Introduction
MicroRNAs (miRNAs), which play important roles in
transformation and carcinogenesis, are potent post-transcriptional
regulators of protein-coding genes and function as tumor oncogenes
or suppressors (1,2). As a member of the oncomiRs, miR-222
has been reported to drive the oncogenesis of many types of
malignancies (3–5), and a recent study demonstrated that
co-suppression of miR-221/222 inhibits glioma cell growth by
targeting the 3′-untranslated region (3′UTR) of p27 mRNA in
vitro and in vivo (4).
However, the mode of action of miR-222 in carcinogenesis has not
been characterized, and the mechanism of the biological function of
miR-222 in oral squamous cell carcinoma (OSCC) remains unknown.
The p53 upregulated modulator of apoptosis (PUMA),
also called Bcl-2 binding component 3 (BBC3), was newly discovered
as a target for activation by p53 in 2001 and was found to possess
a powerful pro-apoptotic effect as a member of the Bcl-2 family
(6–8). Although the specific mechanisms for
inducing apoptosis require further investigation, PUMA is a
promising new target for gene therapy as its pro-apoptotic function
has achieved favorable results (1,2,9).
Studies suggest that PUMA is a direct target of
miR-222 which functions as an endogenous apoptosis regulator in
various common forms of human epithelial cancers (4,5).
However, little is known concerning the role of PUMA in the
targeted treatment of human OSCC. In the present study, we
identified miR-222 as a potent regulator of PUMA, a cell
proliferation inducer and tumor promoter. Inhibition of miR-222
appears to induce cell apoptosis and reduce cell growth and
directly upregulate PUMA expression by targeting the binding sites
in the 3′UTR. The results suggest that miR-222 may play important
roles in regulating the cell biological behavior of OSCC by
targeting PUMA.
Materials and methods
Cell culture and transfection
The OSCC cell lines Tca8113 and UM1 were cultured in
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal bovine serum (FBS), penicillin and streptomycin (100 μg/ml),
and incubated in a humidified 5% CO2 environment at
37°C. The 2′-OMe-oligonucleotides were synthesized and purified by
high-performance liquid chromatography by GenePharma Co. Ltd.
(Shanghai, China). The oligonucleotides were modified by 2′-OMe to
obtain the following sequences: hsa-miR-222 inhibitor (As-miR-222),
5′-ACCCAGUAGCCAGAUGUAG CU-3′; hsa-miR-222 mimics (Pre-miR-222), the
positive-sense strand, 5′-AGCUACAUCUGGCUACUGGGU-3′, and the
antisense strand, 5′-CCAGUAGCCAGAUGUAGCUUU-3′; and the empty
vector, 5′-CAGUACUUUUGUGUAGUACAA-3′. Pre-miR-222, As-miR-222 and
the empty vector were transfected using Lipofectamine™ RNAiMAX
(Invitrogen) according to the recommended protocol, and the
transfection medium was replaced with fresh medium 4–6 h later.
After treatment for 2 days, cells were divided into 4 groups: the
control, empty vector, Pre-miR-222 and As-miR-222 which were used
for subsequent analysis.
RNA preparation and reverse
transcription-polymerase chain reaction (RT-PCR)
After treatment for 48 h, Tca8113 and UM1 cells were
lysed using TRIzol reagent (Invitrogen), and total RNA was
extracted using an RNeasy Mini kit according to the manufacturer’s
instructions. RT-PCR was carried out using a reverse transcription
kit, and the human β-actin gene was used as the internal control.
The PUMA gene primers were as follows:
5′-TGTCGAATAAACGCTTTACAAAC-3′ (forward) and
5′-AACGTTTGTAATGATGGCTTCTG-3′ (reverse). The β-actin primers were
as follows: 5′-GGT CGGAGTCAACGGATTTGGTCG-3′ (forward) and 5′-CCT
CCGACGCCTGCTTCACCAC-3′ (reverse). Relative expression levels were
calculated using the 2−ΔΔCt method (10).
Western blot analysis
After transfection for 48 h, cells of every group
were washed with cold phosphate-buffered saline (PBS) three times
and lysed in lysis buffer. Cell lysates were centrifuged at 14,000
× g for 20 min at 4°C, and the protein concentration was determined
using the Enhanced BCA protein assay kit (Beyotime Institute of
Biotechnology, China) using bovine serum albumin (BSA) as the
standard. Equal amounts of crude protein were mixed with sodium
dodecyl sulphate (SDS) sample buffer and denatured and then
separated on 12% SDS-polyacrylamide gels. After electrophoresis,
gels were transferred to a PVDF membrane by electroblotting. The
membrane was blocked in Tris-buffered saline (TBST) (50 mM Tris-HCl
pH 7.5, 150 mM NaCl, 0.2% Tween) containing 5% skim milk for 1 h at
room temperature. The blots were incubated with the primary
antibodies against PUMA, Bcl-2 and GAPDH overnight at 4°C and
rinsed three times with TBST. The rinsed blots were then incubated
with the secondary antibody for 1 h and washed with TBST. The
membrane was developed by enhanced chemiluminescence and exposed to
AlphaView SA for 1–15 min. The bands of the specific proteins were
quantified after normalization with the density of GAPDH with
ImageJ instrument software.
Immunofluorescence
Tca8113 and UM1 cells were seeded in 24-well plates
with 12-mm-diameter coverslips. After incubation for at least 24 h,
the cells were treated with Pre-miR-222, As-miR-222 or empty vector
for the indicated times. Subsequently, cells were washed three
times with PBS (pH 7.4), fixed in 4% formaldehyde for 20 min and
permeabilized in 0.1% Triton X-100 for 30 min and sequentially
washed three times with PBS. The cells were then incubated with the
indicated primary antibodies (PUMA and Bcl-2) according to the
manufacturer’s instructions, and the primary antibodies were
diluted (1:250) in 1X PBS as recommended by the manufacturer. After
18 h, cells were washed three times with PBS and incubated for 1 h
with each of the corresponding secondary antibodies. Nuclei were
stained with the nuclear stain DAPI. After mounting with the
coverslips on the microscopic slides, cells were examined using
laser scanning confocal microscopy.
Cell migration assay
After transfection, OSCC cells in DMEM/F12 medium
containing 0.1% FBS were transferred to 8-μm pore inserts, and then
placed in companion wells which contained DMEM/F12 medium and 10%
FBS as a chemoattractant. The inserts were removed after a 12-h
incubation, and the non-migrating OSCC cells on the upper surface
were harvested. Cells on the lower surface were fixed and stained
and then counted under a microscope.
Cell viability assay
The OSCC Tca8113 and UM1 cell lines were seeded into
96-well plates with ~4,000 cells/well. After transfection as
described above, 10 μl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
(5 mg/ml) was added to each well at 12, 24, 36, 48, 60 and 72 h
after treatment and incubated for 4 h at 37°C. The supernatant was
discarded, and 100 μl of DMSO was added to each well to dissolve
the precipitate. Optical density (OD) was measured at the
wavelength of 570 nm. The data are expressed as means ± SD, derived
from quintuplicate samples of at least three independent
experiments.
Apoptosis assays
Treated and untreated cells were washed with cold
PBS twice and resuspended in buffer at a concentration of
106/ml. Cells were mixed with 5 μl of FITC-conjugated
Annexin V reagent and 5 μl of propidium iodide (PI). After a 15-min
incubation at room temperature in the dark, samples were analyzed
by flow cytometry.
Statistical analysis
The experimental data are presented as means ± SD
from at least triplicate analyses. Statistical analyses were
performed by one-way analysis of variance (ANOVA) or t-test using
SPSS software 17.0. Statistical significance was set at
P<0.05.
Results
Expression of miR-222 and PUMA in OSCC
cells
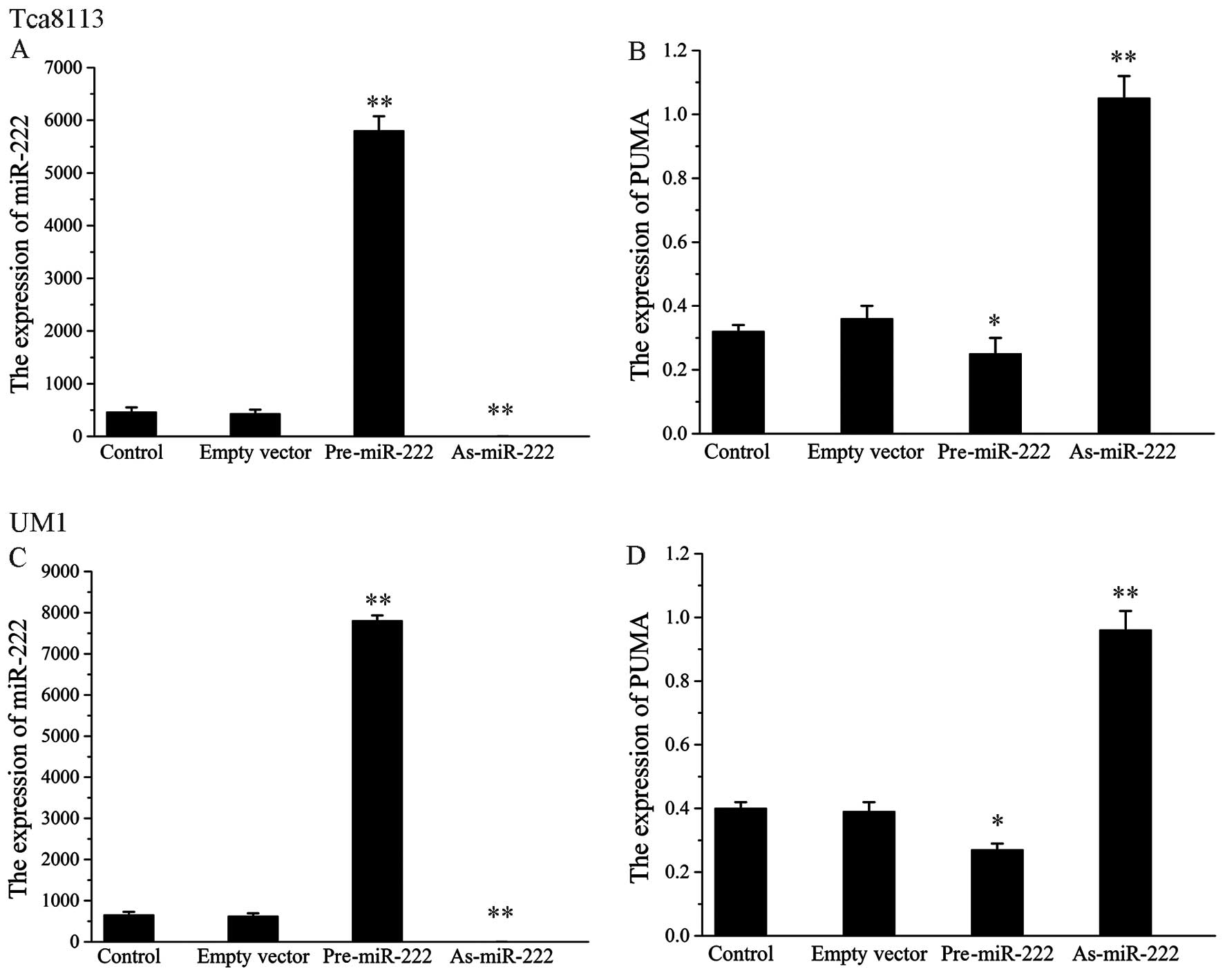
In the transfection groups of Tca8113 and UM1 cells,
As-miR-222 inhibited the expression of miR-222. In order to knock
down endogenous miR-222, chemically engineered oligonucleotides
were synthesized and transfected into Tca8113 and UM1 cells. RT-PCR
analysis showed that As-miR-222 efficiently and specifically
silenced endogenous miR-222. In contrast, the expression of the
PUMA gene in the As-miR-222 group was upregulated to a greater
extent when compared to the control group. However, cells
transfected with Pre-miR-222 exhibited an opposite trend of
expression. No significant differences between the empty vector
transfection group and the control group were noted. The results
showed that miR-222 was negatively correlated with PUMA expression
in the OSCC cell lines. U6 was present as a loading control in the
4 groups (Fig. 1).
Pre-miR-222 and As-miR-222 alter the
expression of apoptotic proteins (Fig.
2)
The expression of apoptosis-related proteins was
measured by western blot analysis in order to detect the molecular
mechanism of the involvement of miR-222 in the apoptosis of Tca8113
and UM1 cells. A significant increase in PUMA, one of the
pro-apoptosis-related proteins, was observed in the Tca8113 and UM1
cells in the As-miR-222 group. In contrast, the expression of Bcl-2
in the As-miR-222 group was significantly downregulated when
compared to that in the control group. The data indicate that
As-miR-222 induces cancer cell apoptosis through suppression of
PUMA and passivation of Bcl-2. Similarly, the Pre-miR-222 group
exhibited increased expression of Bcl-2 and decreased expression of
PUMA.
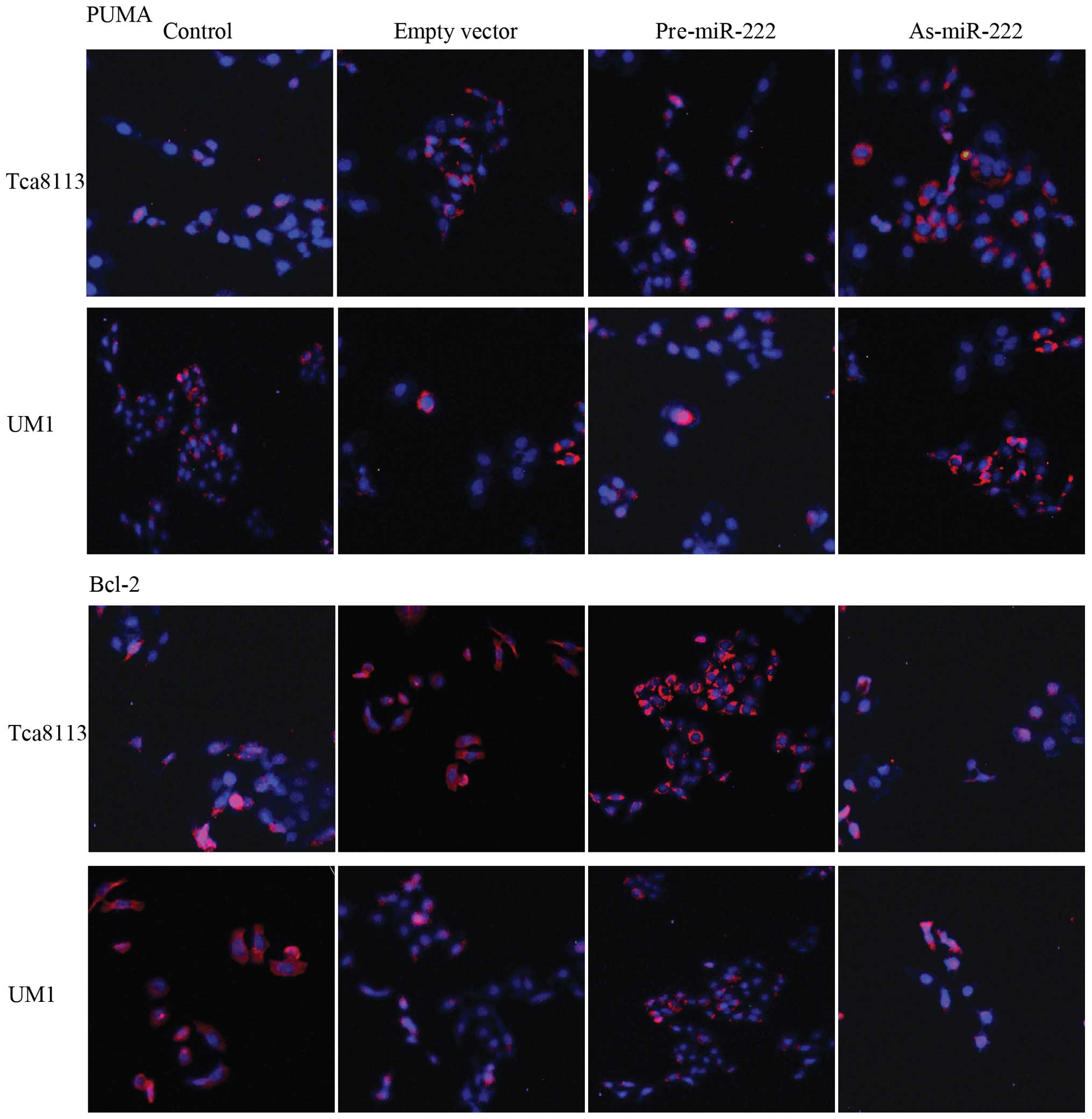
Determination of the expression of PUMA
and Bcl-2 in Tca8113 and UM1 cells
Since PUMA is one of the pro-apoptosis regulators,
overexpression of PUMA has been attributed to the induction of
apoptosis. Therefore, in order to determine the expression of PUMA
in human OSCC Tca8113 and UM1 cells, we performed
immunofluorescence staining and examined the cells under a laser
scanning confocal microscope (Fig.
3). Confocal images of OSCC cells after immunofluorescence
staining showed high red fluorescence of PUMA (revealed by
CY3-conjugate) in the As-miR-222 group, whereas, the Pre-miR-222
group exhibited relatively low red fluorescence suggesting weaker
expression of PUMA. In contrast, confocal images showed that the
expression of Bcl-2 in the As-miR-222 group was significantly
downregulated when compared to that in the control group. However,
in the Pre-miR-222 group the expression of Bcl-2 was upregulated.
The cell nuclei were stained for blue fluorescence (using DAPI
dye). Low expression of PUMA and high expression of Bcl-2 are
important characteristics of various types of cancer including
OSCC.
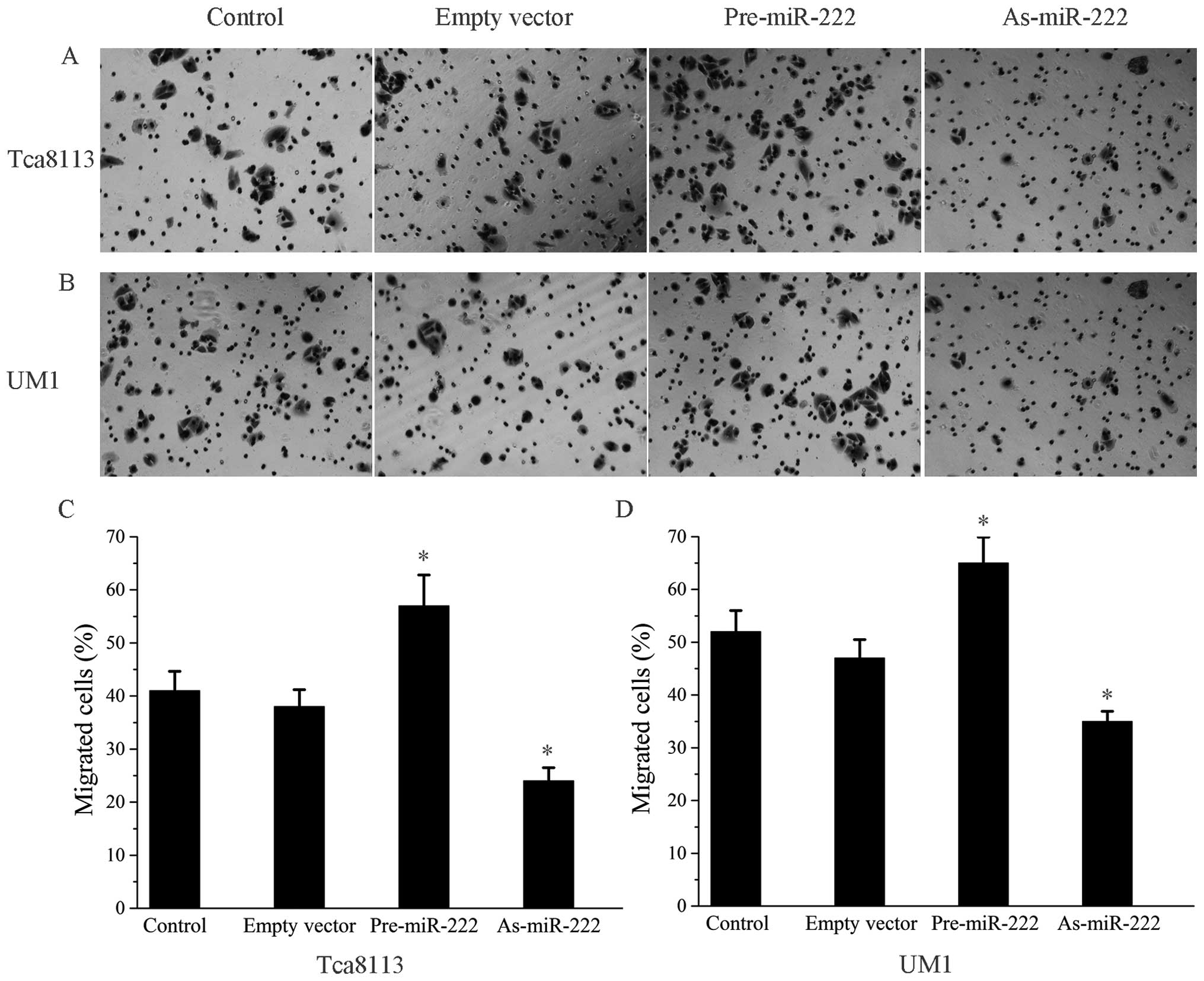
Effect of the alteration of miR-222
expression on the migratory ability of OSCC cells
We examined whether an alteration in miR-222
expression is involved in OSCC cell motility, as cell motility
plays an important role in metastasis. Cells transfected with
Pre-miR-222 or As-miR-222 migrated in a manner which had an obvious
difference to that of the control cells (Fig. 4), indicating that alteration of
miR-222 did affect the migratory ability of the OSCC cells.
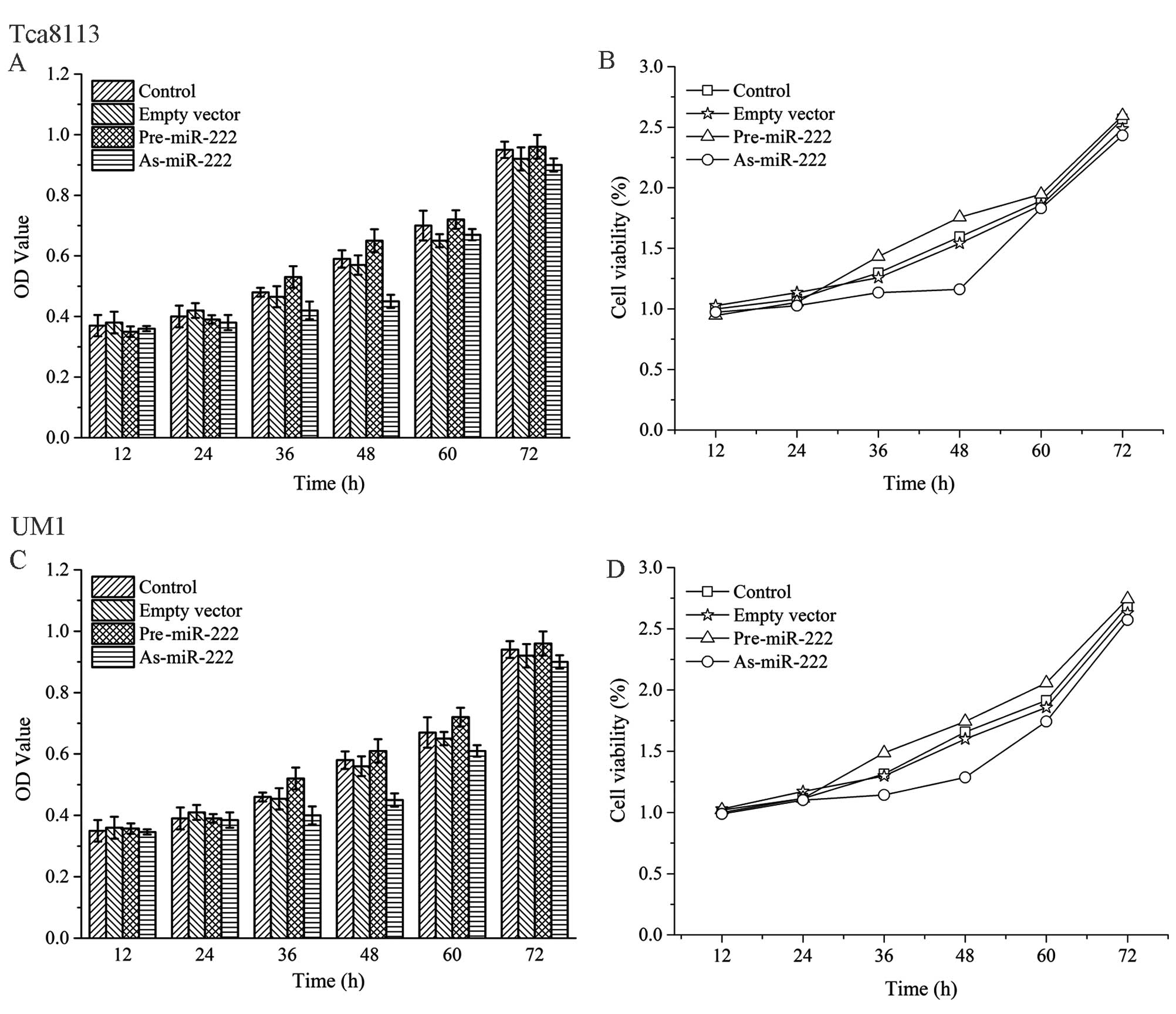
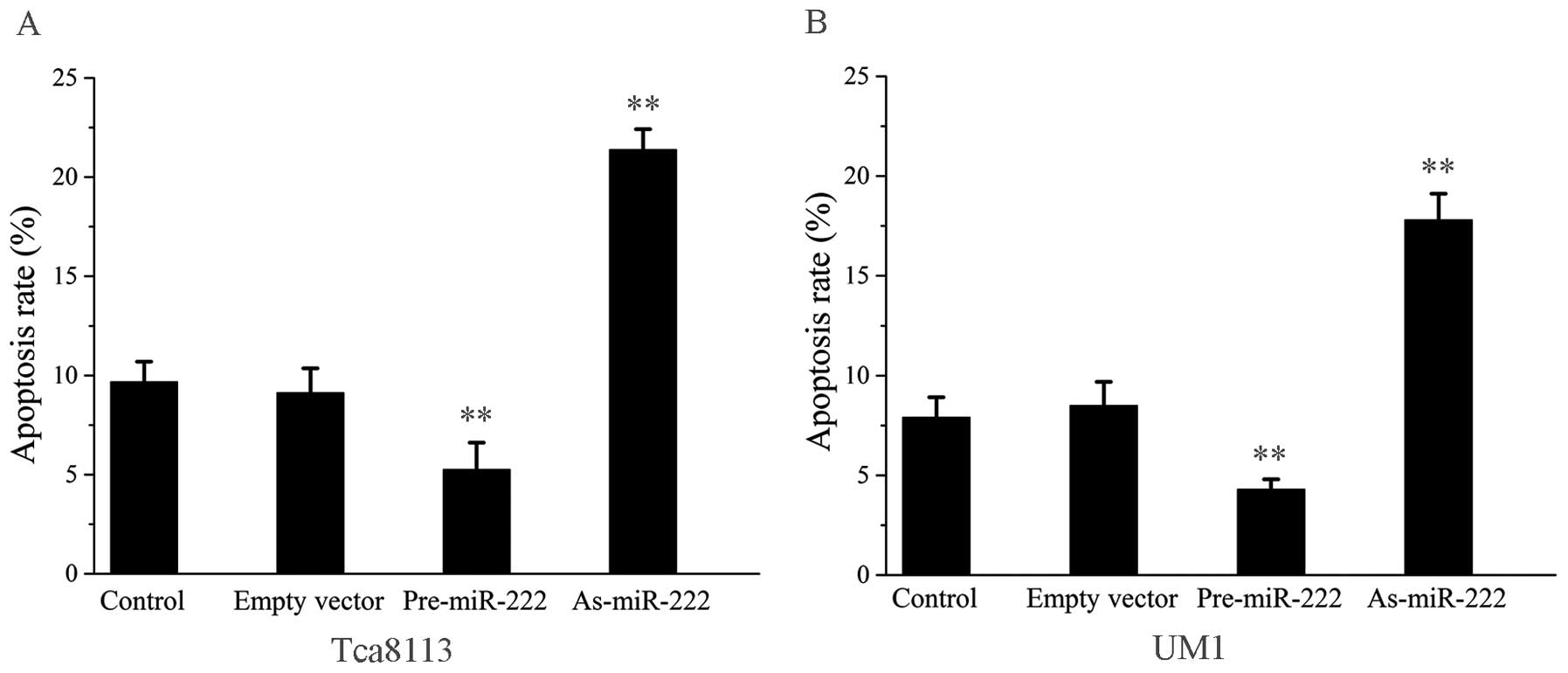
miR-222 influences cancer cell
proliferation and apoptosis
As shown in Fig. 5,
human OSCC Tca8113 and UM1 cells which were treated with As-miR-222
proliferated at a significantly lower rate than that in the other
three groups as evaluated by MTT assay. Apoptosis assays were used
to assess whether As-miR-222 inhibits cell proliferation through
the induction of cell apoptosis. Annexin V-PI analysis showed that
the percentage of apoptotic cells was significantly increased in
the cells treated with As-miR-222 as compared to the other three
groups, suggesting that apoptosis was obviously induced in cells
which were transfected with As-miR-222 (Fig. 6). In the Pre-miR-222 group, both
Tca8113 and UM1 cells showed enhanced proliferative ability and
decreased apoptosis. Thus, an alteration in miR-222 expression may
affect OSCC cell proliferation and apoptosis.
Discussion
Oral squamous cell carcinoma (OSCC) is one of the
most common types of cancer (11,12).
Despite improvements in the treatment of OSCC over the last few
years, new targeted therapies need to be developed as the survival
rate of OSCC patients in the last five years has remained poor
(13,14). Therefore, understanding the
mechanisms of OSCC and identifying new therapeutic options are
clinically significant. As a new strategy, gene therapy was
recently developed and has achieved beneficial results.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that negatively regulate gene expression at the
post-transcriptional level through association with the
3′-untranslated region (3′UTR) of protein-coding genes and
induction of translation inhibition. They play critical roles in
the control of cell proliferation, differentiation, apoptosis and
death (15–19) as well as in the process of OSCC and
head and neck squamous cell carcinoma (HNSCC) (20–22).
As a member of the oncomiRs, miR-222 has a seed sequence, which is
a short evolutionarily conserved region through with miRNAs bind
its target sites in mRNA 3′UTRs, indicating an important role in
coordinated regulation and function. Some studies have reported
that miR-222 may induce cell growth and cell cycle progression by
targeting p27 and p57 (3–5,23,24).
Moreover, several genes have been found to be common targets of
miR-222, such as p27, Bmf and PTEN (4,5,25). One
study showed that miR-222 may inhibit cell apoptosis in human
glioma cells by targeting the pro-apoptotic gene, PUMA (26,27).
In the present study, we report the modulating effect of miR-222 on
the pro-apoptotic gene PUMA by directly targeting the 3′UTR of PUMA
mRNA in OSCC (Tcal8113 and UM1) cell lines. These results may have
implications in the pathogenesis of OSCC.
PUMA, which is also named Bcl-2 binding component 3
(BBC3), is a newly discovered tumor suppressor which was induced by
the p53 tumor suppressor or other apoptotic stimuli and was found
to possess a powerful pro-apoptotic effect. PUMA belongs to the
BH3-only subfamily of the Bcl-2 protein family. This gene encodes
two BH3 domain-containing proteins (PUMA-α and PUMA-β), with
similar activities, and they bind to Bcl-2 and localize to the
mitochondria to induce cytochrome c release to promote
apoptosis (6,28). Similar to all the other BH3-only
proteins, PUMA promotes apoptosis through binding to and
neutralizing pro-survival members of the Bcl-2 family (28). PUMA induces apoptosis through both
p53-dependent and non-p53-dependent pathways and these features
make PUMA a particularly potent effector of apoptosis. In the
present study, upregulation of the pro-apoptosis protein PUMA and
downregulation of the apoptotic protein Bcl-2 were observed
following treatment with As-miR-222. Furthermore, bioinformatics
and luciferase assays showed that miR-222 may modulate PUMA
expression by directly targeting the binding site within the 3′UTR
(5). These findings suggest that
PUMA is probably directly regulated by miR-222.
The integration of various therapeutic strategies is
a trend in current gene therapy to induce synergistic effects
(29,30). In the present study, we demonstrated
that PUMA gene expression in OSCC cell lines was significantly
upregulated at both the mRNA and protein levels by transfection
with As-miR-222. We also found that As-miR-222 transfection may
obviously upregulate PUMA protein expression and significantly
inhibit the growth of Tca8113 and UM1 cells by reducing cell
proliferation and promoting cell apoptosis. However, cells
transfected with Pre-miR-222 exhibited an opposite trend of
expression, and no significant differences between the empty vector
transfection group and control group were noted. The results
indicate that there was a negative correlation between the
expression of PUMA and miR-222, and that the modulation of miR-222
activity may regulate the expression of the PUMA gene, thus
affecting the biological behavior of OSCC cells. PUMA is a novel
target of miR-222 and may be a critical therapeutic target for OSCC
intervention.
Acknowledgements
The present study was supported by the National
Natural Sciences Foundation of China (grant nos. 30973340 and
81272554), the Guangdong Sciences and Technology Project (grant
nos. 2011B050400030 and 2012B031800387), and the Guangdong Natural
Sciences Foundation (grant nos. 9151008901000187 and
S2011020003247).
References
|
1
|
Skaftnesmo KO, Prestegarden L, Micklem DR
and Lorens JB: MicroRNAs in tumorigenesis. Curr Pharm Biotechnol.
8:320–325. 2007. View Article : Google Scholar
|
|
2
|
Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu
Z and You Y: Hsa-mir-181a and hsa-mir-181b function as tumor
suppressors in human glioma cells. Brain Res. 1236:185–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
le Sage C, Nagel R, Egan DA, Schrier M,
Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA,
Farace MG and Agami R: Regulation of the p27Kip1 tumor
suppressor by miR-221 and miR-222 promotes cancer cell
proliferation. EMBO J. 26:3699–3708. 2007.
|
|
4
|
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao
P, Wang G, Zhang A, Jia Z, Han L and Jiang H: Co-suppression of
miR-221/222 cluster suppresses human glioma cell growth by
targeting p27kip1 in vitro and in vivo.
Int J Oncol. 34:1653–1660. 2009.PubMed/NCBI
|
|
5
|
Zhang C-Z, Han L, Zhang A-L, Fu Y-C, Yue
X, Wang G-X, Jin Z-F, Pu P-Y, Zhang Q-Y and Kang C-S: MicroRNA-221
and microRNA-222 regulate gastric carcinoma cell proliferation and
radioresistance by targeting PTEN. BMC Cancer. 10:3672010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakano K and Vousden KH: PUMA, a
novel proapoptotic gene, is induced by p53. Mol Cell. 7:683–694.
2001. View Article : Google Scholar
|
|
7
|
Avila JL, Grundmann O, Burd R and Limesand
KH: Radiation-induced salivary gland dysfunction results from
p53-dependent apoptosis. Int J Radiat Oncol Biol Phys. 73:523–529.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Labi V and Villunger A: PUMA-mediated
tumor suppression: a tale of two stories. Cell Cycle. 9:4269–4275.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang C, Zhang J, Zhang A, Wang Y, Han L,
You Y, Pu P and Kang C: PUMA is a novel target of miR-221/222 in
human epithelial cancers. Int J Oncol. 37:1621–1626.
2010.PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tran N, O’Brien CJ, Clark J and Rose B:
Potential role of micro-RNAs in head and neck tumorigenesis. Head
Neck. 32:1099–1111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chiang WF, Hung PS, Liu SY, Yuan TC, Chang
KW, Chen YP, Liu YC and Lin SC: Increase of ZASC1 gene copy
number in recurrent oral carcinoma. Oral Dis. 17:53–59. 2011.
|
|
13
|
Childs G, Fazzari M, Kung G, Kawachi N,
Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV,
Prystowsky MB, Belbin TJ and Schlecht NF: Low-level expression of
microRNAs let-7d and miR-205 are prognostic markers
of head and neck squamous cell carcinoma. Am J Pathol. 174:736–745.
2009.PubMed/NCBI
|
|
14
|
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY,
Wu KJ, Chiou SH, Lin SC and Chang KW: miR-31 ablates
expression of the HIF regulatory factor FIH to activate the HIF
pathway in head and neck carcinoma. Cancer Res. 70:1635–1644. 2010.
View Article : Google Scholar
|
|
15
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meister G and Tuschl T: Mechanisms of gene
silencing by double-stranded RNA. Nature. 431:343–349. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7
and lin-4 miRNAs results in target mRNA degradation. Cell.
122:553–563. 2005.
|
|
18
|
Giraldez AJ, Mishima Y, Rihel J, Grocock
RJ, Van Dongen S, Inoue K, Enright AJ and Schier AF: Zebrafish
miR-430 promotes deadenylation and clearance of maternal mRNAs.
Science. 312:75–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu L, Fan J and Belasco JG: MicroRNAs
direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA.
103:4034–4039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lo WL, Yu CC, Chiou GY, Chen YW, Huang PI,
Chien CS, Tseng LM, Chu PY, Lu KH, Chang KW, Kao SY and Chiou SH:
MicroRNA-200c attenuates tumour growth and metastasis of
presumptive head and neck squamous cell carcinoma stem cells. J
Pathol. 223:482–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lajer CB, Garnæs E, Friis-Hansen L,
Norrild B, Therkildsen MH, Glud M, Rossing M, Lajer H, Svane D,
Skotte L, Specht L, Buchwald C and Nielsen FC: The role of miRNAs
in human papilloma virus (HPV)-associated cancers: bridging between
HPV-related head and neck cancer and cervical cancer. Br J Cancer.
106:1526–1534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Medina R, Zaidi SK, Liu CG, Stein JL, van
Wijnen AJ, Croce CM and Stein GS: MicroRNAs 221 and 222 bypass
quiescence and compromise cell survival. Cancer Res. 68:2773–2780.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gramantieri L, Fornari F, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L and
Negrini M: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang C, Han L, Zhang A, Yang W, Zhou X,
Pu P, Du Y, Zeng H and Kang C: Global changes of mRNA expression
reveals an increased activity of the interferon-induced signal
transducer and activator of transcription (STAT) pathway by
repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol.
36:1503–1512. 2010.PubMed/NCBI
|
|
27
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ and
Kang CS: MiR-221 and miR-222 target PUMA to induce cell survival in
glioblastoma. Mol Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen L, Willis SN, Wei A, Smith BJ,
Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM and Huang DC:
Differential targeting of prosurvival Bcl-2 proteins by their
BH3-only ligands allows complementary apoptotic function. Mol Cell.
17:393–403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
John-Aryankalayil M, Palayoor ST, Cerna D,
Simone CB II, Falduto MT, Magnuson SR and Coleman CN: Fractionated
radiation therapy can induce a molecular profile for therapeutic
targeting. Radiat Res. 174:446–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marignol L, Robson T, McCarthy HO,
Worthington J, Murray MM, Hollywood D, Lawler M and Hirst DG: The
tissue plasminogen activator gene promoter: a novel tool for
radiogenic gene therapy of the prostate? J Gene Med. 9:1032–1038.
2008. View
Article : Google Scholar : PubMed/NCBI
|