Introduction
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer, comprising 90% of primary liver cancers
(1). In the past decade, HCC has
become one of the most frequent tumors and the most lethal cancer
worldwide (2). More than 80% of HCC
cases are from the Asian and African continents, and >50% of
cases are from mainland China with a vast majority of viral
hepatitis patients (3). Recently,
increasing trends in HCC incidence were reported from several
Western countries, including France, Australia and the USA
(3). Despite new advances in the
management of HCC, treatment of advanced-stage HCC remains a
challenge owing to the complex nature of the disease and the lack
of available therapies (4).
Long-term survival after hepatic resection remains very poor for
the majority of HCC patients who develop recurrence or metastasis
(5). Therefore, targeting invasion
and metastasis is an attractive strategy for HCC therapy. Exploring
the signaling pathways implicated in the pathogenesis of HCC is a
crucial step in the development of new therapeutic strategies for
HCC (6).
Chemokines are a class of small inflammatory or
homeostatic cytokines sharing a common biological activity in
stimulating the migration of different types of cells including
lymphocytes, monocytes, neutrophils, endothelial cells, mesenchymal
stem cells and malignant epithelial cells (7). Chemokines/chemokine receptor network
are critical in shaping the tumor immune response (8). The chemokine receptor CCR9 was
initially identified for its role in the immune system, responsible
for recruiting immune cells (9).
Activation of CCR9 by its selective ligand CCL25 is associated with
pancreatic cancer cell proliferation (10). CCR9 is functionally and
significantly expressed in breast cancer tissue and activation of
this receptor promotes breast tumor cell migration, invasion and
MMP expression, which are key components of breast cancer
metastasis (11). However, the role
of CCR9 in HCC remains unclear. In the present study, our aim was
to evaluate the expression of CCR9 in HCC and to investigate its
clinical significance and potential role in the invasion and
metastasis of HCC.
Materials and methods
Patient samples
A total of 240 cases of paraffin-embedded HCC
samples and their matched non-HCC liver tissues which had been
clinically and histologically diagnosed at the Shandong Provincial
Hospital Affiliated to Shandong University (Shandong, China) from
2000 to 2004 were used. The CCR9 expression from 240 HCC samples
was subjected to immunohistochemistry (IHC). All participants
underwent hepatectomy with a median age of 48 years (range 29–78
years). In addition, fresh specimens with vascular invasion (n=39)
and without vascular invasion (n=26) were also analyzed by
quantitative PCR (qPCR) and western blotting. Prior written consent
from patients and approval by the Institutional Ethics Committee
were obtained.
Follow-up
Follow-up data were obtained by reviewing the
medical records and from direct communication with patients.
Follow-up visits were scheduled postoperatively at intervals of one
month for six months, bimonthly for six months, quarterly for six
months, and semiannually for life. Complete follow-up, ranging from
0 to 83 months, was available for all patients and the median
survival was 34 months.
IHC
Immunohistochemical analysis was carried out to
study CCR9 protein expression in 240 human HCC tissues. In brief,
paraffin-embedded specimens were cut into 4-μm sections and baked
at 65°C for 30 min. The sections were deparaffinized with xylenes
and rehydrated. Sections were submerged into EDTA antigenic
retrieval buffer and microwaved for antigenic retrieval. The
sections were treated with 3% hydrogen peroxide in methanol to
quench the endogenous peroxidase activity, followed by incubation
with 1% bovine serum albumin to block nonspecific binding. Rabbit
anti-CCR9 (1:50; LifeSpan Biosciences) was incubated with the
sections overnight at 4°C. For negative controls, the rabbit
anti-CCR9 antibody was replaced with normal goat serum, or the
rabbit anti-CCR9 antibody was blocked with a recombinant CCR9
polypeptide by co-incubation at 4°C overnight preceding the
immunohistochemical staining procedure. After washing, the tissue
sections were treated with biotinylated anti-rabbit secondary
antibody, followed by further incubation with
streptavidin-horseradish peroxidase complex (both from LifeSpan
Biosciences). The tissue sections were immersed in
3-amino-9-ethylcarbazole and counterstained with 10% Mayer’s
hematoxylin, dehydrated and mounted in crystal mount.
The degree of immunostaining of formalin-fixed,
paraffin-embedded sections was reviewed and scored independently by
two observers, based on both the proportion of positively stained
tumor cells and the intensity of staining (12,13).
The proportion of tumor cells was scored as follows: 0 (no positive
tumor cells), 1 (<10% positive tumor cells), 2 (10–50% positive
tumor cells) and 3 (>50% positive tumor cells). The intensity of
staining was graded according to the following criteria: 0 (no
staining), 1 (weak staining = light yellow), 2 (moderate staining =
yellow brown) and 3 (strong staining = brown). The staining index
was calculated as staining intensity score x proportion of positive
tumor cells. Using this method of assessment, we evaluated the
expression of CCR9 by determining the staining index, which scores
as 0–4, 6 and 9. Cut-off values for CCR9 were chosen on the basis
of a measure of heterogeneity with the log-rank test statistical
analysis with respect to overall survival (OS). An optimal cut-off
value was identified: the staining index score of ≥4 was used to
define tumors as high CCR9 expression and ≤3 as low expression of
CCR9.
qPCR
Total RNA from cells and primary tumor materials was
extracted using the TRIzol reagent (Invitrogen) according to the
manufacturer’s instructions. The extracted RNA (5 μg) from each
sample was used for cDNA synthesis primed with random hexamers.
Real-time PCR primers were designed using the Primer Express v 2.0
software (Applied Biosystems). The primers were: CCR9 forward,
5′-GTGCCTCCCT GAGATCATGT-3′ and reverse, 5′-TGTGCTTTTGGCATCT
TTTG-3′; β-actin forward, 5′-GCTGTATTCCCCTCCATC GT-3′ and reverse,
5′-GCCATGTTCAATGGGGTACT-5′. For PCR amplification of CCR9 cDNA, an
initial amplification using CCR9-specific primers was performed
with a denaturation step at 95°C for 10 min, followed by 35 cycles
of denaturation at 95°C for 60 sec, primer annealing at 58°C for 30
sec and primer extension at 72°C for 30 sec. Upon completion of the
cycling steps, a final extension at 72°C for 5 min was performed
before the reaction was stored at 4°C. Expression data were
normalized to the geometric mean of housekeeping gene β-actin to
control the variability in expression levels. The results were
analyzed using the ΔΔCt method.
Western blot analysis
Total protein was extracted and determined by the
Bradford assay using a commercial kit purchased from the Bio-Rad
Laboratories. Equal quantities of protein were separated
electrophoretically on 10% SDS/polyacrylamide gels and transferred
onto polyvinylidene difluoride membranes (Roche). The membrane was
probed with a 1:500-diluted anti-CCR9 antibody (Abcam). Expression
of CCR9 was determined with horseradish peroxidase-conjugated
anti-rabbit IgG (1:2,000) and enhanced chemiluminescence (Pierce)
according to the manufacturer’s suggested protocols. The membranes
were stripped and reprobed with an anti-β-actin antibody (1:2,000;
Sigma) as a loading control.
Cell lines and vector construction
The cell lines HepG2, Huh7, HEP3B and HCCLM3 were
purchased from the American Type Culture Collection (ATCC) and were
grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum
(FBS) (HyClone). Overexpression of CCR9 human full-length CCR9 cDNA
was amplified by PCR and cloned into a pMSCV-puro retroviral vector
by OriGene Technologies (Rockville, MD, USA). For CCR9 knockdown,
shRNA sequences were cloned into the pSUPER-retro-puro plasmid to
generate pSUPER-retro CCR9 shRNA obtained from OriGene
Technologies.
Cell proliferation and colony formation
assay
The MTT assay was used to assess the cell
proliferation. Briefly, cells were seeded in 96-well plates at a
density of 2×103 cells/well. One plate was taken out at
the same time every day after the cells adhered. Twenty microliters
of MTT (5 mg/ml) were added to each well, and the cells were
incubated for another 4 h. The medium was removed and the formazan
precipitate was solubilized in 150 ml dimethyl sulfoxide. The
absorbance at 490 nm was measured using a microplate reader. All
experiments were performed in triplicate.
Regarding the colony formation, cells were seeded in
6-well plates (2×103 cells/plate) and cultured for 10
days. Subsequently, cells were fixed with ice-cold methanol for 10
min, followed by staining with 1% crystal violet for 1 min.
Cell cycle analysis using the BrdU
HCC cells were seeded on coverslips at a density of
5×104 cells and 24 h post-seeding, the cells were
incubated with BrdU for 1 h, followed by staining with anti-BrdU
antibody (Sigma) according to the manufacturer’s instructions
(Roche Diagnostics). Images were captured using a laser scanning
microscope (Olympus).
Anchorage-independent growth assay
The anchorage-independent growth ability was
determined in soft agar, as previously described (14). Briefly, 3 ml of 0.5% agar in basal
modified Eagle’s medium supplemented with 10% FBS was layered onto
each well of 6-well tissue culture plates. Cells (3×104
cells) suspended in 1 ml normal medium were mixed with 2 ml of 0.5%
agar-basal modified Eagle’s medium supplemented with 10% FBS, and 1
ml of mixture was added into each well over the top of the 0.5%
agar layer. Plates were incubated at 37°C in 5% CO2 for
2 weeks, and the colonies with >32 cells of each were
counted.
Statistical analysis
All statistical analyses were carried out using the
SPSS 17.0 statistical software package. The Mann-Whitney U test was
used to analyze the correlation between CCR9 expression and the
clinicopathological characteristics. Survival curves were plotted
using the Kaplan-Meier method and compared with the log-rank test.
The significance of various variables for survival was analyzed by
the Cox proportional hazards model in the univariate and
multivariate analyses. P<0.05 was considered to indicate a
statistically significant difference.
Results
CCR9 expression in HCC tissues
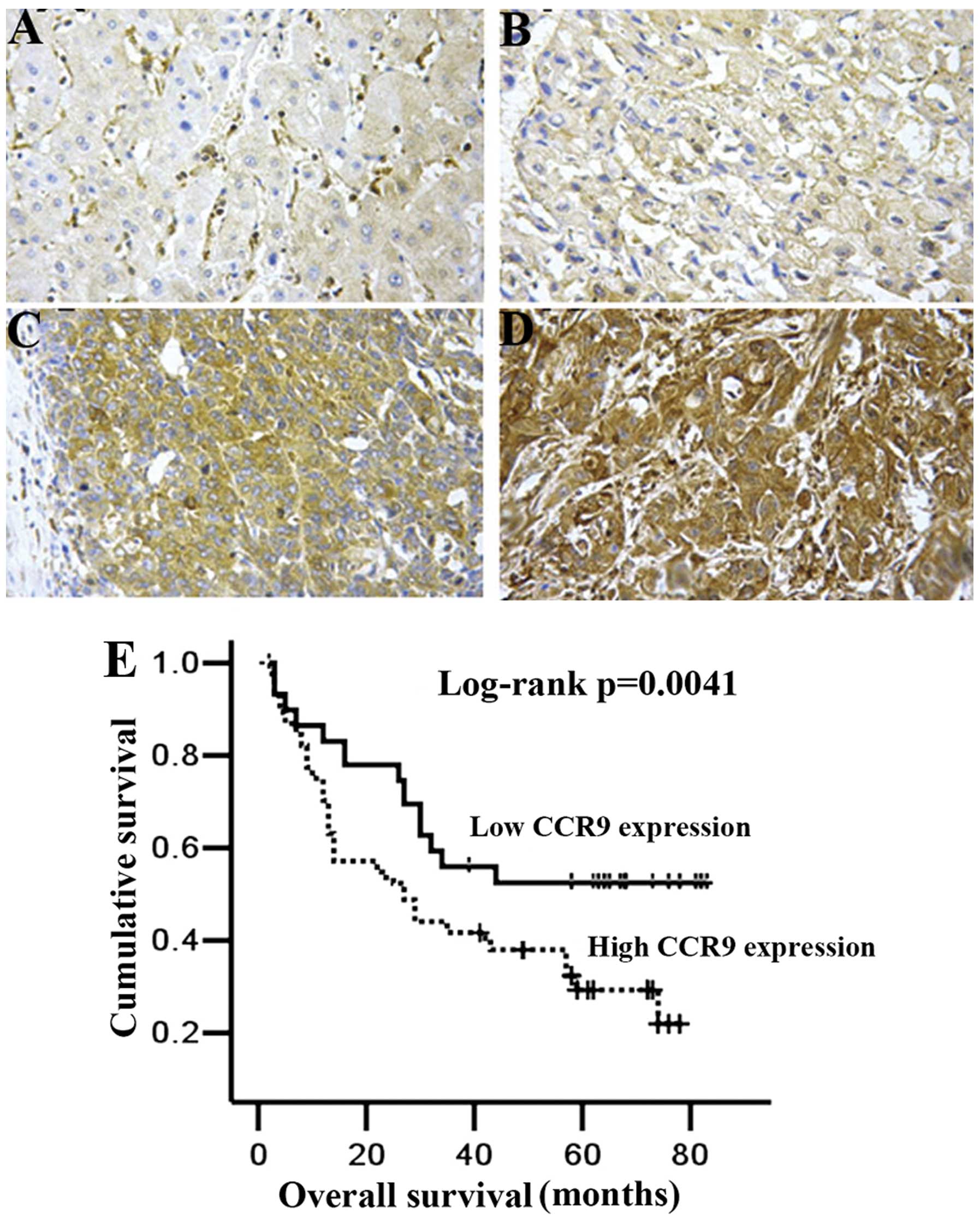
CCR9 expression was detected mainly in the cytoplasm
of tumor cells (Fig. 1). According
to the classification described in the Materials and methods,
immunohistochemical analysis clearly showed that high CCR9
expression was present in 55.8% (134 of 240) of HCC tissues. In the
non-HCC tissues, we observed high CCR9 protein expression only in
9.6% (23 of 240) (P<0.01). These results indicated that CCR9
expression was significantly elevated in HCC samples.
CCR9 expression correlates with
clinicopathological features and prognosis in HCC patients
Immunohistochemical analysis showed that the high
CCR9 levels correlated with tumor node number, high
Edmondson-Steiner grade and vascular invasion (Table I). As indicated in the Materials and
methods, 240 HCC tissues were divided into the group with high CCR9
expression (n=134) and the group with low CCR9 expression (n=106).
As a result, HCC patients with high CCR9 expression had markedly
reduced OS (P=0.0041; Fig. 1E)
compared with patients with low CCR9 expression. Multivariate
analysis showed that CCR9 was an independent prognostic factor for
the OS of HCC patients (Table II,
hazard ratio=2.35, 95% confidence interval=1.09–5.02, P=0.014).
These findings support that CCR9 plays an important role in
HCC.
 | Table ICorrelation between CCR9 expression
and clinicopathological features in hepatocellular carcinoma. |
Table I
Correlation between CCR9 expression
and clinicopathological features in hepatocellular carcinoma.
| Variables | No. of cases | CCR9 expression | P-value |
|---|
|
|---|
| Low | High |
|---|
| Gender | | | | 0.20 |
| Male | 189 | 84 | 105 | |
| Female | 51 | 22 | 29 | |
| Age (years) | | | | 0.26 |
| ≤60 | 198 | 94 | 104 | |
| >60 | 42 | 12 | 30 | |
| Liver cirrhosis | | | | 0.485 |
| Presence | 173 | 67 | 106 | |
| Absence | 67 | 39 | 28 | |
| Capsular
formation | | | | 0.71 |
| Presence | 125 | 61 | 66 | |
| Absence | 115 | 45 | 68 | |
| Tumor size | | | | 0.36 |
| ≤5 cm | 89 | 36 | 53 | |
| >5 cm | 151 | 70 | 81 | |
| Tumor nodule no. | | | | 0.009 |
| Multiple (≥2) | 96 | 35 | 61 | |
| Solitary | 144 | 71 | 73 | |
| Edmondson-Steiner
grade | | | | 0.012 |
| Stage I–II | 131 | 62 | 69 | |
| Stage III–IV | 109 | 44 | 65 | |
| Vascular
invasion | | | | 0.0015 |
| Presence | 133 | 45 | 88 | |
| Absence | 107 | 61 | 46 | |
 | Table IIUnivariate and multivariate analysis
showing overall survival for hepatocellular carcinoma patients. |
Table II
Univariate and multivariate analysis
showing overall survival for hepatocellular carcinoma patients.
| Variables | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| CCR9 | 2.91 | 1.76–6.50 | 0.001 | 2.35 | 1.09–5.02 | 0.014 |
| Gender | 0.62 | 0.28–1.52 | 0.59 | | | |
| Age | 0.82 | 0.48–1.37 | 0.74 | | | |
| Tumor size | 1.69 | 1.13–3.43 | 0.017 | 1.43 | 0.98–2.07 | 0.072 |
| Histologic
grade | 1.36 | 0.86–1.96 | 0.20 | | | |
| Cirrhosis | 0.69 | 0.41–1.14 | 0.14 | | | |
| HBsAg status | 1.44 | 0.56–2.73 | 0.58 | | | |
| Serum AFP | 1.32 | 0.92–2.74 | 0.09 | | | |
| Metastasis | 1.30 | 0.73–2.70 | 0.30 | | | |
| Recurrence | 1.22 | 0.80–2.26 | 0.25 | | | |
Correlation between CCR9 expression and
vascular invasion
We also investigated the relationship between CCR9
expression and vascular invasion in fresh HCC tissues using qPCR
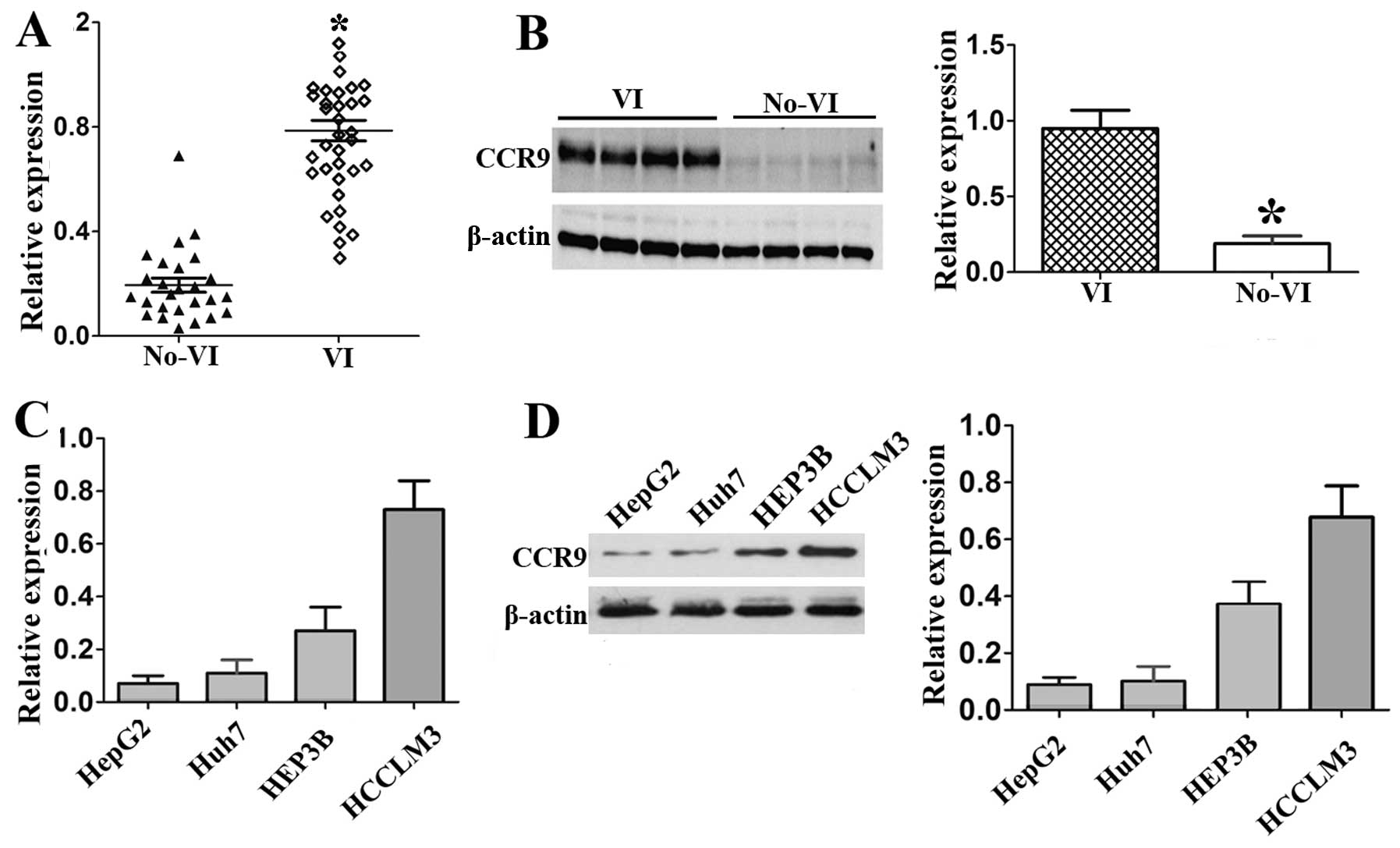
and western blotting. The results showed that CCR9 mRNA level in
HCC tissues with vascular invasion was significantly higher than in
tissues without vascular invasion (P<0.01, Fig. 2A). Consistent with the mRNA
expression, the CCR9 protein expression level was also
significantly elevated in HCC tissues with vascular invasion
(P<0.01; Fig. 2B).
Furthermore, we detected the expression levels of
CCR9 in HCC cell lines. Among the four HCC cell lines, HCCLM3, with
the highly metastatic ability (15), had the highest CCR9 expression at
mRNA and protein levels, followed by HEP3B, Huh7 and HepG2 cells
(Fig. 2C and D). These data suggest
that there may be a correlation between CCR9 and the metastasis
potential of HCC.
Knockdown of CCR9 inhibits the
proliferation of HEP3B and HCCLM3 cells
To further investigate the potential role of CCR9 in
HCC, we assessed the effects of CCR9 silencing on the cell
proliferation of HCC cell lines. Based on the CCR9 expression
levels, we chose HEP3B and HCCLM3 as the study cells.
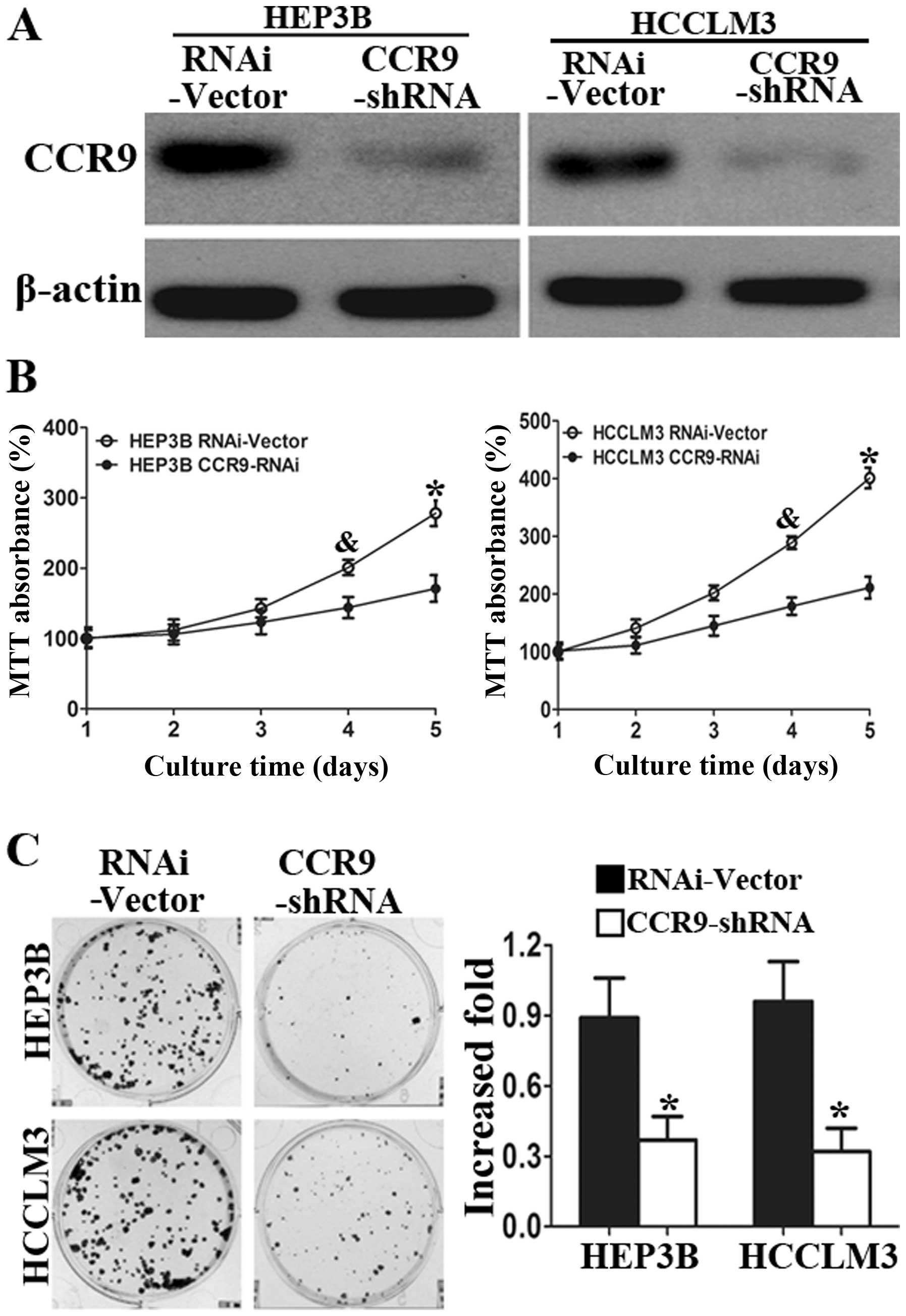
Western blot analysis showed that CCR9 expression in
the two cell lines were knocked down (Fig. 3A). MTT assays showed significant
reduction in cell proliferation of HEP3B-shCCR9 and HCCLM3-shCCR9
cells compared with control cells at 4 and 5 days (Fig. 3B). Silencing of CCR9 also reduced
the colony formation abilities (Fig.
3C) in both cell lines. Herein, we showed that CCR9 silencing
inhibited the proliferation of HCCLM3 and HEP3B cells compared with
control cells.
Overexpression of CCR9 promotes the
proliferation of HCC cells
Furthermore, we investigated the potential role of
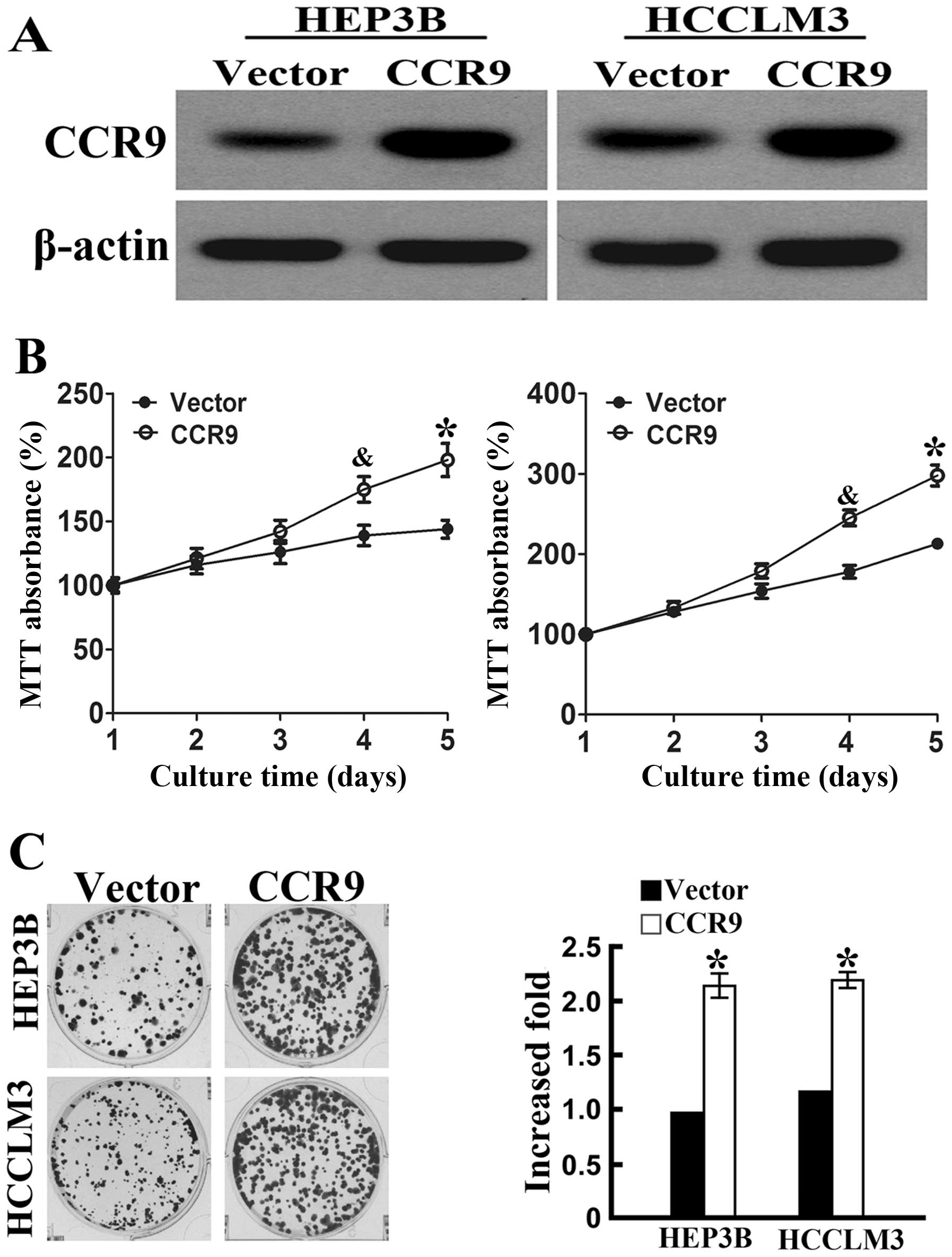
CCR9 overexpression in HCC cell lines. Western blot analysis showed
that stable CCR9-overexpressed cell lines were established
(Fig. 4A). MTT assays showed a
significant increase in cell proliferation of HEP3BCCR9+
cells and HCCLM3CCR9+ cells compared with control cells
at 4 and 5 days (Fig. 4B).
Overexpression of CCR9 also increased the colony formation
abilities (Fig. 4C) in the two cell
lines. Therefore, we showed that CCR9 overexpression promoted the
proliferation of HEP3B and HCCLM3 cells compared with control
cells.
CCR9 promotes cell proliferation by
increasing the ratio of HCC cells at S phase
Since CCR9 promoted cell proliferation, we further
explored the potential mechanisms. The percentage of cells at S
phase was assessed by immunodetection of BrdU. Ectopic expression
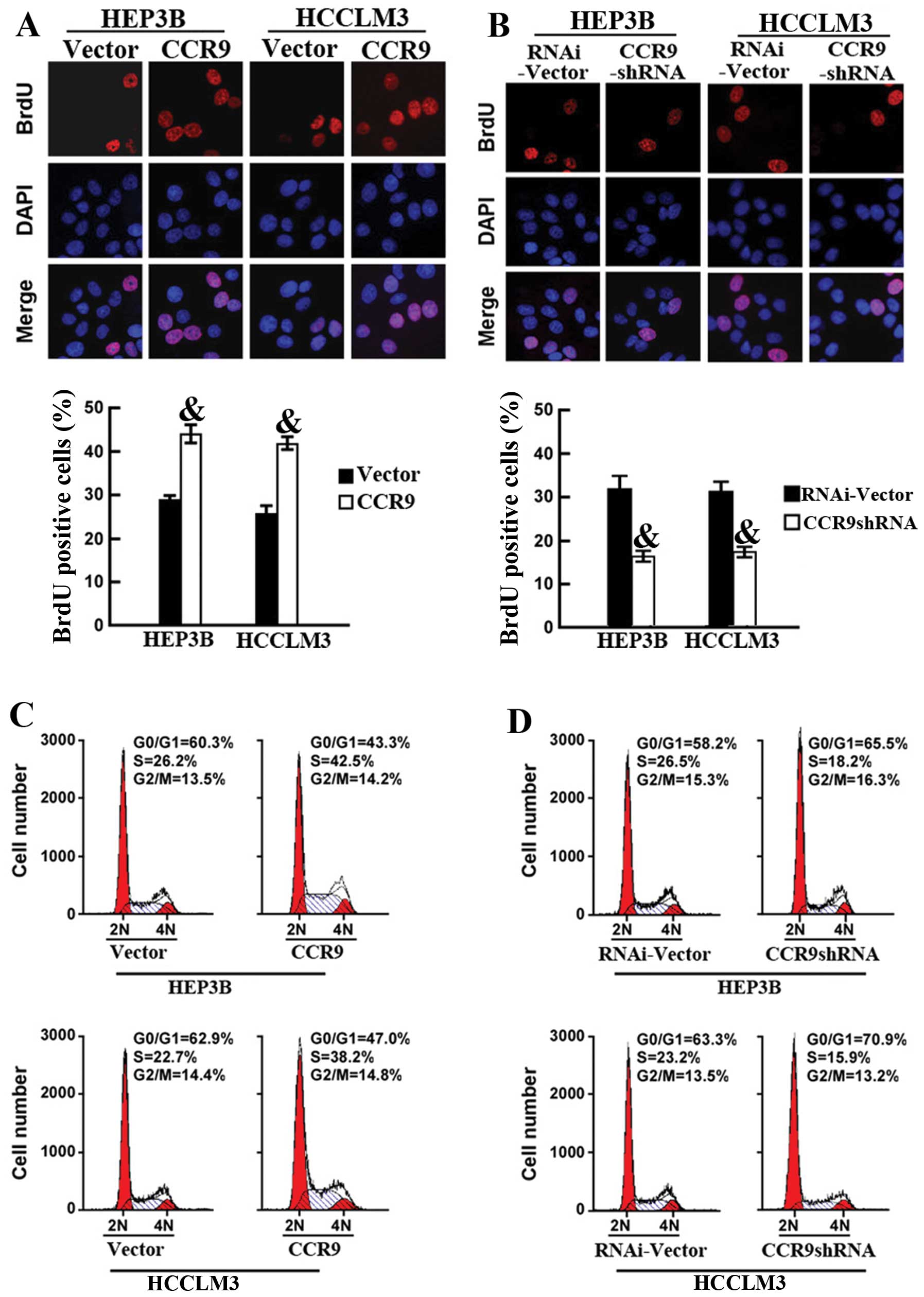
of CCR9 markedly increased the percentage of BrdU-positive cells in
HCC cell lines; 29.4 vs. 43.5% for HEP3B and 27.2 vs. 41.8% for
HCCLM3 (Fig. 5A). Conversely, CCR9
silencing significantly decreased the S phase fraction of
BrdU-positive cells; 32.27 vs. 17.03% for HEP3B cells and 31.23 vs.
17.98% for HCCLM3 cells (Fig. 5B).
Flow cytometric analysis of cell cycle also confirmed that CCR9
overexpression markedly increased the percentage of cells at S
phase and decreased the percentage of cells in the G1/G0 phase
(Fig. 5C). In contrast, CCR9
knockdown increased the percentage of cells in the G1/G0 phase and
decreased the percentage of cells at S phase (Fig. 5D). These results showed that CCR9
promoted cell proliferation by increasing the fraction of HCC cells
at S phase.
CCR9 promotes cell proliferation via
regulation of p21, p27 and cyclin D1
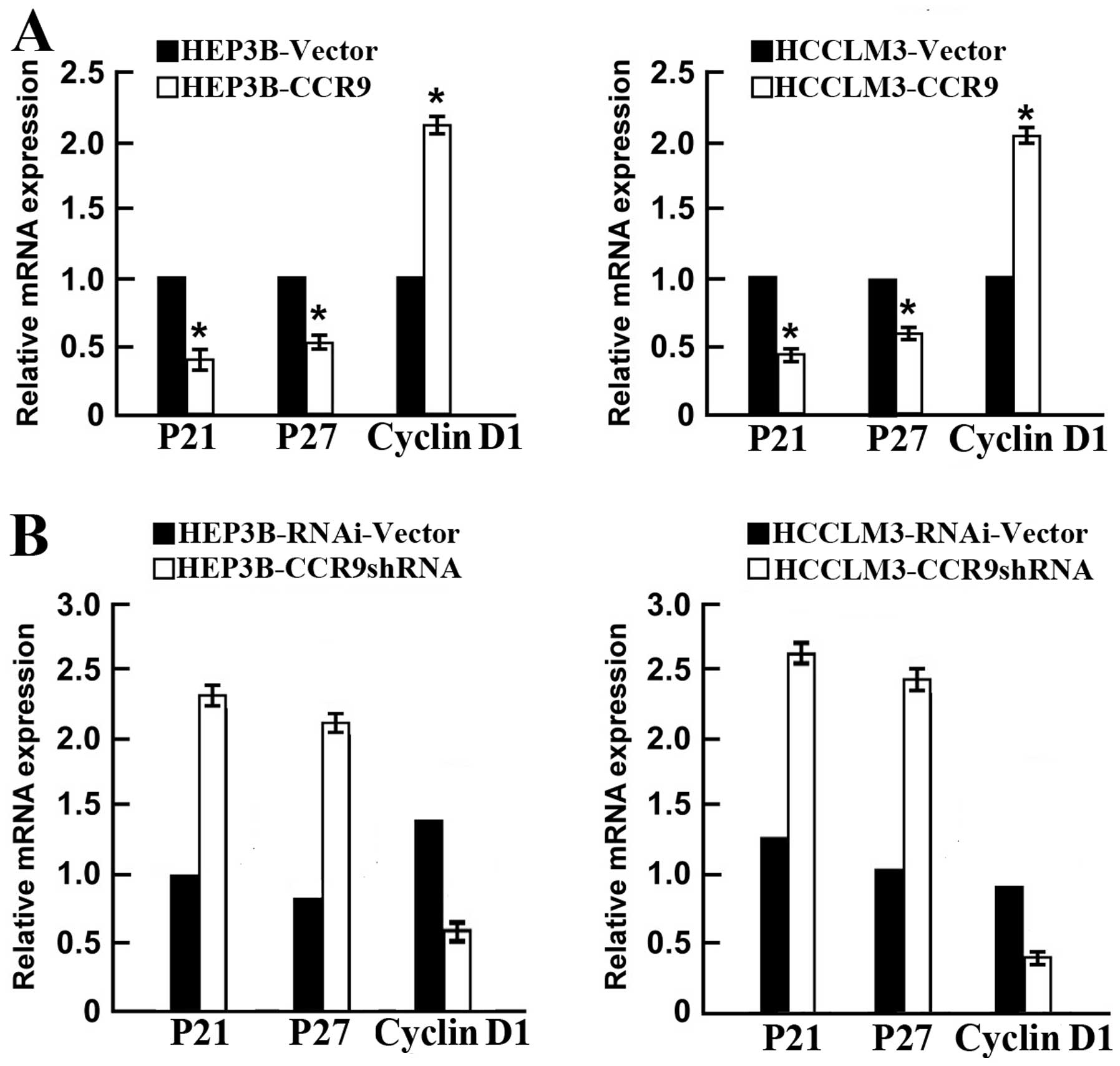
The p21, p27 and cyclin D1 play key roles in the
control of cell cycle associated with tumorigenicity (16,17).
qPCR showed that CCR9 overexpression significantly reduced p21 and
p27, and increased cyclin D1 (Fig.
6A). In contrast, CCR9 silencing markedly enhanced p21 and p27
expression, and inhibited cyclin D1 expression in both cell lines
(Fig. 6B). These results indicated
that CCR9 regulates p21, p27 and cyclin D1 to promote cell
proliferation and tumorigenicity.
Discussion
Recent data show that CCR9+ macrophages
are required for acute liver inflammation in mouse models of
hepatitis (18) and may activate
hepatic stellate cells, promoting liver fibrosis in mice (19). Our results from the present study
show that CCR9 was significantly elevated in HCC tissues. The
increased CCR9 expression was correlated with aggressive features
of HCC. Moreover, survival analysis revealed that high CCR9
expression was associated with poor OS in HCC. Multivariate
analysis revealed that CCR9 was an independent factor for
predicting survival in HCC.
The chemokine receptor CCR9 was initially identified
for its role in the immune system, where it is present on
leukocytes and is critical in T cell development (9). Subsequent data indicated that CCR9
expression is associated with increased cancer cell invasiveness in
melanoma, ovarian, breast and prostate cancer (20–22).
CCR9 shows aberrant expression on pancreatic cancer cells (10) and may be a factor in promoting
pancreatic cancer progression. While the CCL25/CCR9 axis has been
examined in some types of cancer, it remains unknown whether CCR9
plays a role in HCC. In the present study, we investigated
interactions between HCC cell invasiveness and CCR9 expression. We
found that CCR9 overexpression increased the fraction of HCC cells
at S phase, thereby promoting cell growth. Further investigation
indicated that this effect of CCR9 was mediated by regulating p21
and p27 expression as well as cyclin D1 expression at the mRNA
level.
In the present study, we found that upregulation of
CCR9 markedly increased proliferation of both HCC cell lines,
whereas silencing of CCR9 reduced it. Furthermore, soft-agar assay
revealed that the anchorage-independent HCC cell growth was
significantly enhanced upon CCR9 overexpression and inhibited in
case of CCR9 knockdown, suggesting that CCR9 overexpression
promotes the tumorigenicity of HCC cells. Therefore, the biological
roles of CCR9 in HCC metastasis merit further investigation.
In summary, the present study showed that CCR9
expression was markedly elevated in HCC tissue samples and cell
lines, compared to normal control. CCR9 was demonstrated to be a
novel prognostic marker for HCC. Further investigations revealed
that ectopic expression of CCR9 enhanced cell proliferation in HCC
cells, whereas CCR9 silencing impaired cell proliferation, which
was mediated through downregulation of the cell cycle regulators
p21, p27 as well as upregulation of cyclin D1. The results suggest
that CCR9 may be a novel target in HCC treatment. Future studies
are warranted to expand our understanding of CCR9-mediated
signaling in HCC and to develop novel therapeutic agents to target
this pathway in HCC.
Acknowledgements
This study was supported by the Natural Science
Foundation of Shandong Province (no. ZR2012HM079), China.
References
|
1
|
Zhu AX, Duda DG, Sahani DV and Jain RK:
HCC and angiogenesis: possible targets and future directions. Nat
Rev Clin Oncol. 8:292–301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cervello M, McCubrey JA, Cusimano A,
Lampiasi N, Azzolina A and Montalto G: Targeted therapy for
hepatocellular carcinoma: novel agents on the horizon. Oncotarget.
3:236–260. 2012.PubMed/NCBI
|
|
3
|
McClune AC and Tong MJ: Chronic hepatitis
B and hepatocellular carcinoma. Clin Liver Dis. 14:461–476. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Finn RS: Emerging targeted strategies in
advanced hepatocellular carcinoma. Semin Liver Dis. 33(Suppl 1):
S11–S19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou L, Liu J and Luo F: Serum tumor
markers for detection of hepatocellular carcinoma. World J
Gastroenterol. 12:1175–1181. 2006.PubMed/NCBI
|
|
6
|
Psyrri A, Arkadopoulos N, Vassilakopoulou
M, Smyrniotis V and Dimitriadis G: Pathways and targets in
hepatocellular carcinoma. Expert Rev Anticancer Ther. 12:1347–1357.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Viola A and Luster AD: Chemokines and
their receptors: drug targets in immunity and inflammation. Annu
Rev Pharmacol Toxicol. 48:171–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Franciszkiewicz K, Boissonnas A, Boutet M,
Combadière C and Mami-Chouaib F: Role of chemokines and chemokine
receptors in shaping the effector phase of the antitumor immune
response. Cancer Res. 72:6325–6332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Svensson M and Agace WW: Role of
CCL25/CCR9 in immune homeostasis and disease. Expert Rev Clin
Immunol. 2:759–773. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen X, Mailey B, Ellenhorn JD, Chu PG,
Lowy AM and Kim J: CC chemokine receptor 9 enhances proliferation
in pancreatic intraepithelial neoplasia and pancreatic cancer
cells. J Gastrointest Surg. 13:1955–1962. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johnson-Holiday C, Singh R, Johnson E, et
al: CCL25 mediates migration, invasion and matrix metalloproteinase
expression by breast cancer cells in a CCR9-dependent fashion. Int
J Oncol. 38:1279–1285. 2011.
|
|
12
|
Geisler SA, Olshan AF, Weissler MC, et al:
p16 and p53 protein expression as prognostic indicators of survival
and disease recurrence from head and neck cancer. Clin Cancer Res.
8:3445–3453. 2000.PubMed/NCBI
|
|
13
|
Fukuoka J, Fuji T, Shih JH, et al:
Chromatin remodeling factors and BRM/BRG1 expression as prognostic
indicators in non-small cell lung cancer. Clin Cancer Res.
10:4314–4324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao Z, Zhang R, Li J, et al: X-linked
inhibitor of apoptosis protein (XIAP) regulation of cyclin D1
protein expression and cancer cell anchorage-independent growth via
its E3 ligase-mediated protein phosphatase 2A/c-Jun axis. J Biol
Chem. 288:20238–20247. 2013. View Article : Google Scholar
|
|
15
|
Ye QH, Qin LX, Forgues M, et al:
Predicting hepatitis B virus-positive metastatic hepatocellular
carcinomas using gene expression profiling and supervised machine
learning. Nat Med. 9:416–423. 2003. View
Article : Google Scholar
|
|
16
|
Yu Y, Kovacevic Z and Richardson DR:
Tuning cell cycle regulation with an iron key. Cell Cycle.
6:1982–1994. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Macaluso M, Montanari M, Cinti C and
Giordano A: Modulation of cell cycle components by epigenetic and
genetic events. Semin Oncol. 32:452–457. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakamoto N, Ebinuma H, Kanai T, et al:
CCR9+ macrophages are required for acute liver
inflammation in mouse models of hepatitis. Gastroenterology.
142:366–376. 2012.
|
|
19
|
Chu PS, Nakamoto N, Ebinuma H, et al: C-C
motif chemokine receptor 9 positive macrophages activate hepatic
stellate cells and promote liver fibrosis in mice. Hepatology.
58:337–350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amersi FF, Terando AM, Goto Y, et al:
Activation of CCR9/CCL25 in cutaneous melanoma mediates
preferential metastasis to the small intestine. Clin Cancer Res.
14:638–645. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johnson EL, Singh R, Singh S, et al:
CCL25-CCR9 interaction modulates ovarian cancer cell migration,
metalloproteinase expression, and invasion. World J Surg Oncol.
8:622010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Singh S, Singh UP, Stiles JK, Grizzle WE
and Lillard JW Jr: Expression and functional role of CCR9 in
prostate cancer cell migration and invasion. Clin Cancer Res.
10:8743–8750. 2004. View Article : Google Scholar : PubMed/NCBI
|