Introduction
The World Health Organization has reported that
colorectal cancer (CRC) is the third most common cancer worldwide,
accounting for 940,000 million new cases annually and nearly
500,000 deaths each year. On the other hand, the treatment outcome
of CRC patients has recently improved. Patients with previously
untreated metastatic colorectal cancer (mCRC) have demonstrated
substantial improvements, with a median overall survival time now
reaching more than 24 months (1).
One of the factors responsible for this improved outcome may be the
development of systemic chemotherapy, including molecular-targeted
therapy, for mCRC.
Cetuximab is a human-mouse chimeric immunoglobulin
G1 (IgG1) monoclonal antibody for the epidermal growth factor
receptor (EGFR) that has been approved for use in patients with
mCRC expressing EGFR. Some clinical studies examining cetuximab
treatment in patients with mCRC have failed to show a significant
correlation between EGFR expression and the response of patients to
cetuximab therapy (2). The absence
of mutations in KRAS, which is one of the downstream
effectors of the EGFR signaling pathway, appears to be a reliable
marker for predicting the efficacy of cetuximab therapy. However,
some patients with KRAS mutations were recently reported to
benefit from cetuximab therapy (3).
The proposed working mechanism of cetuximab is
thought to include antibody-dependent cell-mediated cytotoxicity
(ADCC) triggered by Fc receptors (Fcγ-R) on natural killer cells,
macrophages and polymorphonuclear leukocytes. ADCC is a
well-recognized immune effector mechanism responsible for the
effect of IgG1. Previous reports have demonstrated the occurrence
of ADCC in CRC cell lines in vitro (2,4).
However, whether ADCC is correlated with the cell surface
expression of EGFR and/or the mutational status of downstream
effectors, such as KRAS and BRAF in CRC remains
unclear.
We demonstrated cetuximab-mediated ADCC in human CRC
cell lines and investigated whether the ADCC activities were
correlated with the cell surface expression levels of EGFR and/or
the mutational status of KRAS and BRAF. Furthermore,
we evaluated cetuximab-mediated ADCC activity using tumor cells
from resected specimens and peripheral blood samples from the same
CRC patients; we then investigated the associations between the
ADCC activities and the cell surface expression levels of EGFR as
well as the mutational statuses of KRAS and BRAF, in
addition to performing an assay using human CRC cell lines.
Materials and methods
Cell lines and cell culture
Nine human CRC cell lines (HT29, HCT15, HCT116,
DLD-1, SW480, SW867, WiDr, CaCo-2 and LoVo) and an epidermoid
carcinoma cell line (A431) with different KRAS and
BRAF mutational statuses were used in the present study.
HT29, SW480, CaCo-2 and A431 cells were cultured in Dulbecco’s
modified Eagle’s medium (DMEM) supplemented with heat-inactivated
10% fetal bovine serum (FBS), 50 U/ml of penicillin/streptomycin
and 4.0 mmol/l of glutamine. The other cell lines were maintained
in complete RPMI-1640 medium with the addition of 10%
heat-inactivated FBS, 100 U/ml of penicillin/streptomycin, and 2.0
mmol/l of glutamine and cultured at 37°C in a 5%
CO2-humidified atmosphere. Adherent cells were removed
using trypsin-EDTA solution [0.05% trypsin and 0.02% EDTA in
phosphate-buffered saline (PBS) without calcium and magnesium].
ADCC assay using human CRC cell
lines
Cetuximab-mediated ADCC activity was evaluated using
a 24-h lactate dehydrogenase (LDH)-releasing assay, the CytoTox 96
non-radioactive cytotoxicity assay kit (Promega, Madison, WI, USA),
according to the manufacturer’s protocol.
Target tumor cells (HT29, HCT116, DLD-1, SW480,
CaCo-2 and A431) with different KRAS and BRAF
mutational statuses (Table I)
(5–10) were seeded at a concentration of
1×105 cells/ml in a 96-well plate. After 24 h, the cells
were exposed to cetuximab or non-specific mouse and human IgG as a
control at concentrations of 0, 10 and 100 μg/ml in a 5%
CO2 incubator for 1 h. Then, the cells were cultured
with peripheral blood mononuclear cells (PBMCs) in a 5%
CO2 incubator for 4 h. The PBMCs were freshly prepared
from healthy human donors, isolated from heparinized peripheral
blood using a Ficoll gradient, and added to wells at each
effector:target (E:T) cell concentration (E:T ratios of 20:1 and
10:1).
 | Table ICell surface expression levels of EGFR
and mutational status of downstream effectors in the cell
lines. |
Table I
Cell surface expression levels of EGFR
and mutational status of downstream effectors in the cell
lines.
| Cell lines | EGFR (%) | KRAS | BRAF | ADCC activity
(%) |
|---|
| A431 | 99.8 | Wild | Mutant | 67 |
| CaCo-2 | 89.6 | Wild | Wild | 70 |
| SW480 | 88.8 | Mutant (G12V) | Wild | 59 |
| DLD-1 | 53.4 | Mutant (G13D) | Wild | 61 |
| HT29 | 6.9 | Wild | Mutant | 26 |
| HCT116 | 0.1 | Mutant (G13D) | Wild | 19 |
Cell lysis was determined by measuring the amount of
released LDH in the culture supernatants. ADCC was evaluated using
the following formula: % Cytotoxicity = [(experimental - effector
spontaneous control - target spontaneous control)/(target maximum
release - target spontaneous control)] × 100.
Flow cytometric analysis
The cell surface expression of EGFR in the CRC cell
lines was quantified using a flow cytometric system (FACSVantage
SE; Becton-Dickinson, San Jose, CA, USA). The binding of cetuximab
to the CRC cell lines was titrated using a flow cytometric
analysis. Biotinylated cetuximab (11) and another anti-EGFR antibody (PE
mouse anti-human EGF receptor; BD Biosciences, San Jose, CA, USA)
were used as primary antibodies. For the analysis using
biotinylated cetuximab, biotin was conjugated to cetuximab using an
adaptation of the method described by Medical & Biological
Laboratories Co., Ltd. (Nagoya, Japan). Then, 1×106
tumor cells were incubated with 1 μg/ml of biotinylated cetuximab
in 1% bovine serum albumin in PBS for 1 h at room temperature. The
cell surface was stained with streptavidin for 15 min at room
temperature in the dark. For the analysis using PE mouse anti-human
EGFR, 1 μg of anti-EGFR antibody per 1×106 tumor cells
was used as the primary antibody. Ten micrograms per milliliter of
fluorescent-labeled anti-mouse IgG was used as the secondary
antibody. The samples were then washed three additional times with
cold PBS, resuspended in 500 μl of PBS, and analyzed using flow
cytometry. For each sample, 2×104 events were acquired.
The analysis was performed by the triplicate determination of at
least three separate experiments. The expression levels were
described as the percentage of positive cells (number of
positive-stained cells × 100/total number of cells).
Ex vivo ADCC assay using tumor cells of
resected specimens and PBMCs from CRC patients
Cetuximab-mediated ADCC was also examined using
tumor cells and PBMCs isolated from the same CRC patients (n=13)
with pathological stage II or III disease (American Joint Committee
on Cancer). A portion of the resected CRC specimens was obtained
from the CRC patient as soon as possible when bowel resection was
performed at our institution. The specimens were cut using scissors
to form single cells and were incubated at a concentration of
1×105 cells/ml in a 96-well plate overnight. At the time
of the experiment, PBMCs were freshly separated from the same CRC
patient. The cells were then exposed to cetuximab or non-specific
mouse and human IgG as a control at concentrations of 0, 10 and 100
μg/ml for 1 h, and the PBMCs were subsequently added to the wells
at each E:T cell concentration (E:T ratios of 20:1 and 10:1). The
subsequent protocol was the same as that used for the in
vitro ADCC assay described above.
Immunohistochemistry
The CRC specimens were immersed in 4% buffered
neutral formalin and fixed for 24 h. Paraffin-embedding was
performed according to standard procedures. Sections (4 μm) were
mounted on coated slides and allowed to dry for 30 min at 60°C and
overnight at 37°C. All of the sections were stained in Estisol 220
(Esti Chem, Køge, Denmark) and rehydrated in graded alcohol
solutions. Endogenous peroxidase was blocked with 3% hydrogen
peroxide. Proteolytic antigen retrieval was performed using 0.1%
protease at room temperature for 20 min. The slides were incubated
in the primary anti-human EGFR antibody (clone DAK-H1-WT; Dako,
Copenhagen, Denmark) for 30 min at room temperature. Visualization
of the reaction was performed using EnVision + DAB (Dako
Cytomation-DK) followed by counterstaining with hematoxylin. All
the staining procedures were manually performed at one time.
Evaluation of immunohistochemical
variables
The expression of EGFR was quantified using a visual
grading system based on the American Society of Clinical
Oncology/College of American Pathologists guidelines for human
epidermal growth factor receptor 2 (HER-2) (12), and the intensity of membranous
staining was graded on a scale of 0 to 3+ (0, percentage of
positive tumor cells <10%, 1+, percentage of positive tumor
cells ≥10% and weak staining; 2+, percentage of positive tumor
cells ≥10% and moderate staining, 3+: percentage of positive tumor
cells ≥10% and strong staining). This evaluation was performed by
two professional pathologists.
DNA extraction and mutation analysis
First, the tumor cells in each tumor block were
histologically evaluated using hematoxylin and eosin staining.
Then, DNA was extracted from formalin-fixed and paraffin-embedded
samples after macrodissection. The presence of KRAS and
BRAF was determined using an allelic discrimination assay
performed on a 7500HT real-time PCR system. The samples were
screened for KRAS mutations located within codons 12 and 13
and BRAF mutations located within codon 600. All of the
mutations were confirmed by direct sequencing (13).
The present study was performed in accordance with
the guidelines of the Declaration of Helsinki, as amended in
Edinburgh, Scotland, in October 2000. The study was approved by the
Institutional Review Board of Keio University Hospital. Written
informed consent was obtained from each patient prior to the
experiments.
Statistical considerations
The correlation between ADCC activity and the cell
surface expression level of EGFR in vitro was evaluated
using a univariate regression analysis. The associations between
the ADCC activities and the cell surface expression levels of EGFR
and/or the KRAS/BRAF mutational status in CRC were analyzed
using a one-factor ANOVA or multiple regression model. P-values
<0.05 were considered to indicate statistically significant
results.
Results
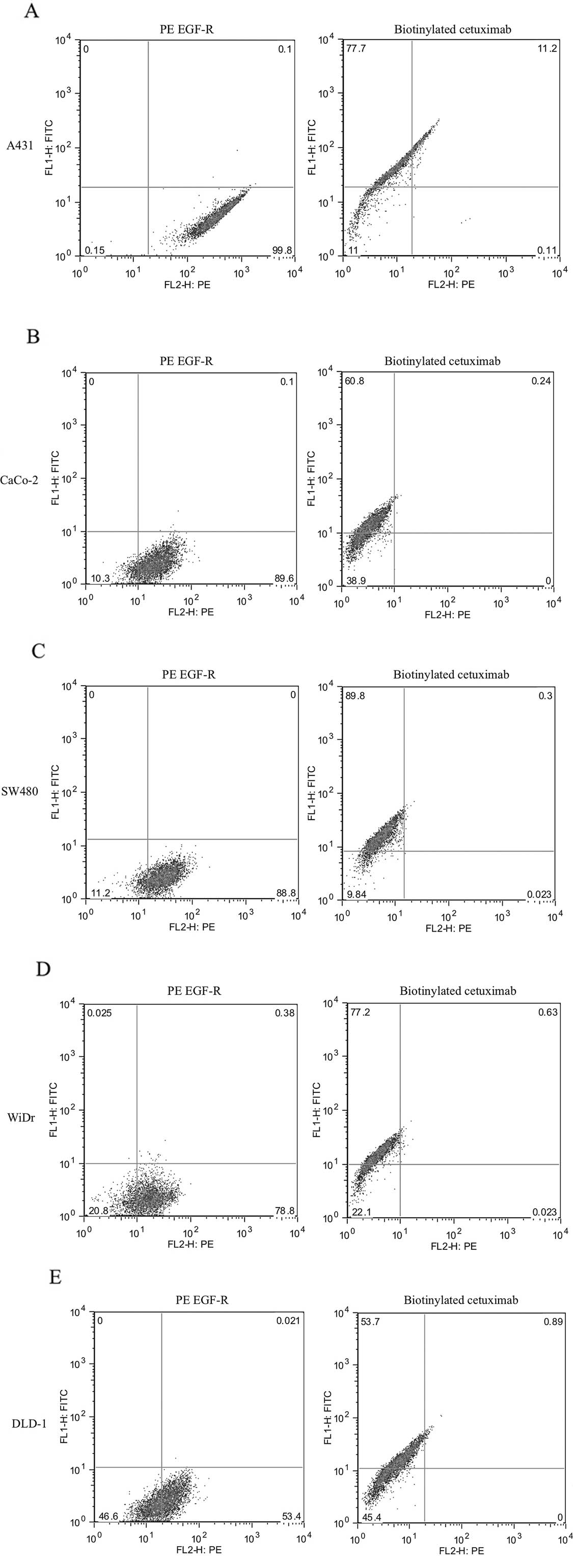
Cell surface expression levels of EGFR in
human CRC cell lines
The cell surface expression levels of EGFR in the
human CRC cell lines were evaluated using flow cytometric analysis
(Fig. 1). Using PE mouse anti-human
EGFR as a primary antibody, the cell surface EGFR expression levels
in the CaCo-2, SW480, WiDr, DLD-1 and SW867 cells were as high as
that of the positive control (A431), whereas the levels in the HT29
and HCT116 cells were relatively low. The EGFR expression level in
each cell line, as detected using biotinylated cetuximab, was
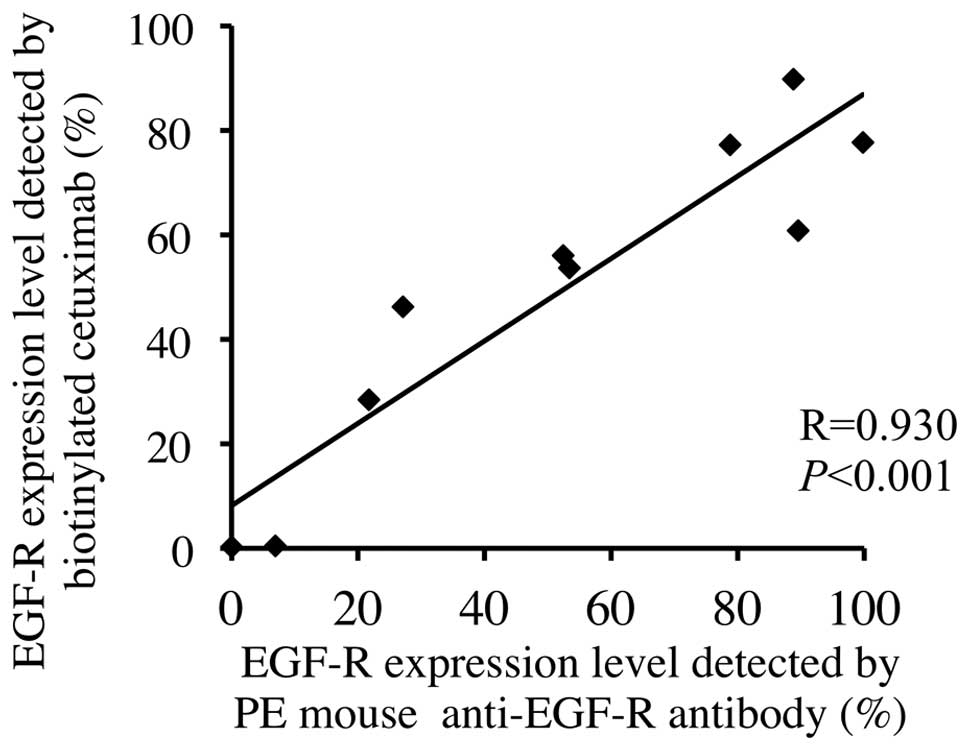
similar to that for the PE mouse anti-human EGFR, and a strong
correlation was observed between these levels, with a correlation
coefficient of 0.930 (P<0.001; Fig.
2).
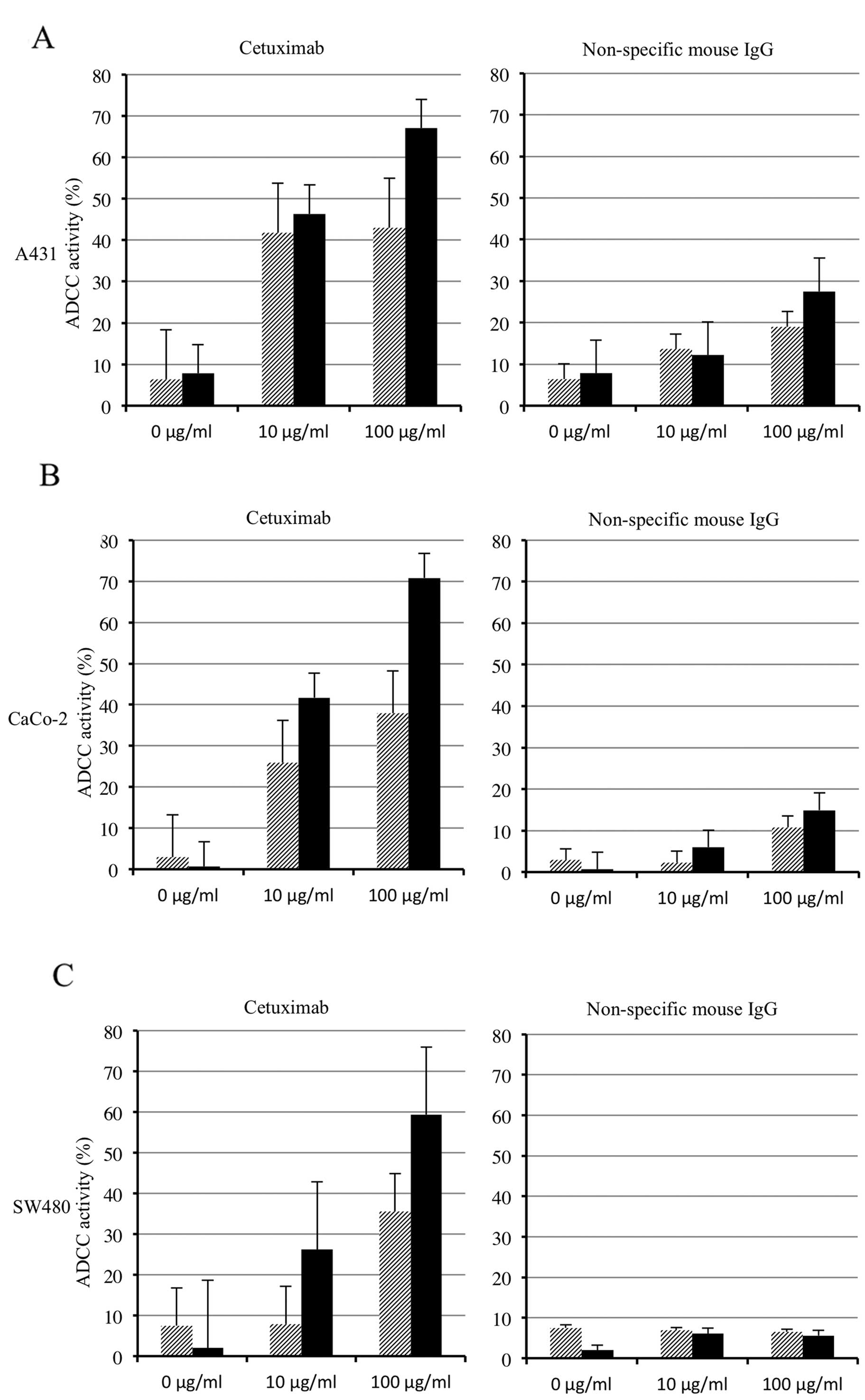
ADCC activities in human CRC cell
lines
Cetuximab-mediated ADCC activities were detected at
various degrees in all of the CRC cell lines, and the highest ADCC
activity in each cell line was detected at a cetuximab
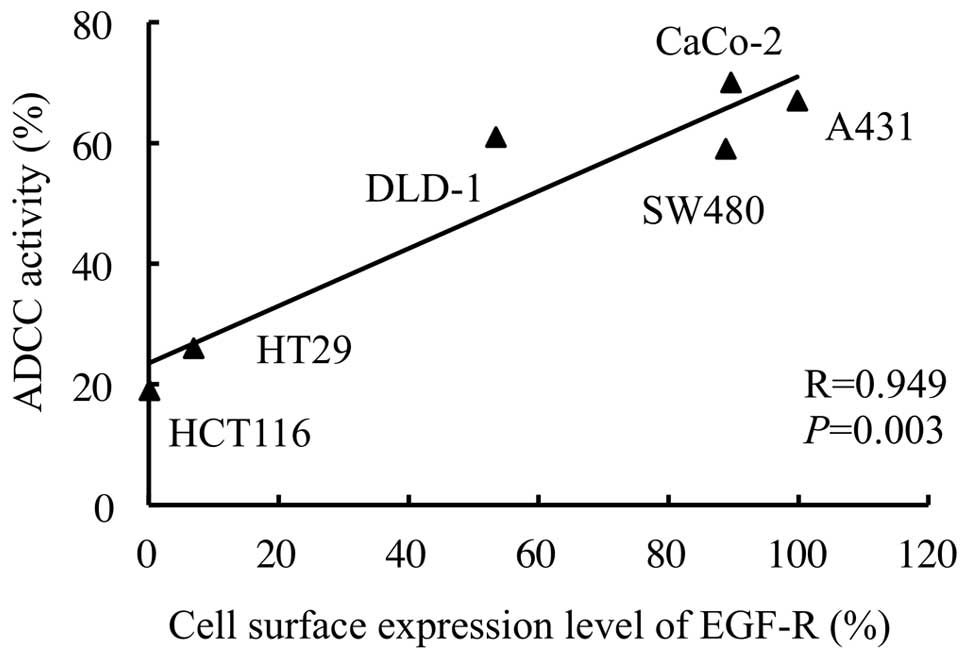
concentration of 100 μg/ml and an E:T ratio of 20:1 (Fig. 3). Under this condition, a strong
correlation was observed between the cell surface expression levels
of the EGFR and ADCC activities in the human CRC cell lines
(correlation coefficient, 0.949; P=0.003; Fig. 4). Furthermore, a multiple regression
analysis using two variables (cell surface expression level of EGFR
and KRAS or BRAF mutation status) revealed that ADCC
activity was significantly associated with the cell surface
expression level of EGFR (standard partial regression coefficient,
0.911; P=0.017), but not with the KRAS/BRAF mutational
status (standard partial regression coefficient, −0.101;
P=0.631).
Cell surface expression levels of EGFR in
resected tumors of CRC patients
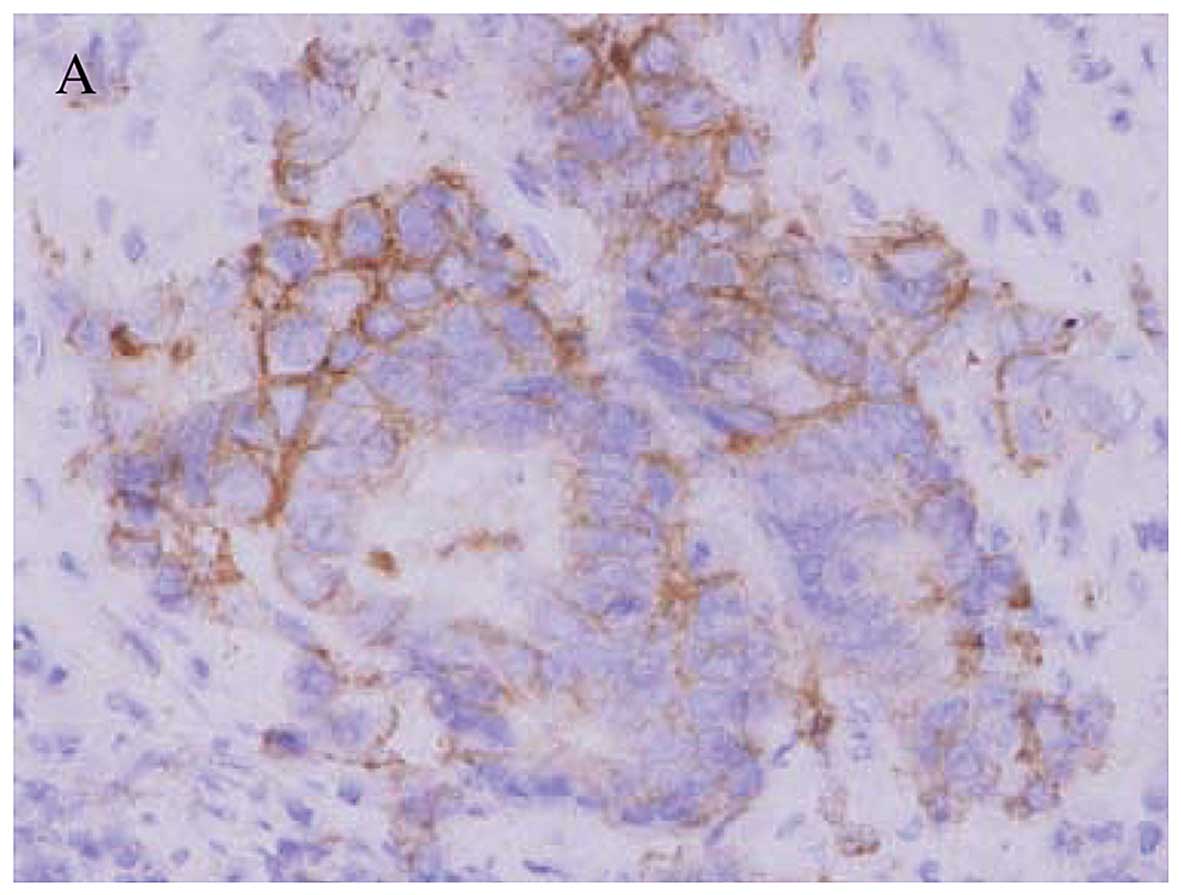
Immunohistochemical staining for EGFR in the
resected tumors is shown in Fig. 5.
Of the 13 patients, a staining score of 3+ was observed in 3
patients (cases A, B and C), a score of 2+ was observed in 3
patients (cases D, E and F), a score of 1+ was observed in 3
patients (cases G, H and I), and no staining was observed in 4
patients (cases J, K, L and M). In all cases, the EGFR staining was
localized in the cell membrane.
ADCC activities in tumor cells from the
CRC patients
Various degrees of cetuximab-mediated ADCC
activities were also detected in the tumor cells from the CRC
patients, and the highest ADCC activity in each patient was also
detected at a cetuximab concentration of 100 μg/ml and an E:T ratio
of 20:1. The profiles, including the percentage of
cetuximab-mediated ADCC activities under this condition and the
cell surface expression levels of EGFR as determined using
immunohistochemistry, for each of the patients are shown in
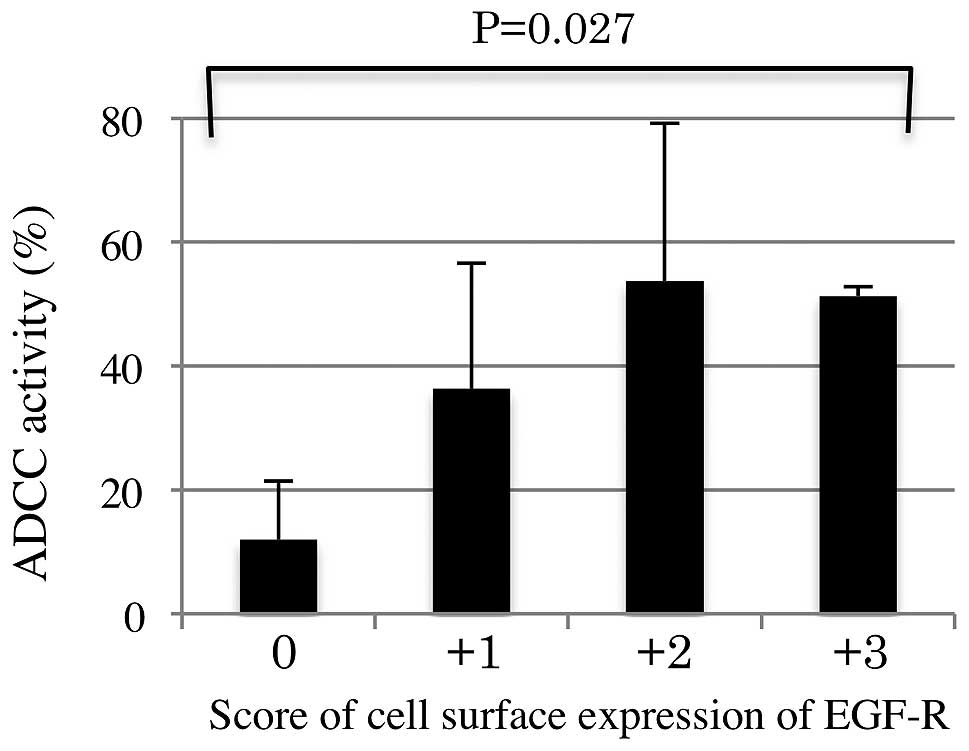
Table II. The cetuximab-mediated
ADCC activities for the tumor cells were higher in CRC patients
with a high expression level of cell surface EGFR, when compared
with patients with a low expression level (P=0.027; Fig. 6). Furthermore, the ADCC activity
level was significantly associated with the cell surface expression
level of EGFR (standard partial regression coefficient: 0.660,
P=0.018), but not with the KRAS/BRAF mutational status
(standard partial regression coefficient, 0.160, P=0.510).
 | Table IIThe profile of 13 patients with
colorectal cancer who underwent surgical resection |
Table II
The profile of 13 patients with
colorectal cancer who underwent surgical resection
| No. | Gender | Location | Stage | EGFR score | KRAS | BRAF | ADCC (%) |
|---|
| 1 | Female | Colon | IIIB | 3+ | Wild | Wild | 53 |
| 2 | Female | Colon | IIIB | 3+ | Wild | Mutant | 51 |
| 3 | Male | Colon | IIIB | 3+ | Mutant (codon12) | Wild | 50 |
| 4 | Female | Rectum | IIIB | 2+ | Mutant (codon12) | Wild | 79 |
| 5 | Male | Colon | IIIB | 2+ | Mutant (codon13) | Wild | 54 |
| 6 | Male | Rectum | II | 2+ | Wild | Wild | 28 |
| 7 | Male | Colon | II | 1+ | Wild | Wild | 59 |
| 8 | Male | Colon | II | 1+ | Wild | Wild | 30 |
| 9 | Female | Rectum | IIIB | 1+ | Wild | Wild | 20 |
| 10 | Female | Colon | II | 0 | Wild | Wild | 26 |
| 11 | Male | Colon | II | 0 | Mutant
(codon12) | Wild | 9 |
| 12 | Male | Rectum | IIIC | 0 | Wild | Wild | 7 |
| 13 | Male | Rectum | II | 0 | Wild | Wild | 6 |
Discussion
At present, antibodies against EGFR, such as
cetuximab and panitumumab, are widely used to treat CRC patients.
One of the proposed mechanisms of action of antibodies against EGFR
is the direct antagonization of the EGF-stimulating activation of
EGFR. These antibodies block the binding of ligands, inhibit EGFR
phosphorylation, induce the internalization of EGFR, and
downregulate the cell surface expression of EGFR. The KRAS
mutational status is well known to be a predictor of the efficacy
of these antibodies, although some CRC patients are unable to
benefit from treatment with anti-EGFR antibodies even when
KRAS mutations are not present. In fact, the mutations of
several downstream effectors of EGFR signaling, such as BRAF, PTEN
and PIK3CA, have been reported in CRC, and the power balance of the
mutational status of these numerous downstream effectors, including
KRAS, is considered to be important for the efficacy of antibodies
against EGFR.
In contrast, the role of ADCC in the antitumor
activity of cetuximab, which is a chimeric human-mouse IgG1
monoclonal antibody against EGFR, has not been fully investigated
in CRC. Although chimeric IgG1 antibodies have been reported to
induce ADCC activity in human effector cells in an efficient manner
(14), the contribution of
cetuximab-mediated ADCC activity to the treatment of CRC patients
remains to be elucidated, and relatively little information
presently exists on this topic, compared with the wide range of
knowledge regarding its role in the inhibition of EGFR signaling.
Therefore, the present study was designed to verify the
associations between the cetuximab-mediated ADCC activity and the
cell surface expression level of the EGFR and to determine whether
the expression level of the EGFR is a marker for the
cetuximab-mediated ADCC activity in vitro using human CRC
cell lines and ex vivo using resected specimens and PBMCs
from CRC patients.
The present study revealed novel and important
findings relevant to cetuximab-mediated ADCC activity in CRC as
follows. First, cetuximab-mediated ADCC activity was correlated
with the cell surface expression level of EGFR, but not with the
mutational statuses of KRAS and BRAF in CRC cell
lines. Second, cetuximab-mediated ADCC activity was also detected
in tumor cells from resected specimens and PBMCs isolated from the
same patients. Third, cetuximab-mediated ADCC activity was also
correlated with the cell surface expression level of EGFR, but not
with the mutational statuses of KRAS and BRAF in the
resected CRC specimens.
The existence of an acquired EGFR ectodomain
mutation (S492R) that prevents cetuximab binding was reported in a
previous study (15). Although the
incidence is considered to be rare, we made a biotinylated
cetuximab to evaluate the cell surface expression level of EGFR in
CRC more precisely (11).
Biotinylated cetuximab is able to recognize the cell surface EGFR,
which is a ligand of cetuximab, even when some mutations in EGFR
are present. As a result, the expression levels detected by other
commercial anti-human EGFR antibodies were strongly correlated with
those detected using biotinylated cetuximab among the CRC cell
lines. Therefore, acquired EGFR ectodomain mutations were thought
to have a minimal effect on the results of the present study.
The average steady state plasma concentration of
cetuximab in cancer patients has been reported to be within the
range of 56–100 μg/ml under the current clinical dose regimen
(16). In the present study, a high
ADCC activity was detected ex vivo using resected CRC
specimens and the patient PBMCs at a cetuximab concentration of 100
μg/ml. Therefore, the efficacy of cetuximab-mediated ADCC should be
obtained in the current clinical usage.
A previous study revealed that low expression of
EGFR might be sufficient for the maximum ADCC activity mediated by
cetuximab in lung cancer cell lines (17), whereas another study revealed that a
positive correlation between cetuximab-mediated ADCC activity and
the cell surface expression level of EGFR was observed in human
lung cancer, human leukemia and human embryonic kidney cell lines
(18). However, the association
with ADCC activity mediated by cetuximab and EGFR expression or the
KRAS mutational status in CRC has been unclear. In the
present study, cetuximab-mediated ADCC activity was strongly and
significantly correlated with the cell surface expression level of
EGFR, but not with the KRAS/BRAF mutational status, in CRC.
In particular, the results of ex vivo experiments using
clinical materials from CRC patients could undoubtedly explain one
of the mechanisms of action of cetuximab in CRC. Although CRC
patients with KRAS mutations are widely known to be able to
obtain little benefit from cetuximab therapy, those patients with
high levels of cell surface EGFR expression may obtain some benefit
from cetuximab treatment. The power balance between the EGFR
signaling status and the expression of EGFR on the cell surface,
which can induce ADCC, may be important for the efficacy of
cetuximab. However, the ADCC activity mediated by cetuximab could
not overcome the power of EGFR signaling accelerated by the
presence of a KRAS/BRAF mutation.
References
|
1
|
Kopetz S, Chang GJ, Overman MJ, et al:
Improved survival in metastatic colorectal cancer is associated
with adoption of hepatic resection and improved chemotherapy. J
Clin Oncol. 27:3677–3683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marechal R, De Schutter J, Nagy N, et al:
Putative contribution of CD56 positive cells in cetuximab treatment
efficacy in first-line metastatic colorectal cancer patients. BMC
Cancer. 10:340–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tejpar S, Celik I, Schlichting M,
Sartorius U, Bokemeyer C and Van Cutsem E: Association of
KRAS G13D tumor mutations with outcome in patients with
metastatic colorectal cancer treated with first-line chemotherapy
with or without cetuximab. J Clin Oncol. 30:3570–3577.
2012.PubMed/NCBI
|
|
4
|
Correale P, Marra M, Remondo C, et al:
Cytotoxic drugs up-regulate epidermal growth factor receptor (EGFR)
expression in colon cancer cells and enhance their susceptibility
to EGFR-targeted antibody-dependent cell-mediated-cytotoxicity
(ADCC). Eur J Cancer. 46:1703–1711. 2010. View Article : Google Scholar
|
|
5
|
Xu H, Yu Y, Marciniak D, Rishi AK, Sarkar
FH, Kucuk O and Majumdar AP: Epidermal growth factor receptor
(EGFR)-related protein inhibits multiple members of the EGFR family
in colon and breast cancer cells. Mol Cancer Ther. 4:435–442.
2005.PubMed/NCBI
|
|
6
|
Yang JL, Qu XJ, Russell PJ and Goldstein
D: Regulation of epidermal growth factor receptor in human colon
cancer cell lines by interferon α. Gut. 53:123–129. 2004.
|
|
7
|
Buck E, Eyzaguirre A, Barr S, et al: Loss
of homotypic cell adhesion by epithelial-mesenchymal transition or
mutation limits sensitivity to epidermal growth factor receptor
inhibition. Mol Cancer Ther. 6:532–541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ikehara N, Semba S, Sakashita M, Aoyama N,
Kasuga M and Yokozaki H: BRAF mutation associated with
dysregulation of apoptosis in human colorectal neoplasms. Int J
Cancer. 115:943–950. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koizumi F, Kanzawa F, Ueda Y, et al:
Synergistic interaction between the EGFR tyrosine kinase inhibitor
gefitinib (‘Iressa’) and the DNA topoisomerase I inhibitor CPT-11
(irinotecan) in human colorectal cancer cells. Int J Cancer.
108:464–472. 2004.
|
|
10
|
Balin-Gauthier D, Delord JP, Rochaix P, et
al: In vivo and in vitro antitumor activity of oxaliplatin in
combination with cetuximab in human colorectal tumor cell lines
expressing different level of EGFR. Cancer Chemother Pharmacol.
57:709–718. 2006. View Article : Google Scholar
|
|
11
|
Shigeta K, Hayashida T, Hoshino Y, et al:
Expression of epidermal growth factor receptor detected by
cetuximab indicates its efficacy to inhibit in vitro and
in vivo proliferation of colorectal cancer cells. PLoS One.
8:e663022013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wolff A, Hammond M, Schwartz J, et al:
American Society of Clinical Oncology/College of American
Pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar
|
|
13
|
De Roock W, Piessevaux H, De Schutter J,
et al: KRAS wild-type state predicts survival and is associated to
early radiological response in metastatic colorectal cancer treated
with cetuximab. Ann Oncol. 19:508–515. 2008.PubMed/NCBI
|
|
14
|
Steplewski Z, Sun LK, Shearman CW, Ghrayeb
J, Daddona P and Koprowski H: Biological activity of human-mouse
IgG1, IgG2, IgG3, and IgG4 chimeric monoclonal antibodies with
antitumor specificity. Proc Natl Acad Sci USA. 85:4852–4856. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Montagut C, Dalmases A, Bellosillo B, et
al: Identification of a mutation in the extracellular domain of the
Epidermal Growth Factor Receptor conferring cetuximab resistance in
colorectal cancer. Nat Med. 18:221–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo FR, Yang Z, Dong H, et al: Correlation
of pharmacokinetics with the antitumor activity of Cetuximab in
nude mice bearing the GEO human colon carcinoma xenograft. Cancer
Chemother Pharmacol. 56:455–464. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kurai J, Chikumi H, Hashimoto K, et al:
Antibody-dependent cellular cytotoxicity mediated by cetuximab
against lung cancer cell lines. Clin Cancer Res. 13:1552–1561.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kimura H, Sakai K, Arao T, Shimoyama T,
Tamura T and Nishio K: Antibody-dependent cellular cytotoxicity of
cetuximab against tumor cells with wild-type or mutant epidermal
growth factor receptor. Cancer Sci. 98:1275–1280. 2007. View Article : Google Scholar : PubMed/NCBI
|