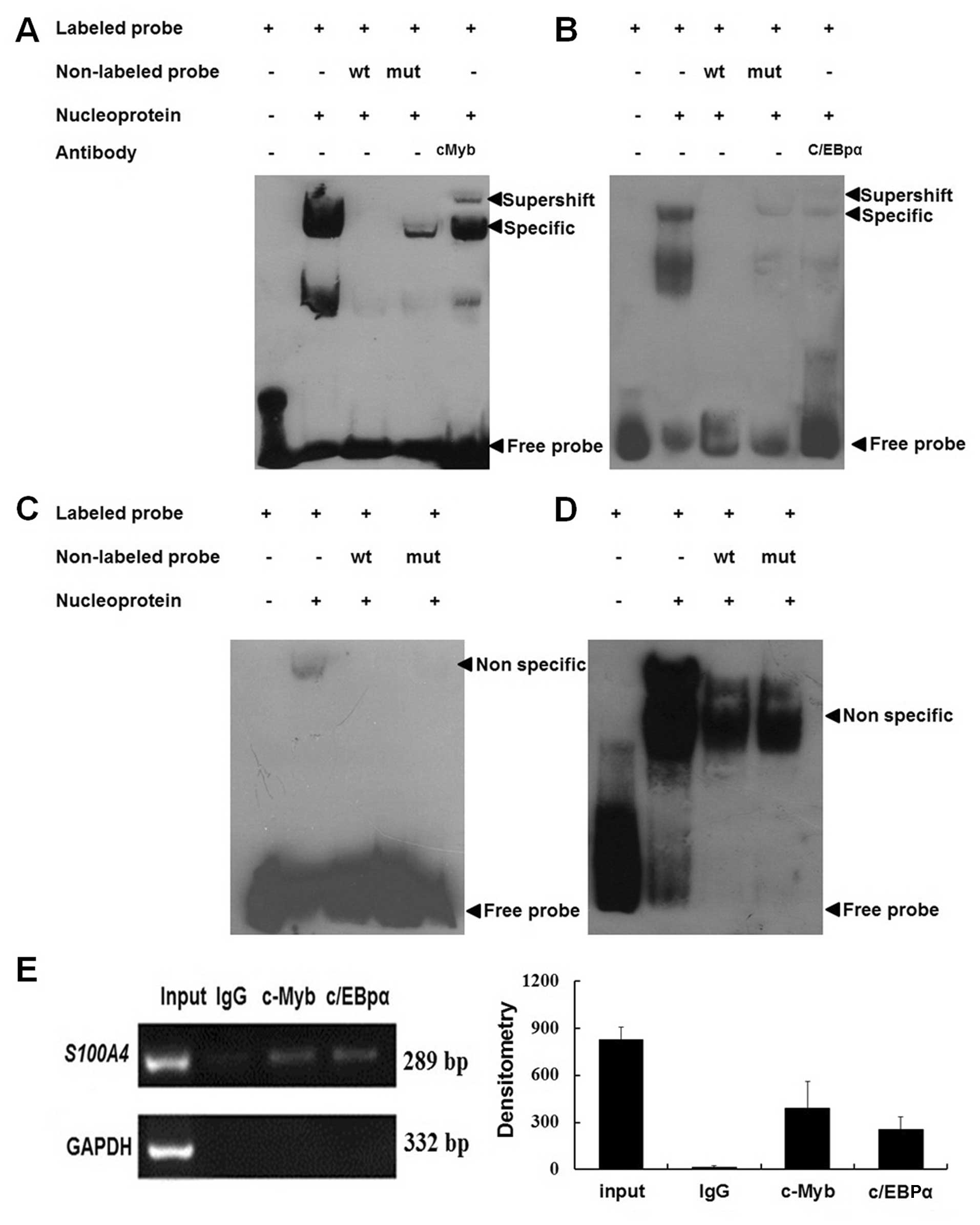
c-Myb and C/EBPα bind to the cis-acting
elements in the S100A4 promoter region −485 – +73
To confirm the binding of the S100A4 gene to
c-Myb, C/EBPα, Ap2 and Msx-1, EMSA and ChIP methods were used. As
shown in Fig. 3A–D, the binding of
c-Myb and C/EBPα, but not Ap2 and Msx-1, to S100A4 was
detected when the wild-type probes were used. The addition of the
cold self-competitors weakened the binding, and the mutant
competitor could not abolish the binding. Supershift bands were
also observed when c-Myb and C/EBPα antibodies were applied. These
results indicated that c-Myb and C/EBPα were able to bind the
S100A4 promoter region −485 – +73 in vitro. ChIP
results showed that both c-Myb and C/EBPα antibodies, but not
non-immune IgG, could immunoprecipitate c-Myb and C/EBPα proteins
binding to the same region in vitro (Fig. 3E).
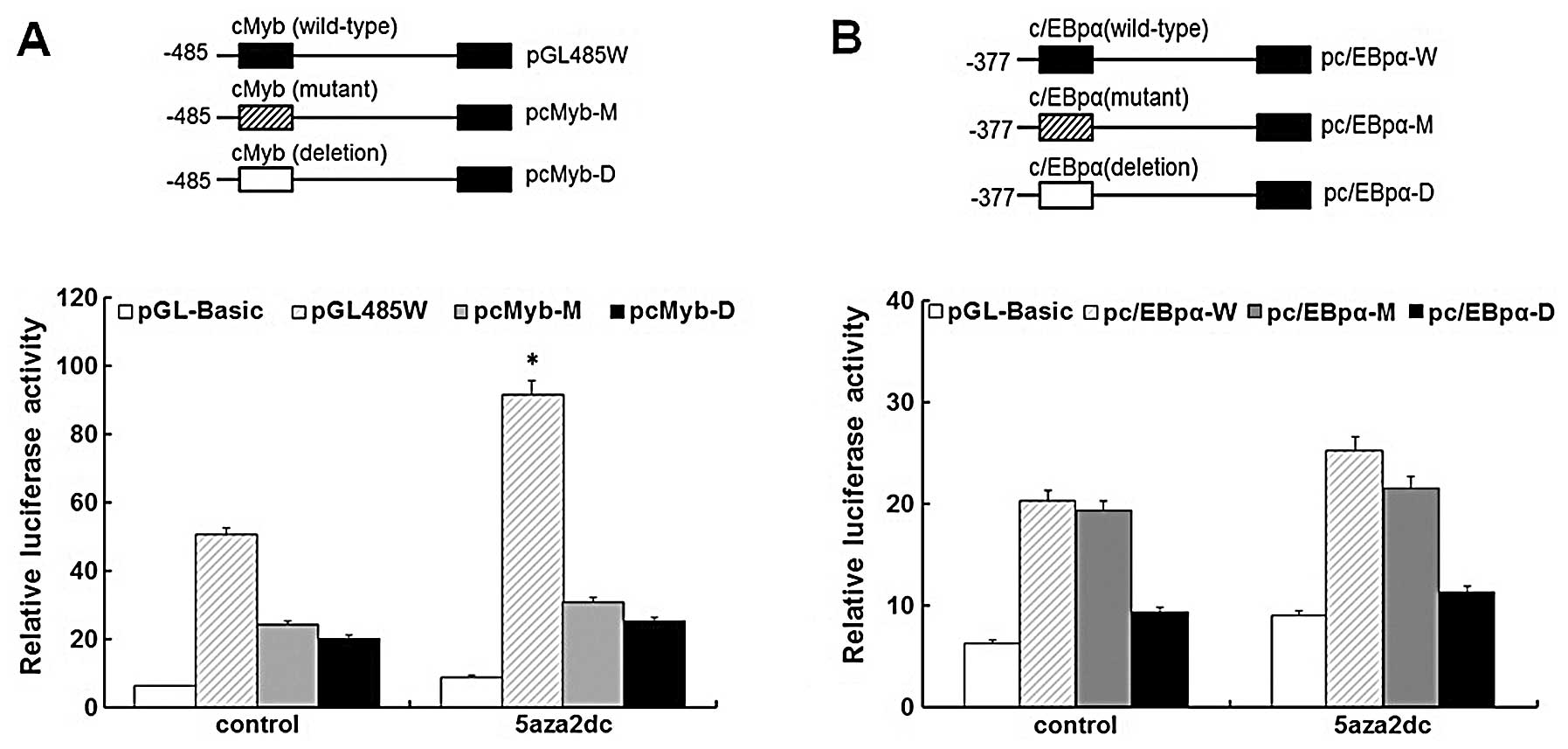
c-Myb is a major trans-acting element of
S100A4 in 5-Aza-induced HEp2 cells
To simulate the effects of demethylation on
methylated sites within the c-Myb and C/EBPα response elements of
S100A4, we constructed the mutant and deletion reporter
vectors of c-Myb and C/EBPα motifs corresponding to the methylated
sites, named pcMyb-M, pcMyb-D, pC/EBPα-M and pC/EBPα-D,
respectively. These vectors were then transiently transfected into
HEp2 cells to evaluate their relative reporter gene activities. The
mutant and deletion fragments (pcMyb-M/pcMyb-D) showed a
significant decrease in the reporter activities compared to the
wild-type construct in the 5-Aza-treated and -untreated groups
(P<0.05), confirming that the mutation and deletion constructs
interfere with the binding of c-Myb to S100A4 (Fig. 4A). Fig.
4A also showed the significant difference in the luciferase
activities of the reported genes between 5-Aza-treated and
-untreated HEp2 cells transfected by the wild-type construct, which
implies that the overexpression of the endogenous c-Myb gene in
HEp2 cells induced by 5-Aza may lead to the overexpression of the
reporter genes in the 5-Aza-treated group. As shown in Fig. 4B, no significant luciferase activity
of the reporter genes on C/EBPα wild-type vectors was observed,
which indicates that 5-Aza does not induce the overexpression of
endogenic C/EBPα in HEp2 cells, even though the C/EBPα-binding
sites in the S100A4 promoter region −485 – +73 were
demethylated in HEp2 cells. There was also no significant
difference in luciferase activity of the reporter genes on C/EBPα
wild-type and mutant-type vectors between the 5-Aza-treated or
-untreated groups, which indicates that the mutant sites reflecting
the methylated ones in C/EBPα do not affect the binding of C/EBPα
to S100A4 in the HEp2 cells. Yet, the C/EBPα deletion vector
showed a significant decrease in the reporter activity when
compared to the wild-type and mutant-type vectors in the
5-Aza-treated and -untreated groups (P<0.05), which suggests
that the C/EBPα response element but not its cytosine methylation
status plays an important role in the transcriptional regulation of
the S100A4 gene.
Discussion
S100A4 is overexpressed in many cancer types
including laryngeal carcinoma, which indicates that this gene plays
an important role in carcinogenesis. As demonstrated in our
previous study, there is a strong correlation between
overexpression and hypomethylation of S100A4 in laryngeal
cancer tissues (15). In addition,
similar phenomena are found in endometrial, pancreatic ductal,
colon and ovarian carcinomas (17–19).
At present, little is known concerning the molecular mechanisms
behind the transcriptional regulation of S100A4
expression-related hypomethylation. In ovarian cancer, the
upregulation of S100A4 expression is associated with
hypomethylation of CpG sites in the first intron of S100A4,
and hypoxia can reduce the methylation of hypoxia-response elements
(HRE) of the S100A4 gene in a time-dependent fashion, in
association with the increased binding of HIF-1α to a
methylation-free HRE (20).
Therefore, we speculate that the methylation-free sites in the
S100A4 promoter region can increase the binding of the
corresponding transcription factors to S100A4 in laryngeal
cancer.
In the present study, we cloned and characterized
over 1.2 kb (−1283 to +73) region lying upstream of the human
S100A4 transcription start site to determine the major
cis-acting elements, particularly those related to the
methylation status in HEp2 cells. As a result, we revealed the
existence of potential positive and negative cis-acting
elements in the corresponding regions of the S100A4
promoter. Further investigation was focused on the region −485 –
+73 as the methylation status of the region significantly affected
the expression of S100A4. Notably, the cytosines in the
region were methylated in the HEp2 cells, and 5-Aza altered the
methylation status (Fig. 2A and B).
In this region, there were several putative cis-acting
elements such as c-Myb, C/EBPα, Ap2 and Msx-1. EMSA and ChIP
results confirmed the binding of c-Myb and C/EBPα to S100A4.
Comparing the luciferase activity of the reporter genes on the
mutant, deletion and wild-type vectors in the HEp2 cells treated by
5-Aza or not, we found that only c-Myb promoted S100A4
expression in the methylation-free HEp2 cells. We thus speculated
that the overexpression of endogenous c-Myb in HEp2 cells induced
by 5-Aza plays a partial role in the S100A4 expression in
the methylation-free HEp2 cells. Nevertheless, the c-Myb response
element in the region −485 – +73 is the major element in the
regulation of S100A4 expression in HEp2 cells when the
methylation status is changed. The c-Myb proto-oncogene product
(c-Myb) plays an important role in the proliferation of
immature hematopoietic, colonic epithelial, and mammary epithelial
cells and also in the differentiation of hematopoietic cells
(21–24). The identified target genes of
c-Myb include c-myc, Bcl-2 and Gbx2, which are involved in
cell cycle control, blockage of apoptosis, and growth and
differentiation control, respectively (25–28).
Presently, no related study on the association of c-Myb with
the development of laryngeal cancer has been reported. Similar to
our study, Wang et al found that methylation of the CpG site
in the WNT5A promoter influences its binding to c-Myb and
5-Aza increased the expression of WNT5A, which suggests that
binding of c-Myb to WNT5A is methylation-sensitive and is inhibited
by the methylation of the CpG site in prostate cancer cells
(29). Campanero et al found
that methylation of the E2F elements in the c-Myc and c-Myb
promoters significantly inhibited the binding of E2F1, and
suggested that methylation of the c-Myb element from the
S100A4 promoter may affect its binding to c-Myb (30). In the present study, S100A4
was confirmed as a novel target of c-Myb and C/EBPα, which enriched
the regulatory network in which c-Myb and C/EBPα were involved.
Fang et al found that 663 transcripts including c-Myb and
S100A4 were regulated by CypB knockdown, and that many of these
gene products contributed to cell proliferation, cell motility and
tumorigenesis, which indicates that the co-overexpression of
c-Myb and S100A4 participates in the pathogenesis of
breast cancer (31). Whether CypB
directly regulates c-Myb and S100A4, respectively, or regulates
S100A4 via c-Myb is not known.
In conclusion, in the present study, we first
revealed that c-Myb and C/EBPα can target the S100A4 gene
through binding the corresponding cis-acting motifs in the
sequence −485 – +73. The overexpression of the S100A4 gene
is caused by demethylation of the c-Myb motif and possible
overexpression of the c-Myb gene in methylation-free HEp2
cells. Whether the overexpression of the c-Myb gene in the
5-Aza-induced HEp2 cells was caused by the demethylation of
c-Myb gene itself or of other upstream genes regulating
c-Myb or both warrants further investigation. The
identification and functional study of potential negative response
elements in the S100A4 promoter region −486 – −530 will be
the focus of subsequent research by our group.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81172577 and 81301766), and the
Natural Science Foundation of Liaoning Province (20092110). J.L.
obtained the grant (81301766), which supports the study of S100A4
on the invasiveness of laryngeal cancer and was approved on August,
2013.
References
|
1
|
Ragin CC, Modugno F and Gollin SM: The
epidemiology and risk factors of head and neck cancer: a focus on
human papillomavirus. J Dent Res. 86:104–114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li DW, Gao S, Shen B and Dong P: Effect of
apoptotic and proliferative indices, P-glycoprotein and survivin
expression on prognosis in laryngeal squamous cell carcinoma. Med
Oncol. 28(Suppl 1): S333–S340. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holgersson G, Ekman S, Reizenstein J, et
al: Molecular profiling using tissue microarrays as a tool to
identify predictive biomarkers in laryngeal cancer treated with
radiotherapy. Cancer Genomics Proteomics. 7:1–7. 2010.PubMed/NCBI
|
|
4
|
Zou J, Yang H, Chen F, et al: Prognostic
significance of fascin-1 and E-cadherin expression in laryngeal
squamous cell carcinoma. Eur J Cancer Prev. 19:11–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Waha A, Felsberg J, Hartmann W, et al:
Frequent epigenetic inactivation of the chaperone SGNE1/7B2
in human gliomas. Int J Cancer. 131:612–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilson IM, Davies JJ, Weber M, et al:
Epigenomics: mapping the methylome. Cell Cycle. 5:155–158. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malone CS, Miner MD, Doerr JR, et al:
CmC(A/T)GG DNA methylation in mature B cell lymphoma
gene silencing. Proc Natl Acad Sci USA. 98:10404–10409.
2001.PubMed/NCBI
|
|
9
|
Esteller M: Cancer epigenetics: DNA
methylation and chromatin alterations in human cancer. Adv Exp Med
Biol. 532:39–49. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Waraya M, Yamashita K, Katoh H, et al:
Cancer specific promoter CpG Islands hypermethylation of HOP
homeobox (HOPX) gene and its potential tumor suppressive role in
pancreatic carcinogenesis. BMC Cancer. 12:3972012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abildgaard MO, Borre M, Mortensen MM, et
al: Downregulation of zinc finger protein 132 in prostate cancer is
associated with aberrant promoter hypermethylation and poor
prognosis. Int J Cancer. 130:885–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lleras RA, Adrien LR, Smith RV, et al:
Hypermethylation of a cluster of Krüppel-type zinc finger protein
genes on chromosome 19q13 in oropharyngeal squamous cell carcinoma.
Am J Pathol. 178:1965–1974. 2011.
|
|
13
|
Rosty C, Ueki T, Argani P, et al:
Overexpression of S100A4 in pancreatic ductal
adenocarcinomas is associated with poor differentiation and DNA
hypomethylation. Am J Pathol. 160:45–50. 2002.
|
|
14
|
Choi JY, James SR, Link PA, et al:
Association between global DNA hypomethylation in leukocytes and
risk of breast cancer. Carcinogenesis. 30:1889–1897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Guo Y, Fu S, Yang M, Sun KL and Fu
WN: Hypomethylation-induced expression of S100A4 increases
the invasiveness of laryngeal squamous cell carcinoma. Oncol Rep.
23:1101–1107. 2010.
|
|
16
|
Burke E and Barik S: Megaprimer PCR:
application in mutagenesis and gene fusion. Methods Mol Biol.
226:525–532. 2003.PubMed/NCBI
|
|
17
|
Xie R, Loose DS, Shipley GL, Xie S,
Bassett RL Jr and Broaddus RR: Hypomethylation-induced expression
of S100A4 in endometrial carcinoma. Mod Pathol.
20:1045–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sato N, Maitra A, Fukushima N, et al:
Frequent hypomethylation of multiple genes overexpressed in
pancreatic ductal adenocarcinoma. Cancer Res. 63:4158–4166.
2003.PubMed/NCBI
|
|
19
|
Nakamura N and Takenaga K: Hypomethylation
of the metastasis-associated S100A4 gene correlates with
gene activation in human colon adenocarcinoma cell lines. Clin Exp
Metastasis. 16:471–479. 1998. View Article : Google Scholar
|
|
20
|
Horiuchi A, Hayashi T, Kikuchi N, et al:
Hypoxia upregulates ovarian cancer invasiveness via the binding of
HIF-1α to a hypoxia-induced, methylation-free hypoxia response
element of S100A4 gene. Int J Cancer. 131:1755–1767.
2012.PubMed/NCBI
|
|
21
|
Mucenski ML, McLain K, Kier AB, et al: A
functional c-myb gene is required for normal murine fetal
hepatic hematopoiesis. Cell. 65:677–689. 1991.
|
|
22
|
Malaterre J, Carpinelli M, Ernst M, et al:
c-Myb is required for progenitor cell homeostasis in colonic
crypts. Proc Natl Acad Sci USA. 104:3829–3834. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Drabsch Y, Hugo H, Zhang R, et al:
Mechanism of and requirement for estrogen-regulated MYB
expression in estrogen-receptor-positive breast cancer cells. Proc
Natl Acad Sci USA. 104:13762–13767. 2007.PubMed/NCBI
|
|
24
|
Bender TP, Kremer CS, Kraus M, Buch T and
Rajewsky K: Critical functions for c-Myb at three checkpoints
during thymocyte development. Nat Immunol. 5:721–729. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakagoshi H, Kanei-Ishii C, Sawazaki T,
Mizuguchi G and Ishii S: Transcriptional activation of the
c-myc gene by the c-myb and B-myb gene
products. Oncogene. 7:1233–1240. 1992.
|
|
26
|
Frampton J, Ramqvist T and Graf T: v-Myb
of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis
in myeloid cells. Genes Dev. 10:2720–2731. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taylor D, Badiani P and Weston K: A
dominant interfering Myb mutant causes apoptosis in T cells. Genes
Dev. 10:2732–2744. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kowenz-Leutz E, Herr P, Niss K and Leutz
A: The homeobox gene GBX2, a target of the myb
oncogene, mediates autocrine growth and monocyte differentiation.
Cell. 91:185–195. 1997.
|
|
29
|
Wang Q, Williamson M, Bott S, et al:
Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer.
Oncogene. 26:6560–6565. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Campanero MR, Armstrong MI and Flemington
EK: CpG methylation as a mechanism for the regulation of E2F
activity. Proc Natl Acad Sci USA. 97:6481–6486. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang F, Flegler AJ, Du P, Lin S and
Clevenger CV: Expression of cyclophilin B is associated with
malignant progression and regulation of genes implicated in the
pathogenesis of breast cancer. Am J Pathol. 174:297–308. 2009.
View Article : Google Scholar : PubMed/NCBI
|