Introduction
Breast cancer is the most common malignant disease
and accounts for 14% of female cancer-related deaths worldwide each
year. Indeed, although diagnostic and therapeutic methods have
greatly improved over the last decade, this cancer remains the
leading cause of cancer-related death among females (1). Therefore, it is essential to develop
more effective methods for its early diagnosis and treatment.
MicroRNAs (miRNAs) are a small class of non-coding
RNAs which regulate gene expression and may play pivotal roles in
the physiological and pathological processes in a variety of
eukaryotic organisms (2). miRNAs
achieve their effects through base-pairing with the 3′-untranslated
region (3′-UTRs) of target mRNAs, which can lead to the
translational repression or mRNA degradation (3,4). It is
known that miRNAs are involved in tumor cell proliferation,
migration, invasiveness and metastasis (5). Moreover, aberrant miRNA expression has
been frequently observed in various types of human tumors. These
reports suggest that miRNAs may function as either tumor-suppressor
genes or oncogenes (6). In human
breast cancer, several miRNAs such as let-7 (7), miR-155 (8) and miR-200 have been shown to be
dysregulated (9). A recent study
showed that miR-195 is significantly downregulated in breast cancer
(10). However, the role that
miR-195-5p plays in the carcinogenesis of breast cancer is still
largely unknown.
Cyclin E1 (CCNE1) belongs to the cyclin family
which, through association with cyclin-dependent kinase 2, controls
the progression of the cell cycle by driving cells from the G1 to
the S phase (11). Previous studies
have shown that CCNE1 is aberrantly expressed and may function as
an oncogene in many types of human cancers (12,13).
Furthermore, significant evidence indicates that breast cancer
patients with higher levels of CCNE1 show a higher mortality rate
when compared with those bearing low CCNE1 levels (14–16).
Thus, CCNE1 has attracted increasing research interest.
In the present study, we initially demonstrated that
the expression level of miR-195-5p in breast cancer specimens was
significantly lower than that in adjacent normal tissues. Cell
functional studies further showed that overexpression of miR-195-5p
inhibited proliferation and colony formation ability, suppressed
cell migration and caused G1 phase arrest by targeting CCNE1 in
MDA-MB-231 breast cancer cells. Therefore, our study suggests that
miR-195-5p may act as a tumor suppressor, and thus may be
considered a potential therapeutic and diagnostic target of breast
cancer.
Materials and methods
Specimens
A total of 40 breast cancer specimens and matched
adjacent normal breast tissues were surgically obtained from
patients at the Department of General Surgery of the Shanghai Tenth
People’s Hospital. Collection of the patient specimens was approved
by the Institutional Ethics Committee of Tongji University. These
samples were snap frozen in liquid nitrogen. All samples were
confirmed as invasive, ductal breast cancer by pathologists. None
of the patients received radiotherapy or chemotherapy prior to
surgery.
Cell culture and transfection
Human MDA-MB-231 breast cancer and HEK293T cells
were purchased from the Chinese Science Institute (Shanghai,
China). The cells were cultured in Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both
from Gibco, USA), penicillin (100 U/ml) and streptomycin (100
μg/ml) (Enpromise, Hangzhou, China) at 37°C with 5% CO2
in saturated humidity. Cells in the logarithmic growth phase (~80%
confluence) were selected for the experiments. Those with over 95%
viability as shown by trypan blue staining were qualified for
further experiments.
miR-195-5p mimics and non-specific negative control
(NC) oligos were purchased from GenePharma (Shanghai, China). The
sequence of the miR-195-5p mimic was 5′-UAGCA GCACAGAAAUAUUGGC-3′
and the sequence of the NC mimic was
5′-UCACAACCUCCUAGAAAGAGUAGA-3′. For transfection, MDA-MB-231 cells
(2×105) were added into each well of a 6-well plate and
cultured with serum- and antiobiotic-free DMEM. When the cell
density achieved 30–40% confluence, Lipofectamine transfection
reagent (Invitrogen, USA) was used to introduce the mimics
according to the manufacturer’s instructions. The ratio of mimics
to Lipofectamine was 1 μg to 3 μl.
miRNA isolation and quantitative
polymerase chain reaction (qPCR)
miRNAs were extracted from the tissues using the
miRcute microRNA isolation kit (Tiangen, Beijing, China) according
to the manufacturer’s instructions. The expression level of
miR-195-5p was detected by the One-Step qRT-PCR method (EzOmics
SYBR qPCR kit). The miR-195-5p primer, U6 primer and EzOmics SYBR
qPCR kit were purchased from Biomics Biotechnologies Inc. (Jiangsu,
China). The U6 primer used as an internal control was:
5′-GTCCTATCCAGTGCAGGGTCC GAGGTGCACTGGATACGACAAAATATGGAAC-3′
(stem-loop primer) 5′-TGCGGGTGCTCGCTTCGCAGC-3′ (sense) and
5′-CCAGTGCAGGGTCCGAGGT-3′ (antisense). Briefly, for amplification
of miR-195-5p, 100 ng RNA was used in a 25-μl reaction system
containing 12.5 μl 2X Master Mix, 0.5 μl 50X SYBR-Green, 0.5 μl
reverse transcription primer (10 μM), 0.5 μl sense and 0.5 μl
antisense primers (10 μM). One Step PCR parameters for miRNA
quantification were as follows: 37°C for 60 min for reverse
transcription, 10 min at 95°C, and then 40 cycles of 20 sec at
95°C, 30 sec at 62°C and 30 sec at 72°C. Each sample was tested in
triplicate.
For RNA analysis, total RNA was isolated from the
cultured cells using TRIzol reagent (Invitrogen, USA) according to
the manufacturer’s instructions. For CCNE1 mRNA detection, reverse
transcription was performed using the PrimeScript RT-PCR kit
(Takara, Shiga, Japan). Real-time PCR was performed using a 7900HT
Fast RT-PCR instrument (Applied Biosystems, Singapore) using
SYBR-Green. GAPDH mRNA levels were used for normalization. The
primer sequences were as follows: CCNE1, 5′-GTGTGGGAGCCAGCCTTG-3′
(sense) and 5′-ATCATCTTCTTTGTCAGGTGTGG-3′ (antisense); GAPDH,
5′-AAGGTCGGAGTCAACGGATT-3′ (sense) and 5′-CTGGAAGATGGTGATGGGATT-3′
(antisense). The PCR parameters for relative quantification were as
follows: 5 min at 94°C, followed by 30 cycles of 30 sec at 94°C, 45
sec at 57°C and 45 sec at 72°C. Each sample was tested in
triplicate, and the fold-change of mRNA expression was calculated
using the 2−ΔΔCt method (17).
Cell proliferation
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT)] assay
Cells were plated at 3,000/well in 96-well plates
(BD Biosciences, USA) and incubated at 37°C. When the cells reached
30–40% confluence, they were transfected with either 50 or 100 nM
miR-195-5p mimics or NC mimics. One group of cells was treated with
lipofectamine alone as a mock control. We then assessed cell
proliferation at 24, 48, 72 and 96 h post-transfection using the
MTT assay. Briefly, 20 μl (5 mg/ml) MTT (Sigma, USA) solution was
added to each well. After a 4-h incubation at 37°C, the supernatant
was removed and 150 μl DMSO was added. After 10 min of agitation
(100 rpm), the absorbance at 490 nm of each sample was measured by
a microplate spectrophotometer. Each experiment was performed in
triplicate and included 6 replicates.
Colony formation assay
Three hundred cells of each group (miR-195-5p, mock
and NC) were plated in a 6-well plate in complete medium. After
incubation at 37°C with 5% CO2 for 7–10 days, or when
the colonies were visible by viewing with the eye, the culture was
terminated. Complete medium was removed, and the plates were washed
twice in phosphate-buffered saline (PBS). The colonies were fixed
in 95% ethanol for 10 min, dried and stained with 0.1% crystal
violet solution for 10 min. Next, each plate was washed three times
with water, and the number of colonies was counted only if the well
contained >50 cells. The experiment was performed three
times.
Transwell migration assay
The Transwell migration assay was performed to
evaluate cell migration ability. First, the filters (Corning,
Lowell, MA, USA) were washed with serum-free DMEM, and placed into
a 24-well plate. The lower chambers contained DMEM with 10% FBS.
For the upper chambers, 3×104 cells were resuspended in
200 μl DMEM with 0.1% BSA. Plates were then incubated at 37°C in 5%
CO2. After 20 h, the cells that migrated through the
membranes were fixed with methanol and stained with crystal violet.
Images of six randomly selected fields-of-view were captured, and
the cells were counted.
Cell cycle assay
miR-195-5p (100 nM), mock and NC cells were
harvested at 48 h after transfection, centrifuged at 1,200 rpm for
10 min and washed three times with cold PBS. Ice-cold 70% ethanol
was subsequently added dropwise, and the cells were fixed at 4°C
overnight. After a 30-min digestion in RNase (0.1 g/l), a total of
250 μl (0.05 g/l) propidium iodide (PI) staining solution was added
to each sample which was then incubated for 30 min at room
temperature (RT) in the dark. The cell cycle was then analyzed by a
flow cytometer (FACSCanto™ II; BD Biosciences).
Dual-luciferase reporter assay
293T cells were seeded in 12-well plates (BD, USA)
in complete medium and incubated at 37°C with 5% CO2.
CCNE1 3′-UTR were cloned into the psiCHECK-2 vector, and
co-transfected with miR-195-5p mimics (100 nM) or NC mimics when
cells reached 80–90% confluence. Thirty-six hours after
transfection, luciferase activity was measured by the
dual-luciferase reporter assay kit (Promega, USA). Briefly, the
cells were washed twice with PBS then lysed by incubation at RT for
15 min with passive lysis buffer (PLB). The supernatants were
collected, and 20 μl of the aliquots was added to 96-well plates.
The firefly luciferase (FL) reporter was measured immediately after
adding Luciferase Assay Reagent II (LAR II). Next, 100 μl of Stop
& Glo® reagent was added to each well to initiate
the Renilla luciferase (RL). The psiCHECK-2 vector that
provides constitutive expression of FL was co-transfected as an
internal control. All experiments were performed three times.
Western blot analysis
Cell protein was extracted using RIPA lysis buffer.
The supernatant was quantified by bicinchoninic acid assay (Pierce,
USA). Next, 25 μg of protein samples was denatured with 5X sodium
dodecyl sulfate (SDS) loading buffer at 95°C for 5 min.
Subsequently, whole protein samples were separated by 10%
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred
onto 0.45-μm nitrocellulose membranes (Beyotime). Following 1 h of
blocking in 5% fat-free milk, the membranes were incubated with the
CCNE1 antibody (1:1,000) and the β-actin antibody (1:1,000) (both
from Epitomics, USA) overnight at 4°C. Blots were then washed and
incubated for 1 h with secondary antibodies. After washing with
PBST, immunoreactive protein bands were detected using the Odyssey
scanning system (LI-COR, Lincoln, NE, USA).
Statistical analysis
Data from at least three separate experiments are
presented as the means ± standard error of the mean (SEM). The
two-tailed t-test was used to draw a comparison between groups.
Differences were considered significant for P-values <0.05.
Results
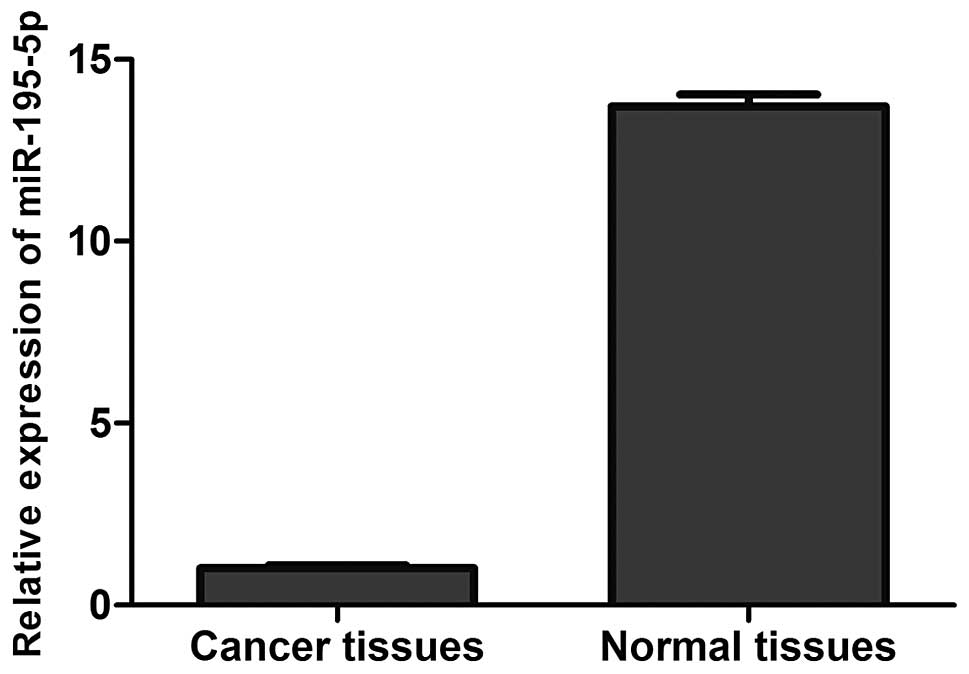
miR-195-5p expression is decreased in
breast cancer specimens
We measured the mRNA expression levels of miR-195-5p
in breast cancer specimens and the adjacent normal tissues by
real-time PCR. As shown in Fig. 1,
compared with the adjacent normal tissues, miR-195-5p expression
was significantly decreased in the breast cancer specimens
(P<0.05).
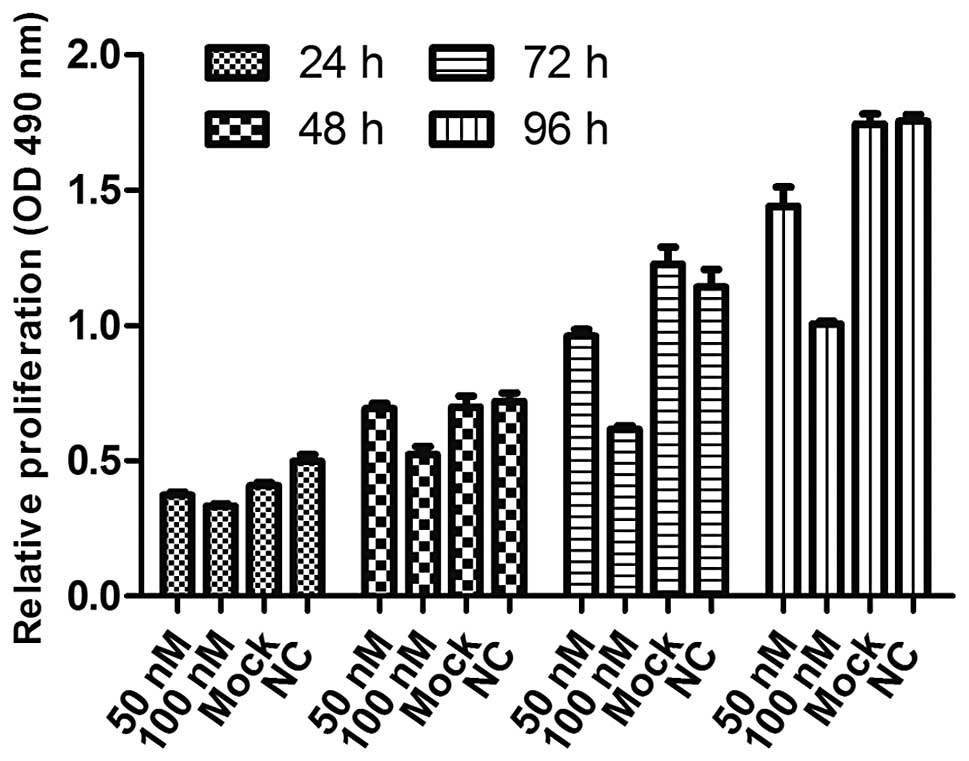
Overexpression of miR-195-5p in
MDA-MB-231 cells inhibits cell proliferation and colony formation
ability
The viability of cells transfected with either 50 or
100 nM miR-195-5p mimics was measured and compared with the mock
and NC transfected cells at 24, 48, 72 and 96 h post-transfection.
We found that the viability of both miR-195-5p mimic groups was
consistently significantly lower than the mock and NC groups in a
time- and dose-dependent manner (Fig.
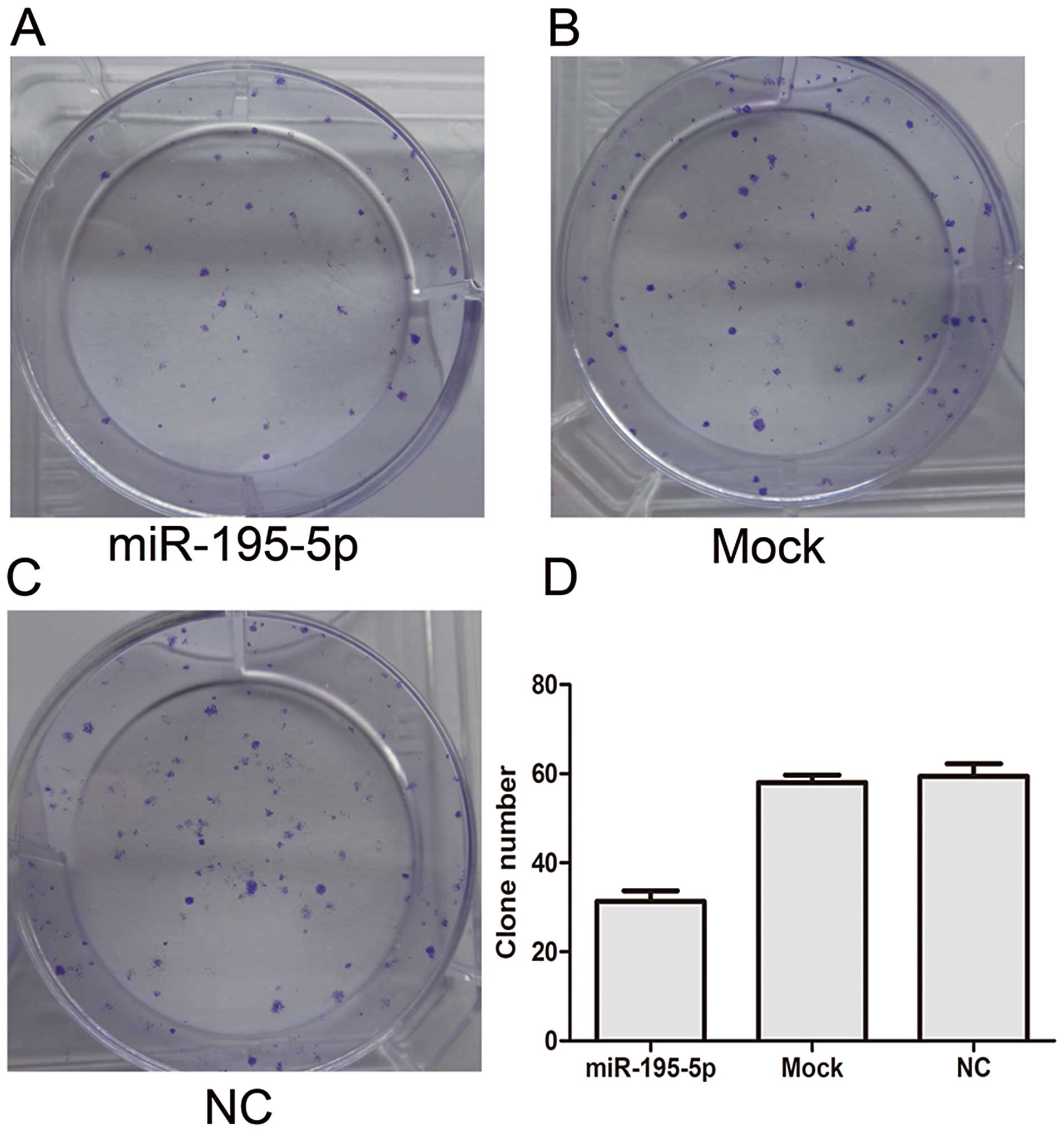
2). Thus, 100 nM was used in the following experiments. As
shown in Fig. 3, the 100 nM
miR-195-5p group exhibited fewer colonies than the mock and NC
groups as determined by the colony formation assay. These results
suggest that transient overexpression of miR-195-5p suppresses the
proliferation and colony formation ability of MDA-MB-231 cells.
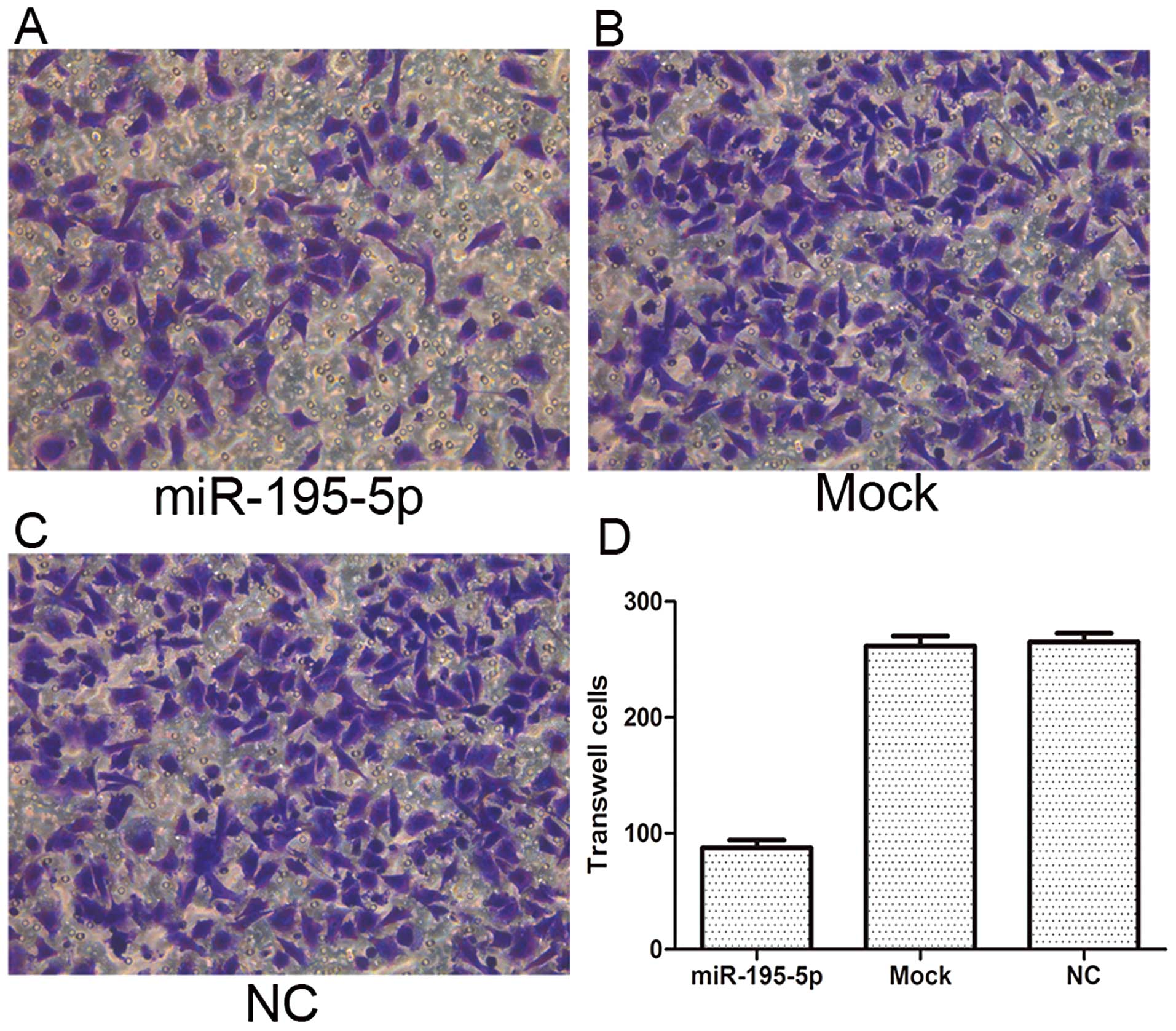
Overexpression of miR-195-5p in
MDA-MB-231 cells inhibits cell migratory ability
The cell migratory ability of the MDA-MB-231 cells
with and without transfection of miR-195-9p mimics was detected by
Transwell migration assay. Our results showed that 20 h after
transfection, the number of migrating cells in the miR-195-5p group
was significantly less than that in either the mock or NC groups
(P<0.05). These data suggest that the migratory ability of
MDA-MB-231 cells may be inhibited by miR-195-5p (Fig. 4).
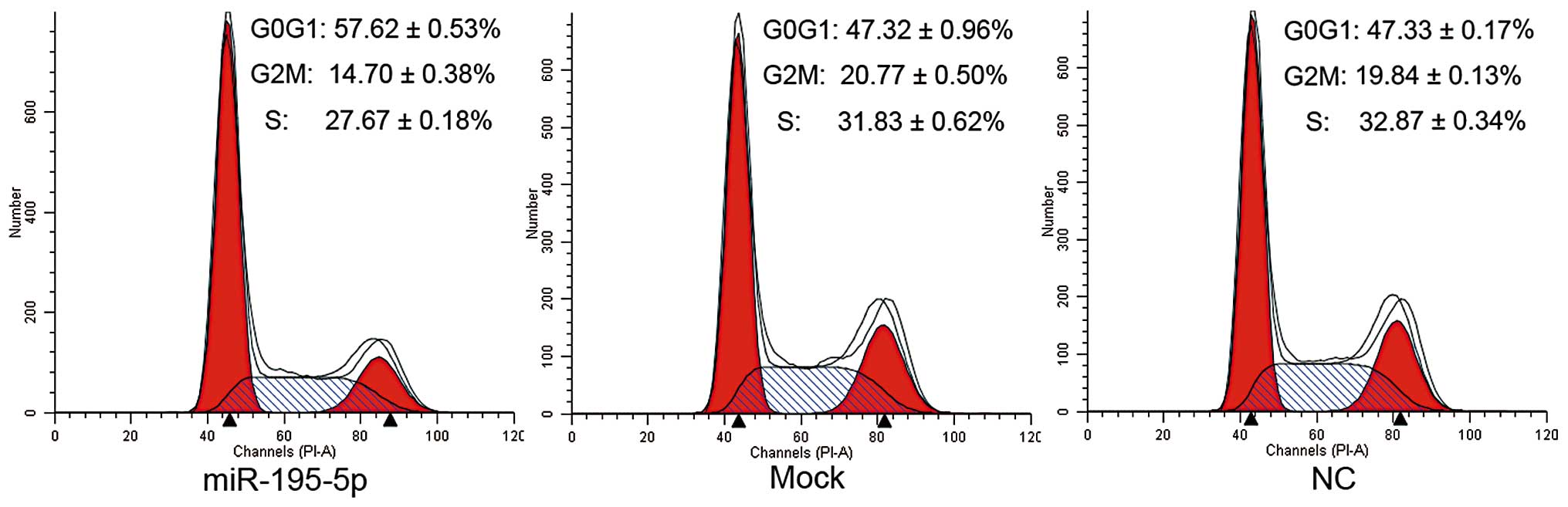
Overexpression of miR-195-5p initiates G1
phase arrest of MDA-MB-231 cells
The cell cycle distribution of the MDA-MB-231 cells
with and without transfection of miR-195-5p mimics was analyzed by
flow cytometry. As shown in Fig. 5,
the percentage of cells remaining in the G1 phase in the miR-195-5p
overexpression group (57.62±0.53%) was significantly greater than
that of the mock (47.32±0.96%) and NC groups (47.33±0.17%,
P<0.05); while the proportion of G2 and S phase cells decreased
in the miR-195-5p group compared with those of the mock and NC
groups (P<0.05). These results indicate that the overexpression
of miR-195-5p prevents cells from entering the S phase through
initiation of G1 phase arrest in MDA-MB-231 cells.
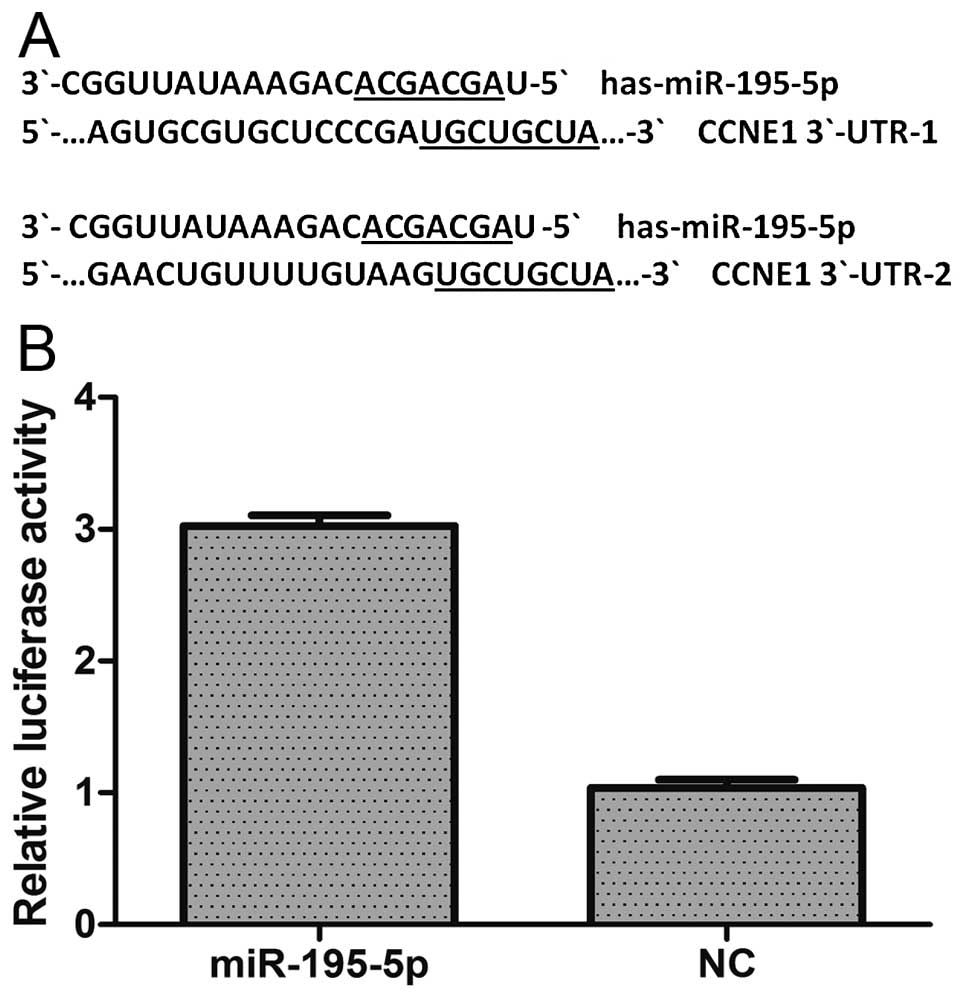
miR-195-5p regulates CCNE1 expression by
targeting its mRNA in MDA-MB-231 cells
To validate the possibility that miR-195-5p may
target CCNE1, we initially searched for putative targets within its
mRNA sequence using three bioinformatic algorithms: miRanda,
TargetScan and miRBase. We found two potential binding sites for
miR-195-5p which were located 247–254 and 485–492 bp downstream
from the 5′ end of the CCNE1 3′-UTR (Fig. 6A). Next we constructed a
psiCHECK-2/CCNE1 3′-UTR vector, which contained the Renilla
luciferase (RL) gene and the 3′-UTR region of CCNE1. This construct
was transfected into 293T cells together with either miR-195-5p or
NC mimics, and the luciferase activity was analyzed. The ratio of
FL/RL was calculated, and showed that the miR-195-5p group had an
~3-fold higher activity than that of the NC group (P<0.05)
(Fig. 6B). These results suggest
that miR-195-5p directly interacts with the CCNE1 3′-UTR in the
psiCHECK-2 reporter plasmid and leads to the degradation of RL
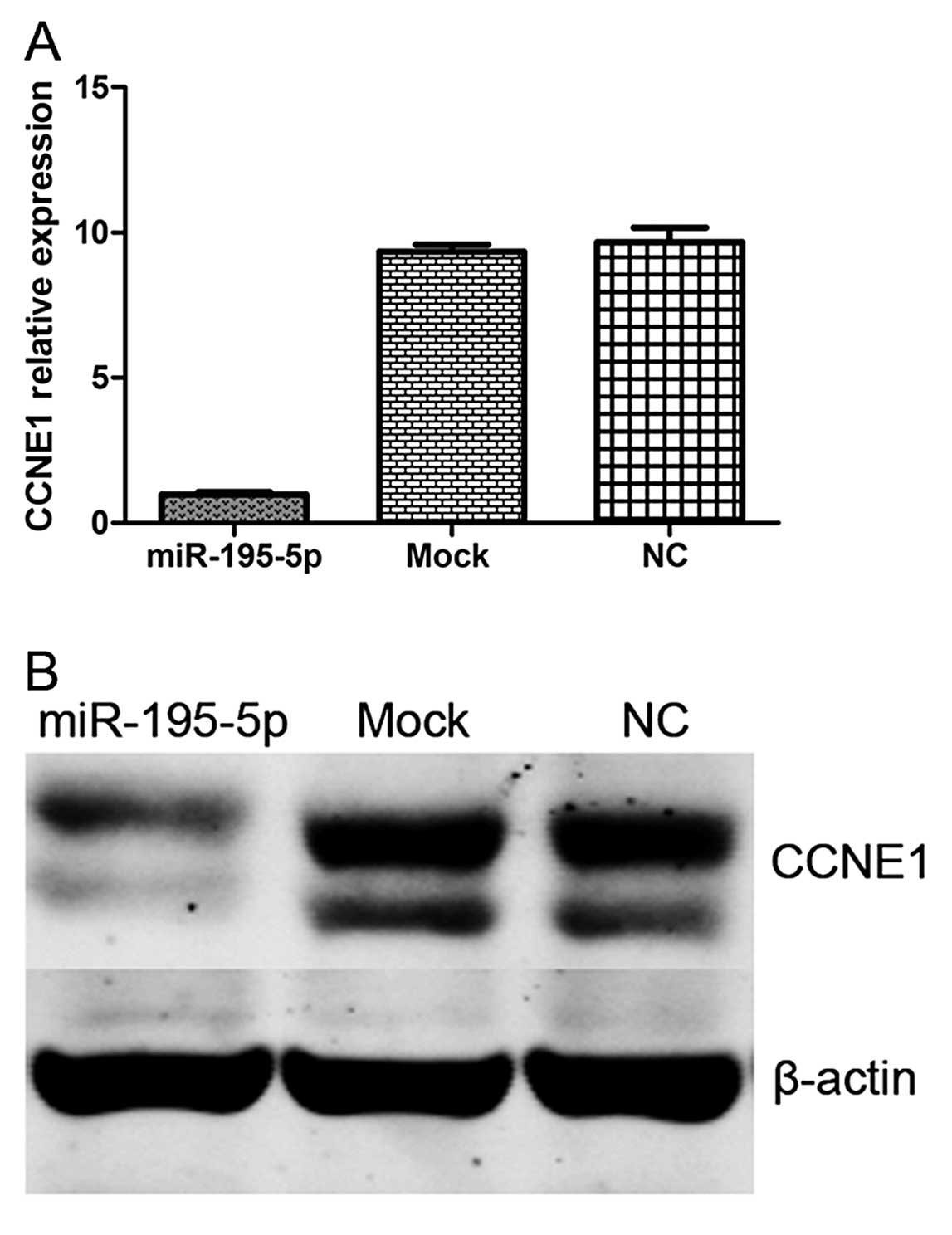
mRNA. Finally, we performed qPCR and western blot analysis of CCNE1
expression in MDA-MB-231 cells with and without transfection of
miR-195-5p mimics, or controls. We demonstrated that overexpression
of miR-195-5p significantly decreased CCNE1 expression at both the
mRNA and protein levels (Fig. 7).
These data further indicate that CCNE1 is a target of
miR-195-5p.
Discussion
In addition to surgery and traditional
chemotherapeutic drugs, several molecularly targeted drugs have
been developed for the treatment of breast cancer. The drugs that
have been assessed for the treatment and prevention of breast
cancer, such as raloxifene, letrozole and exemestane, in
preclinical and clinical studies (18), have resulted in a decline in the
incidence rate of breast cancer. Over the last decades, miRNA
research has become a ‘hot spot’ for research. Recent advances
suggest that dysregulation of miRNAs is a common event in human
cancers (19–22) and that they may thus act as key
regulators of carcinogenesis. Based on these findings, it has been
proposed that more effective targets or targeted drugs for
diagnosing and treating breast cancer may involve miRNAs.
In the present study, we examined the expression of
miR-195-5p in human breast cancer and its potential role in
carcinogenesis. First, through qPCR, we found that the expression
level of miR-195-5p in breast cancer specimens was significantly
lower than that in adjacent normal tissues. This suggests that the
expression of miR-195-5p is associated with the development of
breast cancer, and that it may function as a tumor suppressor.
Indeed, the expression of miRNA-195, which is closely related to
miRNA-195-5p, has been previously reported to be decreased in human
breast cancer. Meanwhile, upregulation of miR-195 expression has
been shown to suppress cell proliferation and invasion by targeting
the Raf-1 and Ccnd1 genes in both ZR-75-30 and MCF7 human breast
cancer cells (10). miRNA-195
includes miRNA-195-5p and miRNA-195-3p, and together they belong to
the miRNA-15 family.
In the present study, we transfected miR-195-5p
mimics into MDA-MB-231 cells to generate its overexpression. This
exogenous overexpression of miR-195-5p significantly inhibited
proliferation and colony formation ability of MDA-MB-231 cells as
measured by MTT and colony formation assays, respectively.
Moreover, cell migration ability was also significantly reduced by
overexpression of miR-195-5p in the MDA-MB-231 cells. Furthermore,
by flow cytometry we found that overexpression of miR-195-5p
prevented cells from entering the S phase and instead caused an
accumulation of cells in the G1 phase.
To ascertain why miR-195-5p exhibited these effects
on the cell function, we investigated putative targets of
miR-195-5p and identified CCNE1, which drives cells from the G1 to
the S phase. Based on three databases, we found that the CCNE1
3′-UTR contains two miR-195-5p matching sites. Notably, the
interaction between miR-195-5p and CCNE1 mRNA has not been
previously reported. To test whether CCNE1 was a real target of
miR-195-5p, we constructed a psiCHECK-2 plasmid containing the
3′-UTR of CCNE1 (psiCHECK-2/CCNE1 3′-UTR). Through dual-luciferase
assays, we confirmed that CCNE1 was a direct target of miR-195-5p.
Additionally, we found that the mRNA and protein levels of CCNE1
were significantly reduced in miR-195-5p-overexpressing cells when
compared with those transfected with either mock or NC, thus
further indicating that CCNE1 is a direct target of miR-195-5p.
In summary, overexpression of miR-195-5p inhibited
the proliferation and colony formation ability, suppressed
migration and caused G1 phase arrest by targeting CCNE1 in
MDA-MB-231 breast cancer cells. All of the data suggest that
miR-195-5p is a tumor suppressor that may inhibit carcinogenesis in
human breast cancer. Therefore, miR-195-5p may be a potential
diagnostic and therapeutic target for breast cancer.
Acknowledgements
This research was supported by the National Natural
Sciences Foundation of China for the project 81272240. Furthermore,
we give special thanks to all the teachers at the Central
Laboratory of the Shanghai Tenth People’s hospital for their
technical assistance.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Harris L, Fritsche H, Mennel R, et al:
American Society of Clinical Oncology 2007 update of
recommendations for the use of tumor markers in breast cancer. J
Clin Oncol. 25:5287–5312. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Santa F, Iosue I, Del Rio A and Fazi F:
microRNA biogenesis pathway as a therapeutic target for human
disease and cancer. Curr Pharm Des. 19:745–764. 2013.PubMed/NCBI
|
|
4
|
Sung H, Jeon S, Lee KM, et al: Common
genetic polymorphisms of microRNA biogenesis pathway genes and
breast cancer survival. BMC Cancer. 12:1952012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: the implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu F, Yao H, Zhu P, et al: let-7
regulates self renewal and tumorigenicity of breast cancer cells.
Cell. 131:1109–1123. 2007. View Article : Google Scholar
|
|
8
|
Mattiske S, Suetani RJ, Neilsen PM and
Callen DF: The oncogenic role of miR-155 in breast cancer. Cancer
Epidemiol Biomarkers Prev. 21:1236–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Radisky DC: miR-200c at the nexus of
epithelial-mesenchymal transition, resistance to apoptosis, and the
breast cancer stem cell phenotype. Breast Cancer Res. 13:1102011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li D, Zhao Y, Liu C, et al: Analysis of
MiR-195 and MiR-497 expression, regulation and role in breast
cancer. Clin Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sauer K and Lehner CF: The role of cyclin
E in the regulation of entry into S phase. Prog Cell Cycle Res.
1:125–139. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakayama N, Nakayama K, Shamima Y, et al:
Gene amplification CCNE1 is related to poor survival and
potential therapeutic target in ovarian cancer. Cancer.
116:2621–2634. 2010.PubMed/NCBI
|
|
13
|
Mao L, Ding J, Perdue A, et al: Cyclin E1
is a common target of BMI1 and MYCN and a prognostic marker for
neuroblastoma progression. Oncogene. 31:3785–3795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Keyomarsi K, Tucker SL, Buchholz TA, et
al: Cyclin E and survival in patients with breast cancer. N Engl J
Med. 347:1566–1575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sgambato A, Camerini A, Collecchi P, et
al: Cyclin E correlates with manganese superoxide dismutase
expression and predicts survival in early breast cancer patients
receiving adjuvant epirubicin-based chemotherapy. Cancer Sci.
100:1026–1033. 2009. View Article : Google Scholar
|
|
16
|
Han JY, Wang H, Xie YT, et al: Association
of germline variation in CCNE1 and CDK2 with breast
cancer risk, progression and survival among Chinese Han women. PLoS
One. 7:e492962012.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCTmethod. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
den Hollander P, Savage MI and Brown PH:
Targeted therapy for breast cancer prevention. Front Oncol.
3:2502013.PubMed/NCBI
|
|
19
|
Huang G, Nishimoto K, Zhou Z, Hughes D and
Kleinerman ES: miR-20a encoded by the miR-17-92 cluster increases
the metastatic potential of osteosarcoma cells by regulating Fas
expression. Cancer Res. 72:908–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li B, Shi XB, Nori D, et al:
Down-regulation of microRNA 106b is involved in p21-mediated cell
cycle arrest in response to radiation in prostate cancer cells.
Prostate. 71:567–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tatarano S, Chiyomaru T, Kawakami K, et
al: Novel oncogenic function of mesoderm development candidate
1 and its regulation by MiR-574-3p in bladder cancer
cell lines. Int J Oncol. 40:951–959. 2012.PubMed/NCBI
|
|
22
|
Li LZ, Zhang CZ, Liu LL, et al: miR-720
inhibits tumor invasion and migration in breast cancer by targeting
TWIST1. Carcinogenesis. Nov 1–2013.(Epub ahead of print).
|