Introduction
Renal cell carcinoma (RCC) is the most lethal of all
urological malignancies, accounting for ~2% of cancers worldwide
(1). In the US, there are ~65,000
new cases of kidney cancer along with 13,500 deaths each year, with
the majority of these cases being related to RCC (2). Over the last four decades, the
incidence of RCC has steadily increased (3). RCC shows less response to chemotherapy
than other urological cancers (4).
Surgical resection for clinically localized disease remains the
mainstay for curative intervention. However, the aggressive and
often insidious nature of RCC is reflected by recurrence rates of
20–40% after nephrectomy for clinically localized disease (5,6). Thus,
there is an urgent need to develop novel therapeutic approaches for
RCC. To achieve this, a deeper understanding of the molecular and
genetic networks that control the initiation and progression of RCC
is imperative.
Yin Yang 1 (YY1) is a ubiquitous and multifunctional
zinc-finger transcription factor member of the Polycomb group
protein family, a group of homeobox gene receptors that play
critical roles in normal biological processes such as
embryogenesis, differentiation, replication and cellular
proliferation (7). YY1 exerts its
effects on genes involved in these processes via its ability to
initiate, activate, or repress transcription depending upon the
context in which it binds. There are several types of YY1-related
diseases, such as viral infection and cancers. YY1 itself was found
to be upregulated in many cancer types, including lymphoma, breast,
prostate, colon, ovarian, cervical, and brain cancers and leukemia
(8). Moreover, in many human cancer
types, YY1 expression levels were found to be significantly
elevated in the metastatic tumor compared to its primary
counterpart, supporting the potential role of YY1 in cancer
development. It was shown that YY1 may regulate by both oncogenes
and tumor-suppressor genes through different mechanisms depending
on the tissue context (9–12), suggesting the dual potential of YY1
as tumor suppressor or oncogene in malignant cells. However, the
role of YY1 in the development of RCC is not fully understood.
microRNAs (miRNAs) are a class of single-stranded,
highly conserved non-coding RNAs of 18–24 nucleotides that regulate
gene expression by messenger RNA (mRNA) degradation or
translational repression (13). To
date, more than 17,000 miRNAs in over 153 species have been
identified (miRBase Sequence Database - release 17; www.miRbase.org), of which ~1,400 are found in humans.
Most miRs are evolutionarily conserved and are often found in
clusters (14). Recent studies have
highlighted the role of miR-34a as a tumor suppressor in a number
of tumor types including prostate cancer, hepatocellular carcinoma,
neuroblastoma and colon cancer (15–19).
Functionally miR-34a was found to affect tumor cell proliferation,
apoptosis, senescence, invasion, metastasis and drug resistance
(19–23).
In the present study, we found that expression of
YY1 was upregulated in RCC samples when compared to its primary
counterpart. We also demonstrated that silencing of YY1 reduced
cell growth and invasion of RCC cells, supporting its oncogenic
role in the development of RCC. Furthermore, we confirmed that YY1
is a target of miR-34a. The elevated expression of YY1 represses
CCAAT/enhancer-binding protein α (C/EBPα), which leads to further
inhibiton of miR-34a. This positive feedback loop promotes YY1
activity, which contributes to the development of RCC.
Materials and methods
Tissue samples
The paired tissue samples from primary tumor and
adjacent non-tumor sites were obtained from 14 RCC patients during
surgery prior to any therapeutic intervention at the Shanghai Tenth
People’s Hospital, Tongji University. All of the samples were
subsequently verified by histology. Informed written consent was
provided by all of the patients. The study protocol was approved by
the Ethics Committee of Shanghai Tenth People’s Hospital. To
identify the differential expression of miR-34a, YY1 and C/EBPα in
tumor specimens versus adjacent non-tumor renal tissue, we pooled
these samples in two subsets. Pool 1 contained 14 primary tumor
samples. Pool 2 was generated from the matched 14 normal renal
tissue samples.
Cell lines and cell culture
RCC cell lines, 786–0 and ACHN, were cultured in
Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal
bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin.
All cells were maintained in 5% CO2 atmosphere at
37°C.
Transfections
Cells were transfected with 50 nM miR-34a mimics and
inhibitors (GenePharma Co., Ltd., Shanghai China), YY1-siRNA
(Sigma-Aldrich Co., LLC, St. Louis, MO, USA), YY1 and C/EBPα
plasmids (Genechem Co., Ltd., Shanghai, China) using Lipofectamine
2000 (Invitrogen Corp., Carlsbad, CA, USA), respectively. RNA and
proteins were harvested 48 h after transfection.
Quantitative RT-PCR
Total RNA was extracted from the tissues or cultured
cells following the indicated treatment using TRIzol (Invitrogen
Corp.) method. qRT-PCR or Stem-loop PCR (24) was performed using the SYBR Green PCR
kit (Takara Biotechnology Co., Ltd., Dalian, China), Real-time PCR
data for mRNA (YY1, C/EBPα) and miRNA (miR34a) are expressed
relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6,
respectively. The primers used were described as follows: YY1
forward, 5′-CCT GGC ATT GAC CTC TCA GAT CCA-3′ and reverse, 5′-GGG
CAA GCT ATT GTT CTT GGA GCA-3′; C/EBPα forward, 5′-AAC ATC GCG GTG
CGC AAG AG-3′ and reverse, 5′-TTC GCG GCT CAG CTG TTC CA-3′. The
stem-loop miR-34a RT primer was 5′-CTC AAC TGG TGT CGT GGA GTC GGC
AAT TCA GTT GAG ACA ACC AG-3′; miR-34a forward, 5′-ACA CTC CAG CTG
GGT GGC AGT GTC TTA GCT GG-3′ and reverse, 5′-TGG TGT CGT GGA GTC
G-3′. All reactions were performed in triplicate. The relative
expression levels of target genes or miRNA were calculated using
the 2−ΔΔCt method (25).
Luciferase reporter assay
The wild-type YY1–3′UTR region, generated by PCR
amplification, was cloned into the pcheck-2 luciferase reporter
plasmid (Promega, Madison, WI, USA). Specific primers were designed
for this system. The forward primer (5′-CCA CTC GAG GCA TCT TCC AGA
AGT GTG AT-3′) was Xhol-site-linked and the reverse primer
Notl-site-linked (5′-CCA GCG GCC GCC ATT CTA CAA CTG AGC ACC
AC-3′). Mutation of the miR-34a binding site was generated by a
PCR-based site-directed mutagenesis method, using wild-type
YY1–3′UTR reporter plasmid as the template. For the reporter assay,
ACHN cells were plated onto 24-well plates and transfected with
wild-type or mutant YY1–3′UTR reporter plasmid and miR-34a mimics
or inhibitors using Lipofectamine 2000. After transfection for 48
h, cells were harvested and assayed with the Dual-Luciferase
Reporter Assay System (Promega) according to the manufacturer’s
instructions. The tests were repeated in triplicate. Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Western blotting
Proteins were resolved on an SDS/PAGE gel (10% gel)
and subjected to immunoblot analysis using monoclonal antibodies
against YY1, C/EBPα or GAPDH (Epitomics, Burlingame, CA, USA). All
of the antibodies were used at working concentration (1:1000) in
PBS-T with 5% non-fat milk. The membrane was further probed with
HRP (horseradish peroxidase)-conjugated rabbit anti-(mouse IgG)
(1:2000 dilution; Santa Cruz Biotechnology), and the protein bands
were visualized using enhanced chemiluminescence (Amersham
Pharmacia).
Migration and invasion assay
To measure the cell migration activity, Transwell
assays were performed using Corning 8.0-μm Transwell®
cell culture inserts (Corning Inc., Corning, NY, USA). Membranes
were coated with purified fibronectin (Sigma) at a concentration of
10 μg/ml. For assessment of invasion, 1:10 diluted Matrigel-coated
Transwell inserts (BD Biosciences, San Jose, CA, USA) were used.
After transfection with either siRNA or expression vectors in
opti-MEM medium for 5 h, cells were collected and resuspended in
serum-free DMEM medium containing 0.1% bovine serum albumin (BSA).
Subsequently, cells (4×105/ml) were seeded in Transwell
chambers. After 24 h of incubation, the cells on the upper surface
of the filter were completely wiped away with a cotton swab. The
cells on the lower surfaces of the membrane were fixed with 100%
methanol, and counted under a microscope.
Cell proliferation assay
Cells were seeded into 96-well plates at an initial
density of 1×105 cells/well. MTT solution (200 μl) (5
mg/ml, Alfa Aesar) was added to each well and incubated for 5 h at
37°C. The supernatant was then discarded, and 100 μl of dimethyl
sulfoxide (DMSO, Sigma) was added to each well to dissolve the
precipitate. After 30 min at room temperature, the plates were
scanned spectrophotometrically with a microplate reader (Beckman
Coulter) set at 595 nm to measure the absorbance.
Statistical analysis
Statistical evaluations were conducted using the
t-test. P-values <0.05 were considered to be statistically
significant.
Results
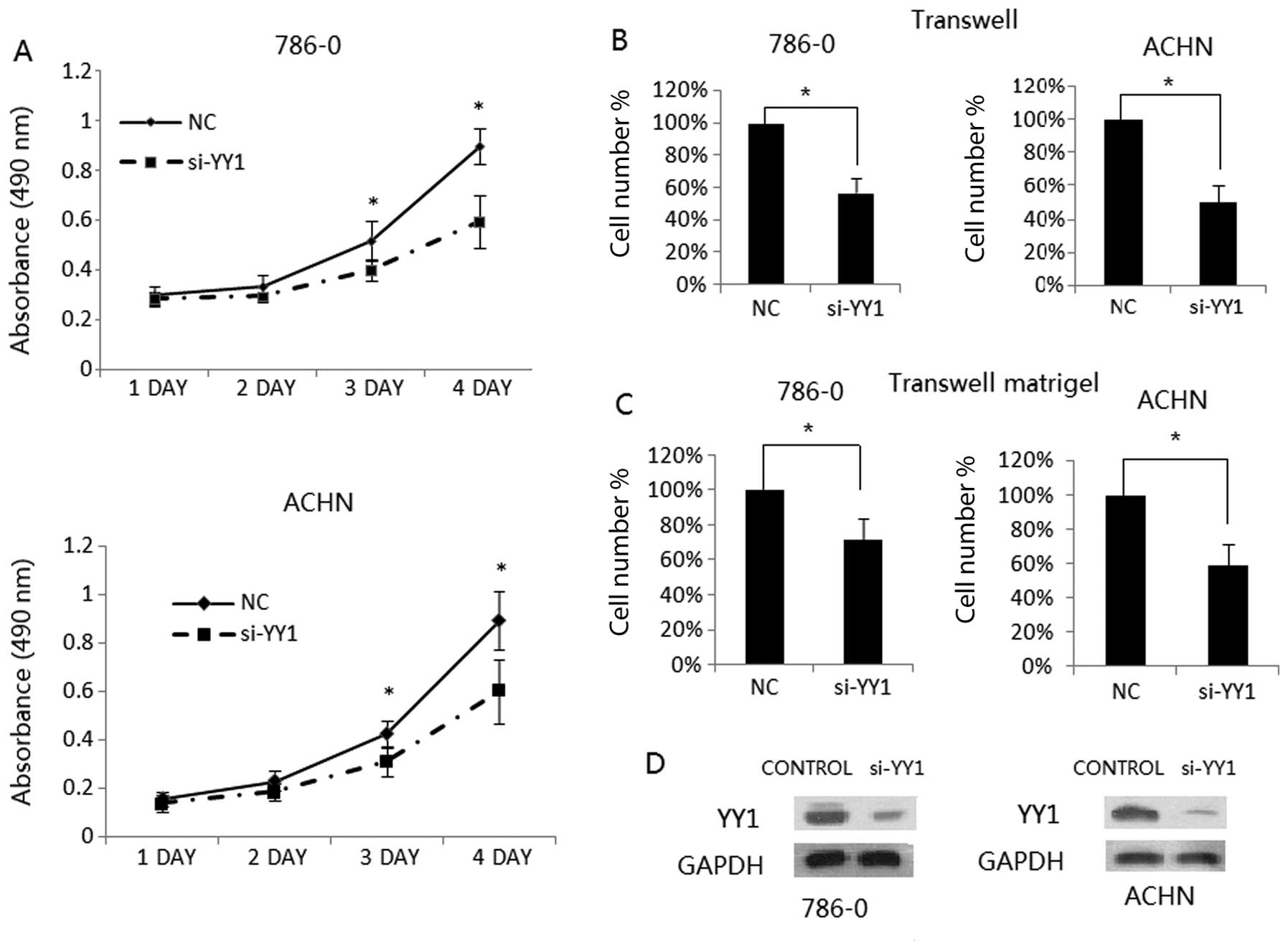
Silencing of YY1 inhibits RCC cell
growth, migration and invasion
To confirm the tumor promotion effect of YY1 in RCC,
we evaluated the impact of YY1 on the growth of RCC cell lines,
786-0 and ACHN. The growth curve of the RCC cell lines transfected
with si-YY control siRNA is shown in Fig. 1A. The cell growth was significantly
decreased in the si-YY1-transfected 786-0 and ACHN cells as
compared with the control siRNA-transfected cells (P<0.05). To
evaluated the impact of YY1 on cell migration and invasion,
Transwell and Matrigel invasion assays were employed. We found that
YY1 knockdown inhibited 786-0 and ACHN cell migration (Fig. 1B). Consistent with this finding, the
Matrigel invasion assay showed that YY1 knockdown significantly
inhibited the invasive capacity of RCC cells (Fig. 1C). These observations suggest that
YY1 plays an important role in promoting cell growth, migration and
invasive potential of RCC cells.
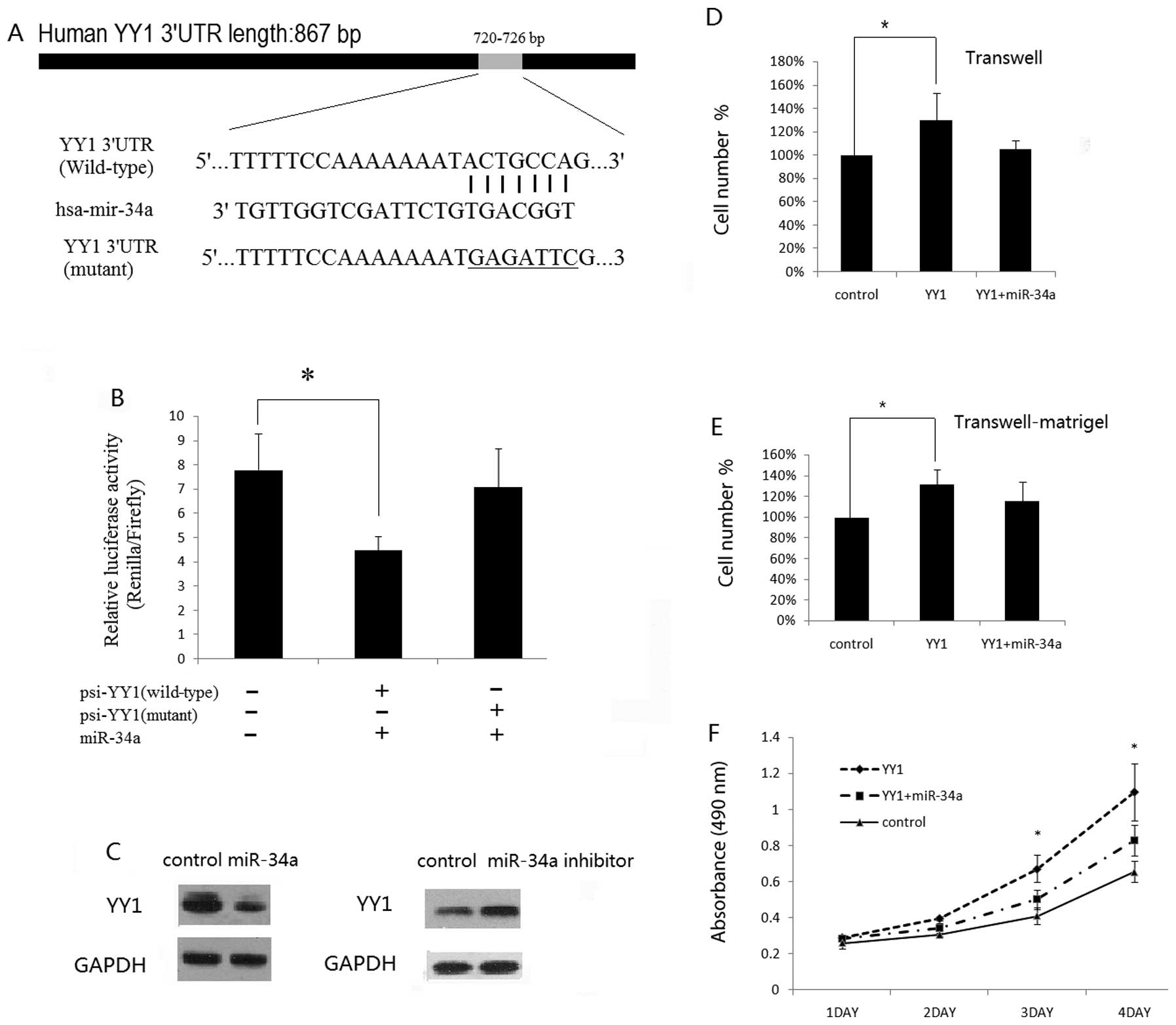
YY1 is a direct target of miR-34a
To associate miRNAs with the regulation of YY1
expression, a bioinformatics search was performed for potential
miRNAs targeting the mRNA of YY1 by using the public database
Targetscan (www.targetscan.org). Noteworthy, there
were several predicated miRNAs to target YY1, including miR-34a.
miR-34a was reported to play a tumor-suppressive role in a number
of tumor types (15–19). We then performed a luciferase
reporter assay to verify that miR-34a directly targets YY1. We
found that co-transfection of miR-34a mimics and wild-type
YY1–3′UTR significantly decreased the luciferase activity in ACHN
cells as compared with the control. However, miR-34a mimics had no
effect on the luciferase activity when co-transfected with mutant
YY1–3′UTR (Fig. 2B). These data
demonstrate a specific inhibitory effect of miR-34a on the 3′UTR of
YY1 through direct interaction. Furthermore, to examine the
potential negative regulatory effect of miR-34a on endogenous YY1,
we transfected miR-34a mimics, miR-34a inhibitors or control mimics
into ACHN cells. In comparison with the miR control, we found the
miR-34a exerted a discernible inhibitory effect on the YY1 protein
level, while an increased protein level of YY1 was found in ACHN
cells following transfection with miR-34a inhibitors (Fig. 2C). These data showed that YY1 is one
of the direct targets of miR-34a.
Ectopic expression of miR-34a decreases
the YY1-induced invasive or proliferative capacity of RCC cells in
vitro
We next determined whether or not miR-34a
overexpression impairs the oncogenic effect of YY1 on cell growth,
migration or invasion of RCC cells. We transfected YY1
overexpression plasmids alone, or co-transfected YY1 plasmids and
miR-34a mimics into ACHN cells. Notably, exogenous YY1 expression
in ACHN cells promoted the migratory, invasive or proliferative
potential of ACHN cells (Fig.
2D–F). Furthermore, constitutively expressed miR-34a rescued
the previously aggressive phenotype in ACHN cells initiated by YY1
(Fig. 2D–F), suggesting that YY1 is
indeed a functional target of miR-34a.
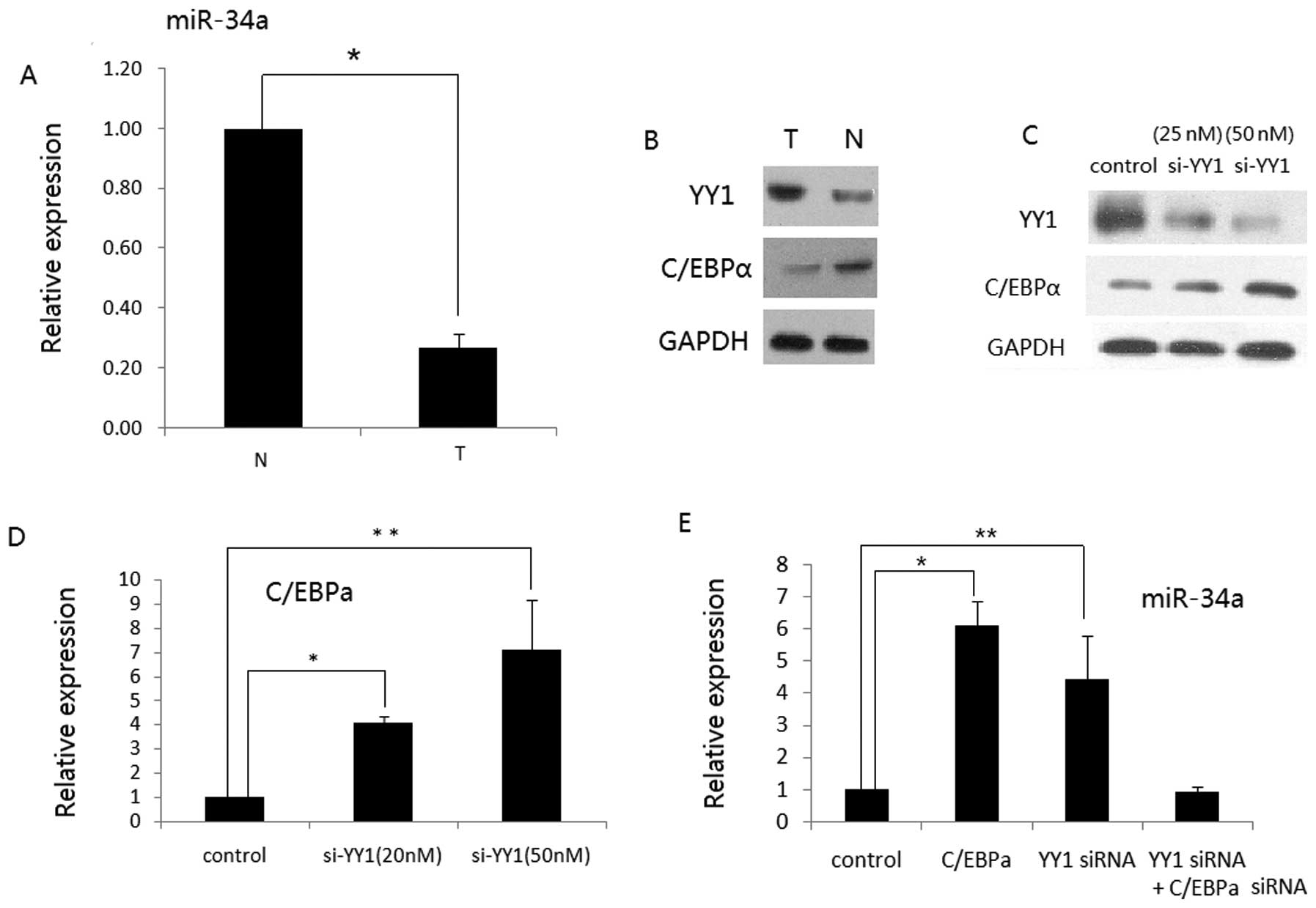
YY1 modulates miR-34a expression via
C/EBPα in human RCC
Since YY1 downregulates C/EBPα expression through a
promoter-dependent manner in HCC cells (26) and C/EBPα leads to upregulation of
miR-34a expression during granulocytic differentiation (27), we aimed to ascertain whether or not
YY1 regulates miR-34a expression via C/EBPα in human RCC. We
observed that miR-34a was downregulated in the pools of cancer
tissues when compared to that of the normal tissues. Moreover,
miR-34a displayed an inverse correlation with YY1, but a positive
correlation with C/EBPα (Fig. 3A and
B), suggesting that a low level of miR-34a may contribute to
the upregulation of YY1 or the elevated expression of YY1 in turn
inhibits miR-34a via C/EBPα suppression. We then revealed that YY1
siRNA was able to induce the expression of C/EBPα both at the mRNA
and protein levels in ACHN cells (Fig.
3C and D). We also found that either C/EBPα overexpression or
YY1 knockdown alone in ACHN cells increased miR-34a expression.
However, when C/EBPα siRNA was transfected into ACHN cells, the
upregulation of miR-34a by si-YY1 was abrogated (Fig. 3E). These data indicate that YY1,
C/EBPα and miR-34a collaborate to form feedforward loops, which
contributes to RCC progression (Fig.
4).
Discussion
In the cancer-associated miRNAs identified to date,
miR-34a has emerged as a robust tumor suppressor with diverse
targets in multiple types of cancers (15–19).
However, the effect of miR-34a and its molecular target in RCC are
still largely unknown. Herein, we report that YY1 is a novel target
of miR-34a. We also found that miR34a was downregulated in RCC
tissues. Furthermore, overexpression of miR-34a impaired the
oncogenic effect of YY1 on cell growth, migration or invasion of
RCC cells, suggesting that miR-34a may serve as a potential
therapeutic target.
YY1 is a 65-kDa multifunctional zinc-finger
transcription factor belonging to the human GLI-Kruppel family of
nuclear proteins (28,29). It can bind to the specific DNA
consensus sequence, 5′-CGCCATNTT-3′, which is present in many
promoters and regulates transcriptional activity by either
activation or repression (28,29).
Therefore, YY1 plays a complicated role in tumor development,
largely depending on tissue context, interaction of partners and
downstream targets. It was shown that YY1 plays an oncogenic role
by activating oncogenes ERBB2 and VEGF, or inhibiting
tumor-suppressor genes p53 and E-cadherin (11,30,31).
It was also shown that YY1 is able to suppress tumorigenesis via
upregulating tumor suppressor genes such as HLJ1 and BRCA1 or by
inhibiting c-myc function by direct interaction (32–34).
In this study, we found that YY1 inhibits C/EBPα
expression in RCC cells. YY1 siRNA was able to induce the
expression of C/EBPα both at the mRNA and protein levels in ACHN
cells. Zhang et al found that YY1 suppresses the expression
of C/EBPα in a promoter-dependent manner in hepatocellular
carcinoma cells (26). It was
reported that YY1 functions as a critical component of epigenetic
regulatory networks. It could promote the methylation of its target
genes by physically interacting with SUZ12 and recruiting DNA
methyltransferases to the promoter of target genes (35). However, Zhang et al did not
find that the downregulation of C/EBPα in HCC cells was due to the
methylation of the promoter despite the upregulation of C/EBPα
expression in HCC cells after Aza treatment. Instead, the
interaction of YY1 with the C/EBPα promoter was suppressed after
Aza treatment. Of note, Girard et al demonstrated that
RARα-PLZF (a fusion protein in acute promyelocytic leukemia)
recruits HDAC1 and causes histone H3 deacetylation at C/EBPα target
loci, thereby decreasing the expression of C/EBPα target genes
(36). Therefore, we hypothesized
that the inhibitory effect of YY1 on C/EBPα may involve a complex
epigenetic network. However, such a hypothesis needs further
investigation.
C/EBPα is a basic leucine zipper transcription
factor that is expressed in many tissues (37). C/EBPα plays an important role in
normal tissue development, namely, in the regulation of cell
proliferation and cell differentiation (38,39).
Several studies here demonstrated that diverse molecular mechanisms
are responsible for C/EBPα inactivation of expression or function
in various types of cancers, including liver cancer (26), leukemia (AML and chronic myelogenous
leukemia) (40,41) and lung cancer (42). It was also shown that C/EBPα
directly regulates miR-34a expression during granulocytic
differentiation (27). Noteworthy,
we found that these two tumor suppressors C/EBPα and miR-34a were
both downregulated in renal tumor tissues. Ectopic expression of
C/EBPα in ACHN cells promoted miR-34a transcription. Furthermore,
YY1 knockdown led to the upregulation of miR-34a through restoring
C/EBPα expression in ACHN cells.
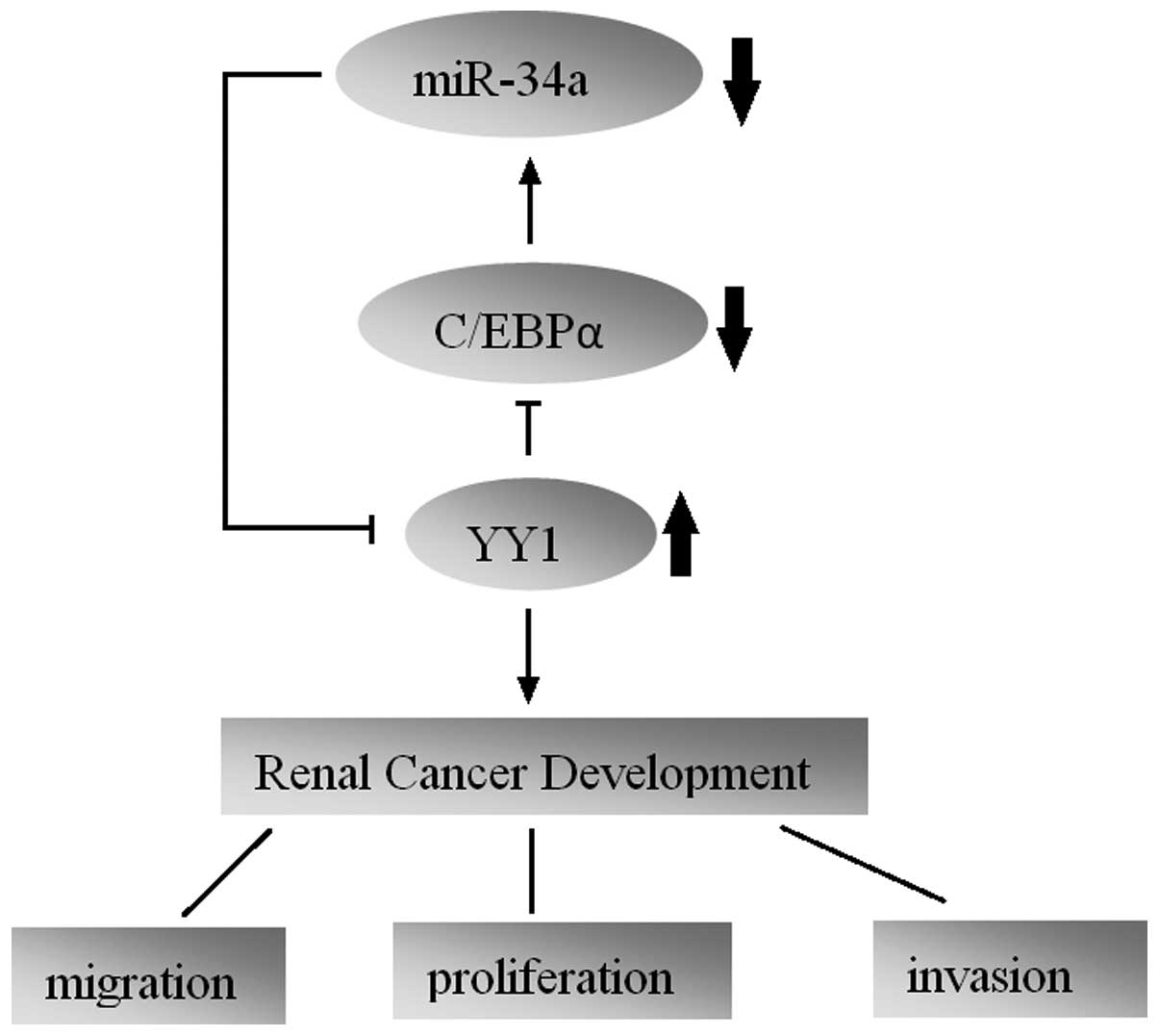
In conclusion, our study first showed a regulatory
circuitry in RCC cells that incorporates protein-coding and miRNA
genes. We found that YY1 is upregulated in human RCC tumors with an
oncogenic function. miR-34a exerts a tumor-suppressive function in
RCC through directly suppressing oncogenic YY1. Furthermore,
repression of C/EBPα caused by YY1 leads to a further reduction in
miR-34a expression, which forms a YY1-C/EBPα-miR-34a positive
feedback loop. Our study highlights the importance of a novel
regulatory circuitry in maintaining the proliferative and
aggressive phenotypes of RCC, which may serve as a potential
therapeutic target for RCC treatment.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81201884 and 81171883).
References
|
1
|
Liou LS, Shi T, Duan ZH, et al: Microarray
gene expression profiling and analysis in renal cell carcinoma. BMC
Urol. 4:92004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Chow WH, Devesa SS, Warren JL and Fraumeni
JF Jr: Rising incidence of renal cell cancer in the United States.
JAMA. 281:1628–1631. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amato RJ: Chemotherapy for renal cell
carcinoma. Semin Oncol. 27:177–186. 2000.PubMed/NCBI
|
|
5
|
Bukowski RM: Natural history and therapy
of metastatic renal cell carcinoma: the role of interleukin-2.
Cancer. 80:1198–1220. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castellano G, Torrisi E, Ligresti G, et
al: The involvement of the transcription factor Yin Yang 1 in
cancer development and progression. Cell Cycle. 8:1367–1372. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zaravinos A and Spandidos DA: Yin yang 1
expression in human tumors. Cell Cycle. 9:512–522. 2010. View Article : Google Scholar
|
|
9
|
Riggs KJ, Saleque S, Wong KK, et al:
Yin-yang 1 activates the c-myc promoter. Mol Cell Biol.
13:7487–7495. 1993.PubMed/NCBI
|
|
10
|
Lee JS, Galvin KM, See RH, et al: Relief
of YY1 transcriptional repression by adenovirus E1A is mediated by
E1A-associated protein p300. Genes Dev. 9:1188–1198. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gronroos E, Terentiev AA, Punga T and
Ericsson J: YY1 inhibits the activation of the p53 tumor suppressor
in response to genotoxic stress. Proc Natl Acad Sci USA.
101:12165–12170. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu S, Murai S, Kataoka K and Miyagishi M:
Cooperative regulation of p73 promoter by Yin Yang 1 and E2F1.
Nucleic Acids Symp Ser. 51:347–348. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brodersen P and Voinnet O: Revisiting the
principles of microRNA target recognition and mode of action. Nat
Rev Mol Cell Biol. 10:141–148. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim VN: MicroRNA biogenesis: coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fujita Y, Kojima K, Hamada N, et al:
Effects of miR-34a on cell growth and chemoresistance in prostate
cancer PC3 cells. Biochem Biophys Res Commun. 377:114–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Guessous F, Zhang Y, et al:
MicroRNA-34a inhibits glioblastoma growth by targeting multiple
oncogenes. Cancer Res. 69:7569–7576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei JS, Song YK, Durinck S, et al: The
MYCN oncogene is a direct target of miR-34a. Oncogene.
27:5204–5213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Welch C, Chen Y and Stallings RL:
MicroRNA-34a functions as a potential tumor suppressor by inducing
apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li N, Fu H, Tie Y, et al: miR-34a inhibits
migration and invasion by down-regulation of c-Met expression in
human hepatocellular carcinoma cells. Cancer Lett. 275:44–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang TC, Wentzel EA, Kent OA, et al:
Transactivation of miR-34a by p53 broadly influences gene
expression and promotes apoptosis. Mol Cell. 26:745–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Kelnar K, Liu B, et al: The
microRNA miR-34a inhibits prostate cancer stem cells and metastasis
by directly repressing CD44. Nat Med. 17:211–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang GL, Zhang XH, Guo GL, et al:
Clinical significance of miR-21 expression in breast cancer:
SYBR-Green I-based real-time RT-PCR study of invasive ductal
carcinoma. Oncol Rep. 21:673–679. 2009.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
26
|
Zhang S, Jiang T, Feng L, et al: Yin
Yang-1 suppresses differentiation of hepatocellular carcinoma cells
through the downregulation of CCAAT/enhancer-binding protein alpha.
J Mol Med. 90:1069–1077. 2012. View Article : Google Scholar
|
|
27
|
Pulikkan JA, Peramangalam PS, Dengler V,
et al: C/EBPalpha regulated microRNA-34a targets E2F3 during
granulopoiesis and is down-regulated in AML with CEBPA mutations.
Blood. 116:5638–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi Y, Seto E, Chang LS and Shenk T:
Transcriptional repression by YY1, a human GLI-Kruppel-related
protein, and relief of repression by adenovirus E1A protein. Cell.
67:377–388. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Galvin KM and Shi Y: Multiple mechanisms
of transcriptional repression by YY1. Mol Cell Biol. 17:3723–3732.
1997.PubMed/NCBI
|
|
30
|
Tong ZT, Cai MY, Wang XG, et al: EZH2
supports nasopharyngeal carcinoma cell aggressiveness by forming a
co-repressor complex with HDAC1/HDAC2 and Snail to inhibit
E-cadherin. Oncogene. 31:583–594. 2012.PubMed/NCBI
|
|
31
|
de Nigris F, Crudele V, Giovane A, et al:
CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during
malignancy. Proc Natl Acad Sci USA. 107:14484–14489.
2010.PubMed/NCBI
|
|
32
|
Lee MH, Lahusen T, Wang RH, et al: Yin
Yang 1 positively regulates BRCA1 and inhibits mammary cancer
formation. Oncogene. 31:116–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Austen M, Cerni C, Luscher-Firzlaff JM and
Luscher B: YY1 can inhibit c-Myc function through a mechanism
requiring DNA binding of YY1 but neither its transactivation domain
nor direct interaction with c-Myc. Oncogene. 17:511–520. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang CC, Tsai MF, Dai TH, et al:
Synergistic activation of the tumor suppressor, HLJ1, by the
transcription factors YY1 and activator protein 1. Cancer Res.
67:4816–4826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Samarakoon R and Higgins PJ: Integration
of non-SMAD and SMAD signaling in TGF-beta1-induced plasminogen
activator inhibitor type-1 gene expression in vascular smooth
muscle cells. Thromb Haemost. 100:976–983. 2008.PubMed/NCBI
|
|
36
|
Girard N, Tremblay M, Humbert M, et al:
RARalpha-PLZF oncogene inhibits C/EBPalpha function in myeloid
cells. Proc Natl Acad Sci USA. 110:13522–13527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schuster MB and Porse BT: C/EBPalpha: a
tumour suppressor in multiple tissues? Biochim Biophys Acta.
1766:88–103. 2006.PubMed/NCBI
|
|
38
|
Lopez RG, Garcia-Silva S, Moore SJ, et al:
C/EBPalpha and beta couple interfollicular keratinocyte
proliferation arrest to commitment and terminal differentiation.
Nat Cell Biol. 11:1181–1190. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramji DP and Foka P:
CCAAT/enhancer-binding proteins: structure, function and
regulation. Biochem J. 365:561–575. 2002.PubMed/NCBI
|
|
40
|
Pabst T and Mueller BU: Complexity of
CEBPA dysregulation in human acute myeloid leukemia. Clin Cancer
Res. 15:5303–5307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Perrotti D, Cesi V, Trotta R, et al:
BCR-ABL suppresses C/EBPalpha expression through inhibitory action
of hnRNP E2. Nat Genet. 30:48–58. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sato A, Yamada N, Ogawa Y and Ikegami M:
CCAAT/enhancer-binding protein-alpha suppresses lung tumor
development in mice through the p38alpha MAP kinase pathway. PloS
One. 8:e570132013. View Article : Google Scholar : PubMed/NCBI
|