Introduction
Epithelial ovarian cancer (EOC) is the most deadly
malignancy of the female reproductive tract in many countries
(1,2). Involvement of steroid hormones,
primarily estrogen, has been associated with EOC. Ample evidence
from epidemiologic, clinical and experimental research has
demonstrated that E2 is responsible for promoting EOC progression
(3–9). Although the effects of E2 on EOC
progression have been extensively studied, the underlying
mechanisms remain unknown and the clinical response to steroid
hormone therapy remains disappointing. Thus, fully identifying the
contributions of E2 to EOC progression is urgently required.
Compelling data have demonstrated that the effects
of E2 on EOC development are mediated by the regulation of target
genes involved in the control of cancer progression. Previous
studies, including ours, have identified a panel of aberrantly
expressed E2-regulated protein-coding genes that are involved in
cellular growth control, such as cyclin D1 and c-myc, and in
cellular metastasis control, such as fibulin-1, cathepsin-D, HIF-1,
nm23-H1, E-cadherin and MMP-2 (4–7,10,11).
Despite these protein-coding genes, undoubtedly, the set of genes
that directly mediate estrogenic effects on EOC progression has not
been fully defined. Therefore, exploration of new E2-regulated
genes is needed, which may help elucidate estrogenic effects on EOC
progression and provide optional therapeutic targets.
The human transcriptome was found to be more complex
than a collection of protein-coding genes, showing extensive
non-coding RNA (ncRNA) expression (12). Long ncRNAs (lncRNAs; >200 nt in
length), initially argued to be spurious transcriptional noise
(13), are emerging as new
regulators in the cancer paradigm. Aberrant expression of lncRNAs
has been reported to be associated with malignant phenotypes in
various human tissues, and some lncRNAs, such as HOTAIR, MALAT-1,
H19, HULC, lincRNA-p21 and MEG3, might also function as tumor
suppressor genes or oncogenes (14–19).
Although several published studies have reported lncRNAs such as
lncRNA-LSINCT5 and HOST2 in EOC (20,21),
to our knowledge, no studies have focused on E2-regulated lncRNAs
in EOC.
Therefore, we sought to identify E2-regulated
lncRNAs in EOC. We found that E2 stimulation of ERα-positive (ERα+)
EOC cells resulted in a panel of differentially expressed lncRNAs,
showing great potential to contribute to cancer progression based
on bioinformatics analyses. Moreover, we found that some candidate
lncRNAs were aberrantly expressed in ERα+ compared to ERα-negative
(ERα−) EOC tissues, and their differential expression was
associated with certain clinicopathological variables and poor
prognosis of ERα+ EOC patients. Our results highlight for the first
time the potential use of lncRNAs as causal link with estrogenic
effects on EOC progression and as surrogate targets to hormone
therapy.
Materials and methods
Cells and treatment
The ovarian cancer cell line SKOV3 was obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
SKOV3 cells were routinely maintained in RPMI-1640 medium
(Gibco-BRL, Gaithersburg, MD, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco) and maintained at 37°C with 5%
CO2. For the E2 induction experiments, cells (plated at
20–30% confluence) were grown for 3 days in phenol red-free
RPMI-1640 (Gibco) containing 5% activated, charcoal-treated foetal
bovine serum (Serana, Bunbury, Australia). Next, the cells were
treated for 24 h with 10−8 M E2 or vehicle alone (DMSO,
0.01% of final volume) as a control.
RNA extraction and microarray
TRIzol (Invitrogen, Carlsbad, CA, USA) was used to
extract total RNA from SKOV3 cells with or without 24-h treatment
with 10−8 M E2. There were three replicates of each
sample; they were purified using an RNeasy Micro kit (cat. #74004;
Qiagen GmbH, Hilden, Germany). An Agilent Bioanalyzer 2100 (Agilent
Technologies, Santa Clara, CA, USA) was used to quantify the RNA
and evaluate its integrity; the 28S:18S ratio was determined, and
an RNA integrity number (RIN) was assigned to each sample. RNA with
no evidence of degradation and no signs of DNA contamination (as
indicated by an RIN ≥7.0 and a 28S:18S ratio ≥0.7) was processed
for further analysis.
The lncRNA and mRNA expression profiles were
obtained using the Glue Grant Human Transcriptome (GG-H) Array,
which was manufactured by Affymetrix and Stanford University. This
array contains 5,869 probes covering 730 non-coding, functional
RNAs and 3,292,929 probes covering 27,670 coding genes collected
from RefSeq, Ensembl and UCSC Known Genes, based on human genome
assembly hg18 (22). The array
experiments and computational analysis were performed according to
the manufacturer’s instructions (Affymetrix, Santa Clara, CA, USA).
Briefly, 0.2 μg of total RNA was amplified and labelled, and 20 μg
of labelled cDNA was loaded onto the array. The array was
hybridized and washed using the GeneChip® Hybridization,
Wash and Stain kit (cat. #900720), Hybridization Oven 645 (cat.
#00-0331-220V) and Fluidics Station 450 (cat. #00-0079). The slides
were scanned in a GeneChip® Scanner 3000 (cat.
#00-00212). The raw data were obtained using Command Console
Software 3.1 with the default settings and were processed using
Affymetrix Power Tools with Robust Multiarray Analysis (RMA) for
background correction, normalization and summarization.
Differentially expressed genes [defined as a fold-change ≥1.5 and a
P-value <0.05 (t-test)] were selected for further study.
Bioinformatics functional analysis of
E2-regulated lncRNAs
Identification of lncRNA-mRNA
targeting pairs
Two procedures were performed to search for the
target mRNAs of lncRNAs. First, UCSC hg18 (http://genome.ucsc.edu/) was used to predict lncRNA
targets. Target genes under cis-regulatory control were
defined as genes whose transcription was regulated by lncRNAs in
nearby genomic locations (≤10 kbp upstream or downstream) (23). Based on mRNA sequence
complementarity and RNA duplex energy prediction,
trans-acting target genes were identified using BLAST
software in the first round of screening (with the parameter e
<1E-5) and RNAplex software for final verification (with the
parameter -e −20) (24).
Additionally, to improve the accuracy of the target prediction, the
predicted lncRNA targets (both cis and trans) were
combined with the differentially expressed mRNAs in the profile.
The resultant overlapping mRNAs were considered the final putative
targets of the differentially expressed lncRNAs. This information
formed the basis for determining the lncRNA-mRNA targeting
pairs.
Gene ontology (GO) and pathway
analysis
For the GO and pathway analyses, the putative
targets were initially inputted into the Database for Annotation,
Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/), which searched
the GO terms to identify the molecular function represented in the
gene profile (25), and then into
the database of the Kyoto Encyclopedia of Genes and Genomes (KEGG;
http://www.genome.ad.jp/kegg/) to
analyze the roles of the targets in molecular pathways (26).
Tissue samples and patient data
The study included 95 patients who underwent surgery
for primary ovarian cancer in the Department of Gynecology,
Obstetrics and Gynecology Hospital of Fudan University between
January 2006 and December 2008. Patients were included based on the
availability of tissue and follow-up data. Patients with borderline
ovarian tumors or with two or more different malignancies were
excluded from the study. None of the patients had received
preoperative radiotherapy, chemotherapy or hormonal therapy. All
EOC tissue samples were frozen immediately after surgery and stored
in liquid nitrogen until use.
Clinical and pathological data of EOC patients were
evaluated by reviewing medical charts and the original pathology
reports. Staging and grading were evaluated in accordance with the
criteria of the International Federation of Gynecologists and
Obstetricians (FIGO) and the World Health Organization (WHO).
Follow-up data were obtained by reviewing the out patient charts,
contacting patients or correspondence. Overall survival (OS) was
calculated from the date of surgery until the date of mortality or
end of follow-up (January 2013). The present study was approved by
the Research Ethics Committee of Fudan University, China. Informed
consent was obtained from all the patients.
Immunohistochemistry
The immunohistochemical study of ERα was performed
using a standard streptavidin-peroxidase method. The endogenous
peroxidase activity was blocked with 3% H2O2
for 10 min. For the antigen retrieval, slides were immersed in 10
mM citrate buffer (pH 6.0) and boiled for 15 min in a microwave
oven. Non-specific binding was blocked by 5% normal goat serum for
10 min. The slides were incubated with a 1:50 dilution of
monoclonal antibody against ERα (Santa Cruz Biotechnology, Santa
Cruz, CA, USA) at 4°C overnight in a moist chamber. The slides were
sequentially incubated with biotinylated goat anti-mouse IgG (1:100
dilution; Santa Cruz Biotechnology) and then
streptavidin-peroxidase conjugate, each for 30 min at room
temperature. Isotope-matched human IgG was used in each case as a
negative control. Finally, the 3,5-diaminobenzidine (DAB) Substrate
kit (Dako) was used for color development followed by Mayer’s
hematoxylin counterstaining. ERα+ cases were defined as tumors with
>10% stained nuclei (27).
Quantitative real-time reverse
transcription polymerase chain reaction (qRT-PCR)
qRT-PCR analysis of lncRNA expression was performed
using FastStart Universal SYBR-Green Master (Rox; Roche) and an ABI
Prism 7900 Real-Time PCR system (Applied Biosystems, Foster City,
CA, USA). Briefly, total RNA was extracted from cells and tissues
and converted to cDNA using an iScript cDNA Synthesis kit (Bio-Rad
Laboratories) according to the manufacturer’s protocol. The PCR
amplifications were performed in a 10-μl (total volume) reaction
that included 1 μl of cDNA template (~5 ng), 5 μl of FastStart
Universal SYBR-Green Master (Rox), 3.6 μl of double-distilled water
and 0.2 μl of each pair of forward and reverse primers (Table I; Sangon Biotech Co., Ltd.,
Shanghai, China). The PCR conditions included an initial
denaturation step at 95°C for 10 min and 40 cycles of 95°C for 15
sec and 60°C for 30 sec. All the experiments were performed in
triplicate, and all the samples were normalized to GAPDH
expression. The expression fold-changes were calculated using the
2−ΔΔCt method.
 | Table IPrimer sequences of the studied
genes. |
Table I
Primer sequences of the studied
genes.
| Gene | Primer | Sequence (5′-3′) |
|---|
| TC1500845 | F: |
ACCACGACTCCCAAGAGGTA |
| R: |
CAGCTGCGATGGTGAGAACT |
| TC0101441 | F: |
CAAGGCAGGTGAGAACGAGT |
| R: |
CTCGACTTAGGGAGCTGCAC |
| TC0100223 | F: |
ATGAGGGCTCTGCTCTATGAATGG |
| R: |
GGCTTGTTCAGTGTCTGTTAAGGGT |
| TC0101686 | F: |
GGCTACTTACATGGTCCAGCA |
| R: |
TAGCATGGAAAGGACCACTGC |
| GAPDH | F: |
TGACTTCAACAGCGACACCCA |
| R: |
CACCCTGTTGCTGTAGCCAAA |
Statistical analysis
The data were processed using SPSS version 16.0
software (SPSS, Inc., Chicago, IL, USA). Comparison of continuous
data was analyzed using the Student’s t-test, whereas categorical
data was analyzed using the Chi-square test and Fisher’s exact test
where appropriate (when the expected frequency was <5). OS
curves were plotted according to the Kaplan-Meier method, with the
log-rank test applied for comparison. Variables were used in
multivariate analysis on the basis of the Cox proportional hazards
model. A P-value <0.05 was considered to indicate a
statistically significant difference (P<0.05).
Results
Identification of E2-regulated lncRNAs in
ERα+ ovarian cancer cells
Identification of E2-regulated lncRNAs
in SKOV3 cells
As our previous studies provided evidence that E2
regulated some protein-coding genes in ERα+ ovarian cancer SKOV3
cells (5–7), we examined whether the expression of
any lncRNAs is also regulated by E2 in SKOV3 cells. In the present
study, SKOV3 cells were treated with 10−8 M E2 for 24 h,
and changes in the lncRNA expression profile were analyzed by
performing a microarray. The microarray data indicated that 115
lncRNAs were significantly dysregulated following E2 treatment,
including 51 upregulated and 64 downregulated lncRNAs (fold-change
≥1.5, P<0.05; data not shown). The top ten relative increased
and decreased E2-regulated lncRNAs are listed in Table II.
 | Table IIThe top ten relative increased and
decreased E2-regulated lncRNAs in SKOV3 cells. |
Table II
The top ten relative increased and
decreased E2-regulated lncRNAs in SKOV3 cells.
| | | Annotations |
|---|
| | |
|
|---|
| Probeset_id | Regulated | Fold-change
(E2/control) | Seqname | Start | End | Strand |
|---|
| TC0500815 | Upregulated | 3.419413635 | chr5 | 1043143 | 1050457 | − |
| TC0101441 | Upregulated | 3.275356271 | chr1 | 202377159 | 202378011 | + |
| TC0901107 | Upregulated | 3.269435942 | chr9 | 89871170 | 89871958 | − |
| TC0301101 | Upregulated | 3.258219523 | chr3 | 37825199 | 37878275 | − |
| TC1900181 | Upregulated | 3.247328231 | chr19 | 10820078 | 10841404 | + |
| TC1201706 | Upregulated | 3.232226492 | chr12 | 131237621 | 131240193 | − |
| TC0601086 | Upregulated | 3.186614845 | chr6 | 29802359 | 29824805 | − |
| TC1500845 | Upregulated | 3.183351441 | chr15 | 38773376 | 38774597 | − |
| TC0300769 | Upregulated | 3.143316689 | chr3 | 169450147 | 169514658 | + |
| TC0X00076 | Upregulated | 2.989272086 | chrX | 18821244 | 18823011 | + |
| TC0501141 | Downregulated | 0.300418806 | chr5 | 90642594 | 90645975 | − |
| TC1201365 | Downregulated | 0.301520979 | chr12 | 64556922 | 64561625 | − |
| TC0101686 | Downregulated | 0.30390326 | chr1 | 244341256 | 244343791 | + |
| TC1200811 | Downregulated | 0.31213872 | chr12 | 125146739 | 125152812 | + |
| TC0300928 | Downregulated | 0.318626574 | chr3 | 197148023 | 197150980 | + |
| TC1900906 | Downregulated | 0.325966672 | chr19 | 61499140 | 61513631 | + |
| TC0801241 | Downregulated | 0.328971081 | chr8 | 128289293 | 128300515 | − |
| TC1400428 | Downregulated | 0.333717978 | chr14 | 88886369 | 88900796 | + |
| TC0201596 | Downregulated | 0.335402449 | chr2 | 69870840 | 69879634 | − |
| TC0100223 | Downregulated | 0.342972038 | chr1 | 21465570 | 21466331 | + |
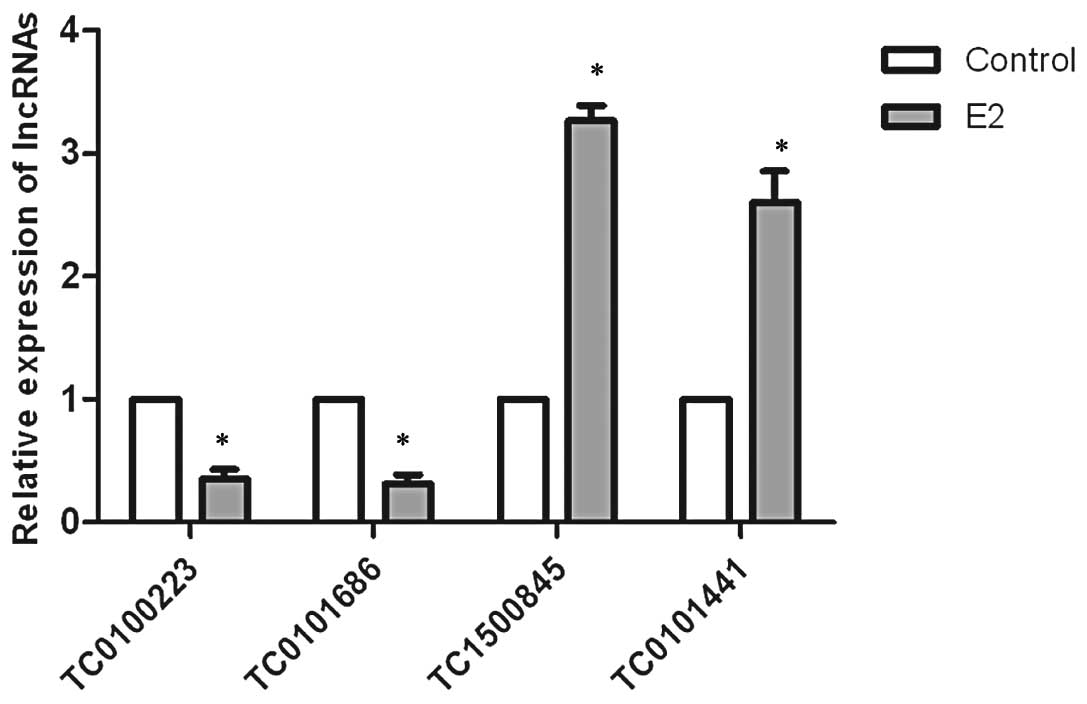
To confirm the microarray findings, we examined the
expression of four lncRNAs selected from the top ten relative
increased and decreased E2-regulated lncRNAs using qRT-PCR. The
results revealed that the expression levels of TC0100223 and
TC0101686 were significantly downregulated by E2, whereas TC1500845
and TC0101441 were significantly upregulated by E2 in SKOV3 cells,
consistent with the microarray results (Fig. 1).
Putative targets of E2-regulated lncRNAs
and their functional analysis
Based on the overlap between the targets predicted
by bioinformatics and the differentially expressed mRNAs detected
in the microarray, we constructed 55 E2-regulated lncRNA-target
mRNA pairs (Table III). The GO
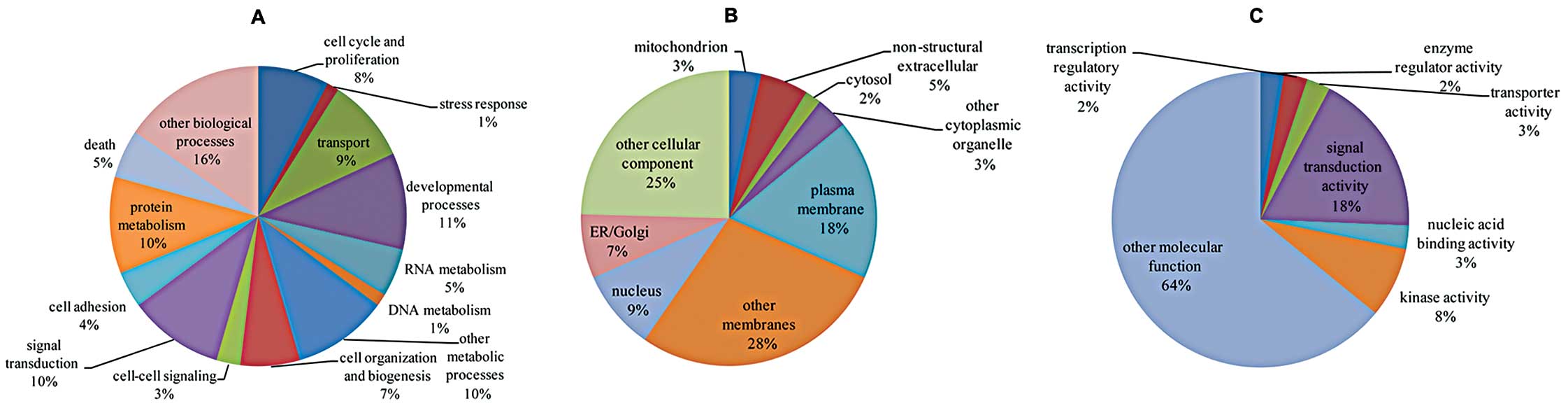
(Fig. 2) and pathway (Table IV) analyses showed that a set of
E2-regulated lncRNAs that mapped to the target mRNAs were
correlated with several cellular processes and pathways known to be
related to cancer progression, such as cell cycle and
proliferation, developmental processes, cell adhesion, cell death,
MAPK signaling, Hedgehog signaling, Jak-STAT signaling and cancer
pathways, suggesting their great potential to contribute to cancer
progression.
 | Table IIIlncRNA-target mRNA pairs regulated by
E2 in SKOV3 cells. |
Table III
lncRNA-target mRNA pairs regulated by
E2 in SKOV3 cells.
| Information of
lncRNAs | lncRNA-mRNA
pairs | Information of
mRNAs |
|---|
|
|
|
|---|
| Start | End | Strand | Seqname | lncRNA
Probeset_id | mRNA Symbol | Type | Start | End | Strand | Seqname |
|---|
| 43821718 | 43827655 | + | chr20 | TC2000321 | CD40 | cis | 44180313 | 44366257 | + | chr20 |
| 43821718 | 43827655 | + | chr20 | TC2000321 | UBE2C | cis | 43874662 | 43879003 | + | chr20 |
| 63310787 | 63321606 | − | chr19 | TC1901868 | ZNF544 | cis | 63432092 | 63480673 | + | chr19 |
| 63310787 | 63321606 | − | chr19 | TC1901868 | ZSCAN4 | cis | 62872115 | 62882317 | + | chr19 |
| 63310787 | 63321606 | − | chr19 | TC1901868 | ZNF417 | cis | 63110053 | 63119796 | − | chr19 |
| 63310787 | 63321606 | − | chr19 | TC1901868 | ZNF460 | cis | 62483670 | 62496635 | + | chr19 |
| 61499140 | 61513631 | + | chr19 | TC1900906 | ZNF460 | cis | 62483670 | 62496635 | + | chr19 |
| 10820078 | 10841404 | + | chr19 | TC1900181 | CDC37 | cis | 10362809 | 10375271 | − | chr19 |
| 10820078 | 10841404 | + | chr19 | TC1900181 | QTRT1 | cis | 10673106 | 10805160 | + | chr19 |
| 75925981 | 75926392 | + | chr17 | TC1700826 | KIAA1618 | cis | 75849262 | 75925295 | + | chr17 |
| 29738362 | 29740074 | − | chr16 | TC1600989 | QPRT | cis | 29597859 | 29616810 | + | chr16 |
| 70157455 | 70163889 | + | chr16 | TC1600575 | LOC652737 | cis | 69398791 | 69457733 | − | chr16 |
| 38773376 | 38774597 | − | chr15 | TC1500845 | TYRO3 | cis | 39638524 | 39658826 | + | chr15 |
| 38773376 | 38774597 | − | chr15 | TC1500845 | IVD | cis | 38484978 | 38515438 | + | chr15 |
| 67751133 | 67757033 | + | chr15 | TC1500441 | PAQR5 | cis | 67378348 | 67486098 | + | chr15 |
| 21379262 | 21379796 | + | chr14 | TC1400062 | ABHD4 | cis | 22136986 | 22151097 | + | chr14 |
| 11599456 | 11603583 | + | chr12 | TC1200137 | TAS2R7 | cis | 10845399 | 10846493 | − | chr12 |
| 7154170 | 7159091 | + | chr12 | TC1200076 | CDCA3 | cis | 6824224 | 6830686 | − | chr12 |
| 7154170 | 7159091 | + | chr12 | TC1200076 | CLSTN3 | cis | 7174234 | 7202795 | + | chr12 |
| 7154170 | 7159091 | + | chr12 | TC1200076 | ING4 | cis | 6629707 | 6642565 | − | chr12 |
| 7154170 | 7159091 | + | chr12 | TC1200076 | PTPN6 | cis | 6926001 | 6940741 | + | chr12 |
| 6966612 | 7033762 | + | chr12 | TC1200074 | CDCA3 | cis | 6824224 | 6830686 | − | chr12 |
| 6966612 | 7033762 | + | chr12 | TC1200074 | CLSTN3 | cis | 7174234 | 7202795 | + | chr12 |
| 6966612 | 7033762 | + | chr12 | TC1200074 | ING4 | cis | 6629707 | 6642565 | − | chr12 |
| 6966612 | 7033762 | + | chr12 | TC1200074 | PTPN6 | cis | 6926001 | 6940741 | + | chr12 |
| 5383589 | 5384883 | + | chr12 | TC1200045 | NTF3 | cis | 5473527 | 5474725 | + | chr12 |
| 58457692 | 58582501 | − | chr11 | TC1101435 | STX3 | cis | 59279108 | 59326752 | + | chr11 |
| 18821244 | 18823011 | + | chrX | TC0X00076 | GPR64 | cis | 18917348 | 19050676 | − | chrX |
| 66450847 | 66456323 | − | chr9 | TC0900981 | PIK3C2B | trans | 2.03E+08 | 202730566 | − | chr1 |
| 12264367 | 12270292 | + | chr8 | TC0800087 | DLC1 | cis | 12985243 | 13506486 | − | chr8 |
| 12264367 | 12270292 | + | chr8 | TC0800087 | CTSB | cis | 11737442 | 11763147 | − | chr8 |
| 1.28E+08 | 1.28E+08 | − | chr7 | TC0701620 | OPN1SW | cis | 1.28E+08 | 128203087 | − | chr7 |
| 1.28E+08 | 1.28E+08 | − | chr7 | TC0701620 | TSPAN33 | cis | 1.29E+08 | 128595907 | + | chr7 |
| 1.28E+08 | 1.28E+08 | − | chr7 | TC0701620 | SMO | cis | 1.29E+08 | 128640619 | + | chr7 |
| 1.42E+08 | 1.42E+08 | + | chr7 | TC0700794 | TAS2R5 | cis | 1.41E+08 | 141137635 | + | chr7 |
| 1.4E+08 | 1.41E+08 | + | chr7 | TC0700753 | TAS2R5 | cis | 1.41E+08 | 141137635 | + | chr7 |
| 29802359 | 29824805 | − | chr6 | TC0601086 | KIAA1949 | cis | 30752146 | 30763651 | − | chr6 |
| 1.5E+08 | 1.5E+08 | − | chr5 | TC0501415 | TNIP1 | cis | 1.5E+08 | 150446914 | − | chr5 |
| 1.5E+08 | 1.5E+08 | − | chr5 | TC0501415 | SLC36A1 | cis | 1.51E+08 | 150852132 | + | chr5 |
| 1.5E+08 | 1.5E+08 | − | chr5 | TC0501415 | ANXA6 | cis | 1.5E+08 | 150517636 | − | chr5 |
| 1.5E+08 | 1.5E+08 | − | chr5 | TC0501415 | CCDC69 | cis | 1.51E+08 | 150583899 | − | chr5 |
| 1043143 | 1050457 | − | chr5 | TC0500815 | LPCAT1 | cis | 1514544 | 1577092 | − | chr5 |
| 1.15E+08 | 1.15E+08 | − | chr4 | TC0401208 | ARSJ | cis | 1.15E+08 | 115120306 | − | chr4 |
| 37825199 | 37878275 | − | chr3 | TC0301101 | XYLB | cis | 38363244 | 38431471 | + | chr3 |
| 1.35E+08 | 1.35E+08 | + | chr3 | TC0300635 | AMOTL2 | cis | 1.36E+08 | 135576450 | − | chr3 |
| 1.24E+08 | 1.24E+08 | + | chr3 | TC0300552 | STXBP5L | cis | 1.22E+08 | 122621336 | + | chr3 |
| 1.14E+08 | 1.14E+08 | + | chr2 | TC0200609 | PAX8 | cis | 1.14E+08 | 113752969 | − | chr2 |
| 2.46E+08 | 2.46E+08 | − | chr1 | TC0103436 | ZNF670 | cis | 2.45E+08 | 245308738 | − | chr1 |
| 2.44E+08 | 2.44E+08 | + | chr1 | TC0101686 | ZNF670 | cis | 2.45E+08 | 245308738 | − | chr1 |
| 2.02E+08 | 2.02E+08 | + | chr1 | TC0101441 | PIK3C2B | cis | 2.03E+08 | 202730566 | − | chr1 |
| 2.02E+08 | 2.02E+08 | + | chr1 | TC0101441 | ATP2B4 | cis | 2.02E+08 | 201979832 | + | chr1 |
| 53566493 | 53569511 | + | chr1 | TC0100566 | C1orf163 | cis | 52925096 | 52936964 | − | chr1 |
| 53566493 | 53569511 | + | chr1 | TC0100566 | CC2D1B | cis | 52588855 | 52604453 | − | chr1 |
| 21465570 | 21466331 | + | chr1 | TC0100223 | ECE1 | cis | 21417664 | 21544621 | − | chr1 |
| 21465570 | 21466331 | + | chr1 | TC0100223 | RAP1GAP | cis | 21795301 | 21868437 | − | chr1 |
 | Table IVTarget mRNA-related pathways in SKOV3
cells. |
Table IV
Target mRNA-related pathways in SKOV3
cells.
| Term | Count | Genes |
|---|
| Pentose and
glucuronate interconversions | 1 | XYLB |
| Valine, leucine and
isoleucine degradation | 1 | IVD |
| Inositol phosphate
metabolism | 1 | PIK3C2B |
| Nicotinate and
nicotinamide metabolism | 1 | QPRT |
| Metabolic
pathways | 4 | IVD, QPRT, XYLB,
PIK3C2B |
| MAPK signaling
pathway | 1 | NTF3 |
|
Phosphatidylinositol signaling system | 1 | PIK3C2B |
| Ubiquitin mediated
proteolysis | 1 | UBE2C |
| SNARE interactions
in vesicular transport | 1 | STX3 |
| Hedgehog signaling
pathway | 1 | SMO |
| Adherens
junction | 1 | PTPN6 |
| Jak-STAT signaling
pathway | 1 | PTPN6 |
| Natural killer cell
mediated cytotoxicity | 1 | PTPN6 |
| T cell receptor
signaling pathway | 1 | PTPN6 |
| B cell receptor
signaling pathway | 1 | PTPN6 |
| Neurotrophin
signaling pathway | 1 | NTF3 |
| Pathways in
cancer | 1 | SMO |
| Basal cell
carcinoma | 1 | SMO |
Expression of several candidate lncRNAs
in ERα+ EOC tissues
In order to confirm the potential of some
E2-regulated lncRNAs to contribute to cancer progression, we
initially selected the four lncRNAs (TC0100223, TC0101686,
TC1500845 and TC0101441) as candidates and tested their expression
levels in EOC tissues. Considering the fact that ERα is the main
form expressed in malignant ovarian tumors and as ERα has been
reported to promote poor prognosis in EOC patients (28,29),
we determined whether the expression of in vitro
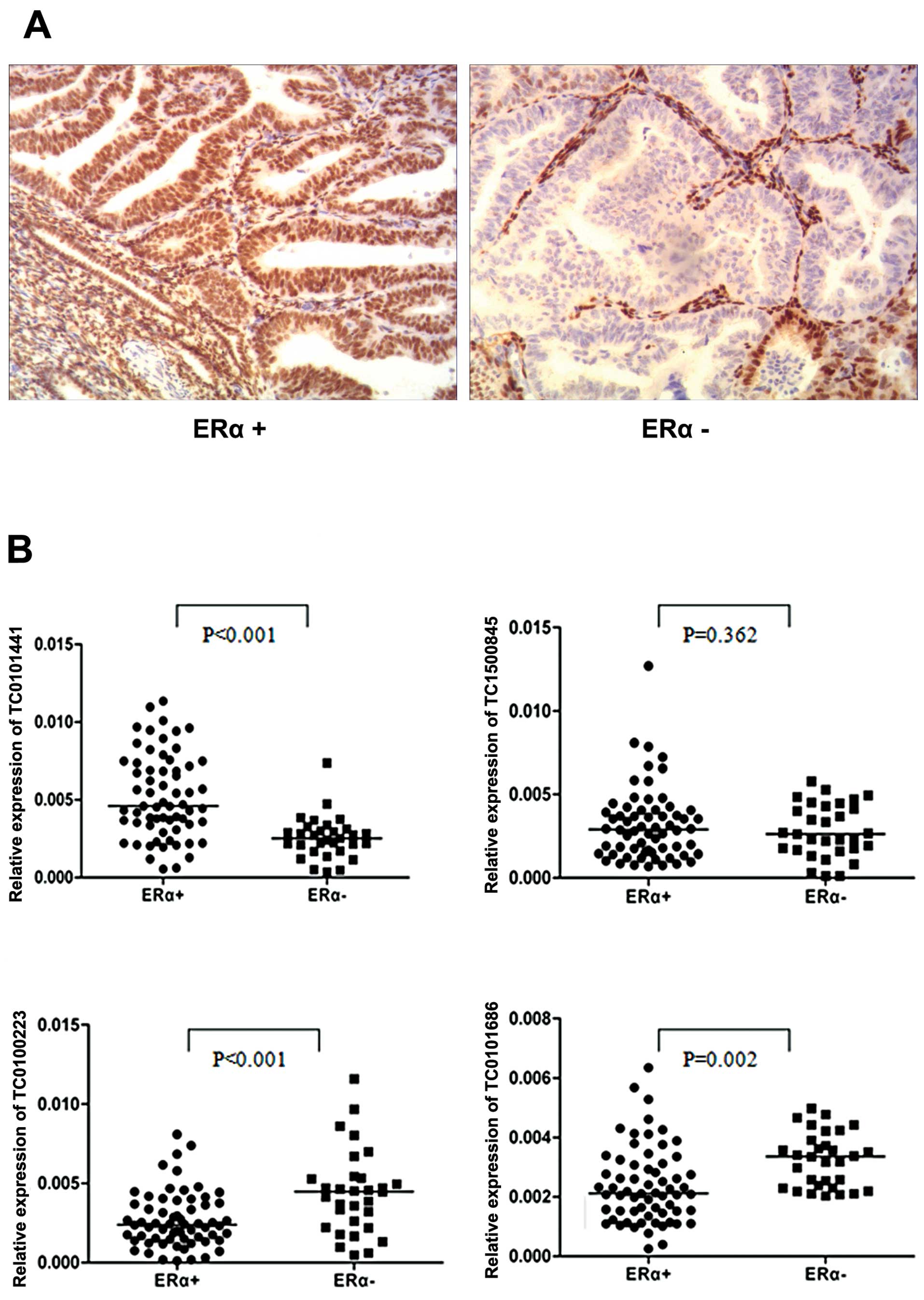
E2-regulated lncRNAs, detected in ERα+ ovarian cancer cells, could
discriminate between ERα+ and ERα− EOC tissues. Based on the
qRT-PCR assay, we found that ERα+ tissues had lower expression of
TC0100223 and TC0101686 and higher expression of TC0101441
(Fig. 3, ERα+, n=64 vs. ERα−, n=31,
P<0.01; Fig. 3A shows the
representative immunohistochemistry results of ERα expression in
EOC tissues). In contrast, TC1500845 was not differentially
expressed between ERα+ and ERα− EOC tissues. These results may be
suggestive of the potential clinical significance of TC0100223,
TC0101686 and TC0101441 in ERα+ EOC.
Association of lncRNA expression with
clinicopathological characteristics in ERα+ EOC
According to the median value which was used as the
cut-off (30), specific lncRNA
expression in ERα+ EOC tissues, equal or more than median value was
defined as high lncRNA group, and less than median value was
defined as low lncRNA group. As shown in Table V, low-expression of TC0100223 and
TC0101686 and high-expression of TC0101441 were closely related to
ERα+ EOC tissues with advanced FIGO stage and/or high histological
grade (P<0.05), suggesting that aberrant expression of the three
candidate lncRNAs is associated with a more malignant ovarian
cancer phenotype.
 | Table VAssociation of lncRNA expression with
clinicopathological variables in ERα-positive EOC patients. |
Table V
Association of lncRNA expression with
clinicopathological variables in ERα-positive EOC patients.
| | High TC0101441
expression | Low TC0100223
expression | Low TC0101686
expression |
|---|
| |
|
|
|
|---|
| Variables | Cases (N) | n (%) | P-value | n (%) | P-value | n (%) | P-value |
|---|
| Age (years) |
| <50 | 25 | 14 (56.0) | 0.442 | 10 (40.0) | 0.2 | 15 (60.0) | 0.2 |
| ≥50 | 39 | 18 (46.2) | | 22 (56.4) | | 17 (43.6) | |
| Histological
subtype |
| Serous | 49 | 23 (46.9) | 0.376 | 24 (49.0) | 0.768 | 22 (44.9) | 0.14 |
| Other | 15 | 9 (60.0) | | 8 (53.3) | | 10 (66.7) | |
| FIGO stage |
| I–II | 24 | 5 (20.8) | <0.001 | 8 (33.3) | 0.039 | 7 (29.2) | 0.01 |
| III–IV | 40 | 27 (67.5) | | 24 (60.0) | | 25 (62.5) | |
| Histological
grade |
| G1–G2 | 27 | 6 (22.2) | <0.001 | 9 (33.3) | 0.023 | 11 (40.7) | 0.206 |
| G3 | 37 | 26 (70.3) | | 23 (62.2) | | 21 (56.8) | |
| Ascites |
| >100 | 20 | 11 (55.0) | 0.59 | 7 (35.0) | 0.106 | 12 (60.0) | 0.281 |
| ≥100 | 44 | 21 (47.7) | | 25 (56.8) | | 20 (45.5) | |
Association of lncRNA expression with
prognosis of ERα+ EOC patients
We investigated whether the expression of TC0100223,
TC0101686 and TC0101441 correlated with the postoperative survival
of ERα+ EOC patients. Among the 64 ERα+ EOC patients, 38 died
during follow-up. In univariate analysis, OS was associated with
the FIGO stage, histological grade and expression of TC0100223 and
TC0101441 (P<0.05; Table VI).
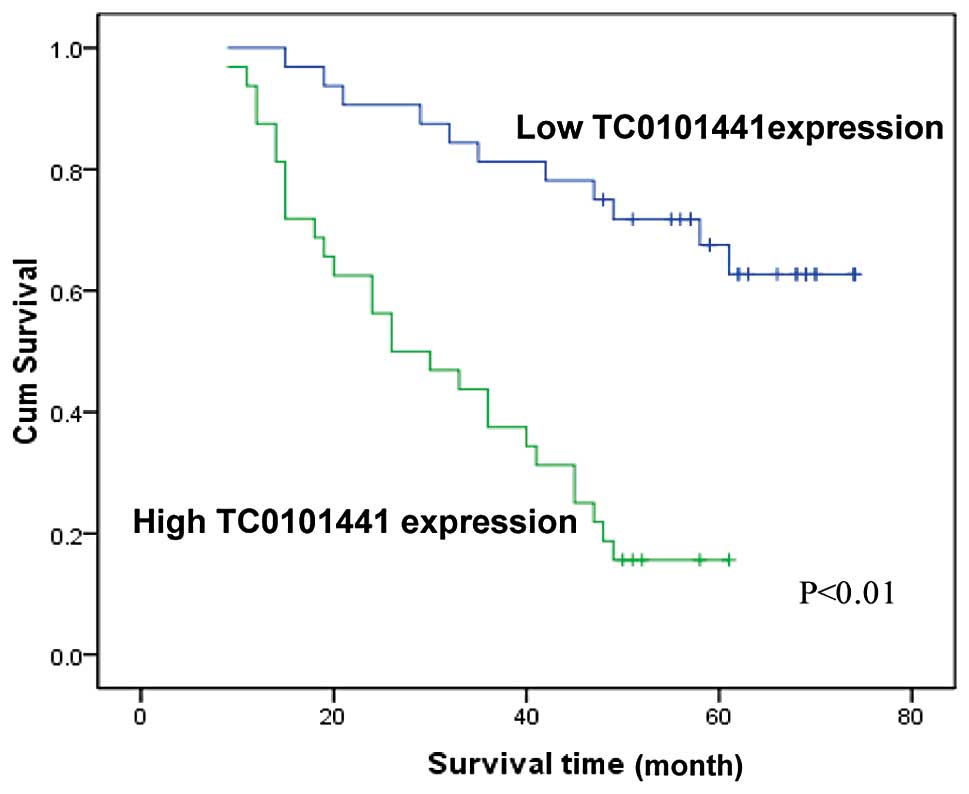
Multivariate analysis further confirmed that high TC0101441
expression, advanced FIGO stage and high histological grade were
independent factors for evaluation of OS in ERα+ EOC patients
(P<0.05, Table VI; Fig. 4 shows the OS curves according to
TC0101441 expression). Thus, it was concluded that of the three
candidate lncRNAs, TC0101441 could be used as an independent
prognostic factor for OS of ERα+ EOC patients.
 | Table VIUnivariate analysis for overall
survival in ERα-positive EOC patients. |
Table VI
Univariate analysis for overall
survival in ERα-positive EOC patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Overall
survival | | Overall
survival |
|---|
|
| |
|
|---|
| Variables | Mean ± SE
(months) | P-value | β | SE | Wald | P-value | Exp (β) | 95% CI |
|---|
| Age (years) |
| <50 | 45.52±4.21 | 0.939 | - | - | - | - | - | - |
| ≥50 | 47.49±3.98 | | - | - | - | - | - | - |
| Histological
subtype |
| Serous | 45.99±3.53 | 0.496 | - | - | - | - | - | - |
| Other | 52.30±5.71 | | - | - | - | - | - | - |
| FIGO stage |
| I–II | 69.63±2.42 | | - | - | - | - | - | - |
| III–IV | 33.03±2.73 | <0.001 | 2.305 | 0.635 | 13.168 | <0.001 | 10.022 | 2.886–34.803 |
| Histological
grade |
| G1–G2 | 64.81±2.97 | | - | - | - | - | - | - |
| G3 | 32.62±2.90 | <0.001 | 0.991 | 0.481 | 4.238 | 0.04 | 2.693 | 1.049–6.915 |
| Ascites |
| <100 | 44.71±5.95 | 0.77 | - | - | - | - | - | - |
| ≥100 | 48.67±3.48 | | - | - | - | - | - | - |
| TC0101686
expression |
| Low | 44.45±4.23 | 0.325 | - | - | - | - | - | - |
| High | 50.32±4.28 | | - | - | - | - | - | - |
| TC0100223
expression |
| Low | 37.48±3.48 | 0.018 | - | - | - | - | - | - |
| High | 54.40±3.99 | | 0.019 | 0.352 | 0.003 | 0.957 | 0.981 | 0.492–1.956 |
| TC0101441
expression |
| Low | 60.88±3.48 | | - | - | - | - | - | - |
| High | 32.16±3.04 | <0.001 | 0.817 | 0.402 | 4.122 | 0.042 | 2.263 | 1.029–4.979 |
Discussion
In the present study, we identified a series of
differentially expressed E2-regulated lncRNAs in ERα+ ovarian
cancer SKOV3 cells using a microarray. Bioinformatics functional
analyses indicated that a fraction of these lncRNAs had the
potential to contribute to cancer progression. Furthermore, in
order to confirm that some E2-regulated lncRNAs are related to the
development of ERα+ EOC, we tested the expression of several
candidate lncRNAs in EOC tissues. The results showed that some
candidate lncRNAs were aberrantly expressed in ERα+ compared to
ERα− EOC tissues, and their differential expression was associated
with certain clinicopathological variables and poor prognosis of
ERα+ EOC. To the best of our knowledge, this is the first study to
report E2-regulated lncRNAs in ERα+ EOC, the results of which may
provide insight into the estrogenic effects on EOC progression and
the design of new target therapies based on a perspective of
lncRNA.
It is known that the ovary is a main source and
target tissue of E2 in women. The action of E2 on ovarian tissue is
believed to be mediated by the two ERs, ERα and ERβ. ERβ is highly
expressed in normal epithelial ovarian cells and benign tumors,
whereas ERα is expressed to a much greater extent in malignant
ovarian tumors (28). Several
studies thus far have revealed the contributions to EOC progression
by multiple E2/ERα-regulated target protein-coding genes, such as
cyclin D1 and c-myc (which are involved in cellular growth control)
and fibulin-1 and cathepsin-D (which are involved in cellular
motility and invasion) (10,11).
Our previous studies also showed that E2 promoted metastasis and
invasion in ERα+ ovarian cancer SKOV3 cells by regulating HIF-1,
nm23-H1, E-cadherin and MMP-2 (5–7).
Despite these protein-coding genes, however, the exact effects of
E2 on EOC development remain unclear. In the present study, we
identified 115 E2-regulated lncRNAs in ERα+ SKOV3 cells using a
microarray, and subsequent bioinformatics analyses indicated that a
subset of these lncRNAs had the potential to contribute to cancer
progression. These findings may extend our current knowledge
regarding E2 regulation of protein-coding genes in EOC progression
to include lncRNAs.
lncRNAs, initially argued to be spurious
transcriptional noise, are emerging as new regulators in the cancer
paradigm. Misregulation of lncRNAs has been documented in many
types of human cancer. For example, DDC and PCGEM are overexpressed
in prostate cancer compared to normal prostate tissue, implicating
their roles in tumorigenesis (31,32).
Increased expression of MALAT1 indicates a poorer clinical outcome
of lung cancer patients (15).
HOTAIR is overexpressed in primary breast tumors and metastases,
and elevated HOTAIR expression is an indispensable predictor of
eventual metastasis and mortality (14). Inspired by these lines of evidence
of lncRNA roles in cancer biology, we hypothesized that certain
E2-regulated lncRNAs detected in SKOV3 cells in the current study
may also have the potential to contribute to EOC progression. To
address this hypothesis, we initially tested two upregulated
(TC1500845 and TC0101441) and two downregulated lncRNAs (TC0100223
and TC0101686) in EOC tissues, the results of which showed a
significant correlation of overexpressed TC0101441 and
low-expressed TC0100223 and TC0101686 with ERα+ compared to ERα−
EOC. Moreover, low-expression of TC0100223 and TC0101686 and
overexpression of TC0101441 were found to be related to ERα+ EOC
tissues with advanced FIGO stage and/or high histological grade.
Most importantly, multivariate survival analysis revealed that
TC0101441 was an independent prognostic factor for overall
survival. Taken together, our findings suggest that the aberrant
expression of certain E2-regulated lncRNAs is associated with
malignant cancer phenotypes and poor clinical outcome of ERα+ EOC
patients. Hence, these results may also lead us to consider that
E2-regulated lncRNAs can be used as candidate biomarkers for EOC
prognosis and therapy. Clearly, further studies are required to
elucidate the roles and mechanisms by which these lncRNAs promote
EOC development in detail.
In conclusion, the present study provided the first
evidence that E2 can modulate a panel of lncRNAs in ERα+ EOC cells.
Some aberrantly expressed lncRNAs, including TC0100223, TC0101686
and TC0101441, are correlated with the advanced cancer phenotypes.
Of note, TC0101441 was an independent factor for poor prognosis of
ERα+ EOC patients. Collectively, encouraged by the involvement of
these E2-regulated lncRNAs in ERα+ EOC progression, our data
highlight the utility of considering lncRNA expression in the
future understanding of estrogenic effects on EOC progression and
in the design of new target therapies.
Acknowledgements
The authors are grateful to the department managers
who provided the microarray services at the National Engineering
Center for Biochip at the Shanghai Biotechnology Corporation. The
project described was supported by a Grant Number 81370689 from the
National Natural Science Foundation of China.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Tsilidis KK, Allen NE, Key TJ, Dossus L,
Kaaks R, Bakken K, Lund E, Fournier A, Dahm CC, Overvad K, Hansen
L, Tjønneland A, Rinaldi S, Romieu I, Boutron-Ruault MC,
Clavel-Chapelon F, Lukanova A, Boeing H, Schütze M, Benetou V,
Palli D, Berrino F, Galasso R, Tumino R, Sacerdote C,
Bueno-de-Mesquita HB, van Duijnhoven FJ, Braem MG, Onland-Moret NC,
Gram IT, Rodríguez L, Duell EJ, Sánchez MJ, Huerta JM, Ardanaz E,
Amiano P, Khaw KT, Wareham N and Riboli E: Menopausal hormone
therapy and risk of ovarian cancer in the European prospective
investigation into cancer and nutrition. Cancer Causes Control.
22:1075–1084. 2011. View Article : Google Scholar
|
|
4
|
Park SH, Cheung LW, Wong AS and Leung PC:
Estrogen regulates snail and slug in the down-regulation of
E-cadherin and induces metastatic potential of ovarian cancer cells
through estrogen receptor alpha. Mol Endocrinol. 22:2085–2098.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hua K, Din J, Cao Q, Feng W, Zhang Y, Yao
L, Huang Y, Zhao Y and Feng Y: Estrogen and progestin regulate
HIF-1α expression in ovarian cancer cell lines via the activation
of Akt signaling transduction pathway. Oncol Rep. 21:893–898.
2009.
|
|
6
|
Hua K, Feng W, Cao Q, Zhou X, Lu X and
Feng Y: Estrogen and progestin regulate metastasis through the
PI3K/AKT pathway in human ovarian cancer. Int J Oncol. 33:959–967.
2008.PubMed/NCBI
|
|
7
|
Ding JX, Feng YJ, Yao LQ, Yu M, Jin HY and
Yin LH: The reinforcement of invasion in epithelial ovarian cancer
cells by 17β-estradiol is associated with upregulation of Snail.
Gynecol Oncol. 103:623–630. 2006.PubMed/NCBI
|
|
8
|
Spillman MA, Manning NG, Dye WW, Sartorius
CA, Post MD, Harrell JC, Jacobsen BM and Horwitz KB:
Tissue-specific pathways for estrogen regulation of ovarian cancer
growth and metastasis. Cancer Res. 70:8927–8936. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Laviolette LA, Garson K, Macdonald EA,
Senterman MK, Courville K, Crane CA and Vanderhyden BC:
17β-estradiol accelerates tumor onset and decreases survival in a
transgenic mouse model of ovarian cancer. Endocrinology.
151:929–938. 2010.
|
|
10
|
Twal WO, Czirok A, Hegedus B, Knaak C,
Chintalapudi MR, Okagawa H, Sugi Y and Argraves WS: Fibulin-1
suppression of fibronectin regulated cell adhesion and motility. J
Cell Sci. 114:4587–4598. 2001.PubMed/NCBI
|
|
11
|
Rochefort H, Chalbos D, Cunat S, Lucas A,
Platet N and Garcia M: Estrogen regulated proteases and
antiproteases in ovarian and breast cancer cells. J Steroid Biochem
Mol Biol. 76:119–124. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P,
Kong B, Li R, West RB, van de Vijver MJ, Sukumar S and Chang HY:
Long non-coding RNA HOTAIR reprograms chromatin state to promote
cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
Attardi LD, Regev A, Lander ES, Jacks T and Rinn JL: A large
intergenic noncoding RNA induced by p53 mediates global gene
repression in the p53 response. Cell. 142:409–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: a tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Silva JM, Boczek NJ, Berres MW, Ma X and
Smith DI: LSINCT5 is over expressed in breast and ovarian cancer
and affects cellular proliferation. RNA Biol. 8:496–505. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rangel LB, Sherman-Baust CA, Wernyj RP,
Schwartz DR, Cho KR and Morin PJ: Characterization of novel human
ovarian cancer-specific transcripts (HOSTs) identified by serial
analysis of gene expression. Oncogene. 22:7225–7232. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu W, Seok J, Mindrinos MN, Schweitzer AC,
Jiang H, Wilhelmy J, Clark TA, Kapur K, Xing Y, Faham M, Storey JD,
Moldawer LL, Maier RV, Tompkins RG, Wong WH, Davis RW and Xiao W:
Human transcriptome array for high-throughput clinical studies.
Proc Natl Acad Sci USA. 108:3707–3712. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia H, Osak M, Bogu GK, Stanton LW,
Johnson R and Lipovich L: Genome-wide computational identification
and manual annotation of human long noncoding RNA genes. RNA.
16:1478–1487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tafer H and Hofacker IL: RNAplex: a fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang C, Han L, Zhang A, Yang W, Zhou X,
Pu P, Du Y, Zeng H and Kang C: Global changes of mRNA expression
reveals an increased activity of the interferon-induced signal
transducer and activator of transcription (STAT) pathway by
repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol.
36:1503–1512. 2010.PubMed/NCBI
|
|
27
|
Yang XY, Xi MR, Yang KX and Yu H:
Prognostic value of estrogen receptor and progesterone receptor
status in young Chinese ovarian carcinoma patients. Gynecol Oncol.
113:99–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pujol P, Rey JM, Nirde P, Roger P,
Gastaldi M, Laffargue F, Rochefort H and Maudelonde T: Differential
expression of estrogen receptor-α and -β messenger RNAs as a
potential marker of ovarian carcinogenesis. Cancer Res.
58:5367–5373. 1998.
|
|
29
|
Darb-Esfahani S, Wirtz RM, Sinn BV,
Budczies J, Noske A, Weichert W, Faggad A, Scharff S, Sehouli J,
Oskay-Ozcelik G, Zamagni C, De Iaco P, Martoni A, Dietel M and
Denkert C: Estrogen receptor 1 mRNA is a prognostic factor in
ovarian carcinoma: determination by kinetic PCR in formalin-fixed
paraffin-embedded tissue. Endocr Relat Cancer. 16:1229–1239. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, Zhang L, Wang F, Sun SH
and Zhou WP: Long noncoding RNA associated with microvascular
invasion in hepatocellular carcinoma promotes angiogenesis and
serves as a predictor for hepatocellular carcinoma patients’ poor
recurrence-free survival after hepatectomy. Hepatology.
56:2231–2241. 2012.PubMed/NCBI
|
|
31
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: a new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 9:5975–5979. 1999.PubMed/NCBI
|
|
32
|
Srikantan V, Zou Z, Petrovics G, Xu L,
Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino
K, Buzard GS, Mostofi FK, McLeod DG, Moul JW and Srivastava S:
PCGEM1, a prostate-specific gene, is overexpressed in
prostate cancer. Proc Natl Acad Sci USA. 97:12216–12221. 2000.
View Article : Google Scholar
|