Introduction
Head and neck squamous cell carcinoma (HNSCC) is one
of the six most common types of cancer in the world, with ~500,000
new cases annually. Oral squamous cell carcinoma (OSCC) is the most
common among the pathological types. The five-year survival rate
after diagnosis for OSCC remains low at ~50%, which is considerably
lower than that for other cancers, including colorectal, cervical
and breast cancer (1,2). To develop more optimized and more
effective drug targets for OSCC treatment, a deeper understanding
of the molecular mechanisms underlying oncogenesis and accurate
identification of targets for therapeutic intervention are
critical.
MicroRNAs (miRNAs) are classes of small, highly
conserved non-coding RNAs that control gene expression by binding
to the seed sequence at the 3′-untranslated region (3′UTR) of
targets, resulting in translational repression or mRNA degradation
(3). miRNAs are regulators in human
carcinogenesis, embryonic development and cell senescence. Previous
studies have shown that the abundance of miRNAs is abnormally
expressed in human cancer tissues, especially in the epithelium
origin; for instance, miR-21 is characterized as an onco-miRNA
(4). Several genes have been
validated to be direct targets of miR-21 in human cancer.
Programmed cell death 4 (PDCD4) is upregulated, whereas the
glioblastoma cell is transfected with antisense miR-21
oligonucleotides and PDCD4′s 3′UTR carrier binding sites of miR-21
(5). Phosphatase and tensin homolog
(PTEN) are direct targets of miR-21, and reduction of miR-21 can
suppress cancer cell proliferation via strengthened PTEN expression
in hepatocellular carcinoma (6). In
addition, miR-21 downregulates the tissue inhibitor of
metalloproteinase-3 (TIMP-3) to influence cancer cell migration and
invasion ability (7).
Signal transducer and activator of transcription
(STAT) proteins are the principal signaling proteins of cytokines
and growth factors in mammals. STAT3 responds to signals, such as
IL-6, IL-10 and VEGF. STAT3 is vital in tumorigenesis and
cancer-induced immunosuppression (8,9).
Evidence has shown that STAT3 activates miR-21 to promote cancer
cell growth (10–12). However, the precise biological
function and molecular mechanism of STAT3/miR-21 network in OSCC
remains unknown.
In the present study, we analyzed the correlation
between miR-21 and STAT3 expression in OSCC samples. Results showed
that the overexpression of miR-21 and STAT3 was related to lymph
node metastasis and accumulative survival rate. We showed that
blockage of STAT3 suppressed cancer cell growth and migration in
OSCC, accompanying the increasing expressions of PDCD4, PTEN and
TIMP-3 mediated by transcriptional inhibition of miR-21.
Materials and methods
Cell culture and reagents
Human TSCCA and TCA8113 OSCC cell lines were
purchased from China Center for Type Culture Collection (Wuhan,
China), and maintained in Dulbecco’s modified Eagle’s medium (DMEM)
(Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibco,
USA), 2 mM glutamine, 100 units of penicillin/ml and 100 mg of
streptomycin/ml (Sigma, USA) at 37°C with 5% CO2. WP1066
(Calbiochem, Germany) and IL-6 (Sigma) were dissolved in dimethyl
sulfoxide (DMSO; Solarbo, China). WP1066 was added to cells at a
final concentration of 5 μM for 48-h treatment (10). IL-6-treated cells were referenced
from a previous report (13).
Human tissue samples
The present study was approved by the Tianjin
Medical University Institutional Review Board for human use. Ninety
OSCC tissue samples were collected at the Department of
Otorhinolaryngology and Maxillofacial Oncology at Tianjin Medical
University Cancer Institute and Hospital. Tissue and clinical
information were obtained as part of the study. Clinical diagnoses
were reviewed to classify the pathological grade of the samples
according to the American Joint Committee on Cancer TNM staging
classification for the lip and oral cavity (2010). A total of 45
cases of high and moderate differentiated OSCC tumor samples, 45
cases of poorly differentiated OSCC tumor samples and 10 cancer
adjacent relative normal margin tongue tissue samples were included
in the current research. All the tumor and tumor margin tissues
were paraffin-embedded for immunohistochemistry (IHC) staining and
in situ hybridization.
miR-21 detection by in situ
hybridization
Using antisense locked nucleic acid (LNA)-modified
oligonucleotides probe, in situ hybridization was performed
using an in situ hybridization kit (Boster, China). In
situ hybridization detection of miR-21 in HNSCC samples and
xenograft SCC sections was performed (14). LNA/DNA oligonucleotides contained
locked nucleic acids at eight consecutive centrally located bases
(indicated by the underlined part) and had the following sequences:
LNA-miR-21, 5′-TCAACATCAGTCTGATAAGCTA-3′. Nuclei were
counterstained using the DAPI karyotyping kit (GenMed, USA) and
were visualized using FluoView confocal laser scanning microscopes
FV1000 (Olympus, Japan).
qRT-PCR analysis of miR-21 expression and
RT-PCR for miR-21 targets
qRT-PCR analysis of miR-21 expression in
OSCC-cultured cells was performed (14). Total RNA was extracted by TRIzol
(Invitrogen, USA). Both RT and PCR primers were purchased from
GenePharma (Shanghai, China). U6 RNA was used for normalization.
Relative quantification was conducted using amplification
efficiencies derived from cDNA standard curves. Relative gene
expressions were obtained. Quantitative analysis of change in
expression levels was performed using a real-time PCR machine (7500
ABI; USA). MicroRNA Assay kit (Applied Biosystems) was used for
detection of miR-21. The protocol was performed for 40 cycles at
95°C for 3 min, 95°C for 15 sec and 60°C for 30 sec. Data were
shown as fold-change (2−ΔΔCt) and initially analyzed
using Opticon Monitor analysis software version 2.02 (MJ Research,
USA).
Exactly 5 μl of the total 20 μl of
reverse-transcribed product was used for PCR (30 cycles of 92°C for
15 sec, 55°C for 30 sec and 72°C for 2 min) in 1X PCR buffer
containing 1.5 mmol/l MgCl2, 250 μmol/l deoxynucleotide
triphosphates, 0.5 units of Taq polymerase (Life Technologies,
USA), 100 ng of primers for PDCD4 (forward primer, 5′-ATG GAT GTA
GAA AAT GAG CAG-3′ and reverse primer, 5′-TTA AAG TCT TCT CAA ATG
CCC-3′); TIMP3 (forward primer, 5′-GAG AGT CTC TGT GGC CTT AAG CTG
G-3′ and reverse primer, 5′-CTG GGA AGA GTT AGT GTC CAA GGG-3′);
PTEN (forward primer, 5′-ATT CCT CCA ATT CAG GAC CCA C-3′ and
reverse primer, 5′-CCT GGT ATG AAG AAC GTA TTT ACC C-3′), and GAPDH
(forward primer, 5′-TCG TGG AAG GAC TCA TGA CC-3′ and reverse
primer, 5′-TCC ACC ACC CTG TTG CTG TA-3′) (SBS Bio, Beijing,
China). The reaction products were analyzed on 2% agarose gels
containing ethidium bromide; cDNA synthesis was verified by
detection of β-actin transcript.
Cancer cell clone formation, cell cycle,
apoptosis and Transwell chamber assays
Cancer cell clone formation, cell cycle, apoptosis
and Transwell chamber assays were performed as described before
(15).
Luciferase assay
The pMIR-21-Report-Luc reporter plasmid was
purchased from Signosis (Signosis, USA). The mature miR-21 binding
sequence was subcloned into a firefly luciferase-based reporter
construct immediately downstream of Luc coding sequence. TSCCA and
TCA8113 cells were plated (2×106 cells/well) in 6-well
plates. After transfection, the cells were split into 96-well
plates (4×103 cells/well) in duplicate and harvested for
luciferase assays 24 h later using a Luciferase Assay kit (Promega,
USA).
Western blot analysis
Parental and WP1066-treated cells were washed with
ice-cold phosphate-buffered saline (PBS) three times. The cells
were solubilized in 1% Nonidet P-40 lysis buffer (20 mM Tris, pH
8.0, 137 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM
CaCl2, 1 mM MgCl2, 1 mM phenylmethylsulfonyl
fluoride, 1 mM sodium fluoride, 1 mM sodium orthovanadate and a
protease inhibitor mixture). A total of 40 mg lysates were
subjected to SDS-PAGE on 8% SDS-acrylamide gel. Separate proteins
were transferred to PVDF membranes (Millipore, USA) and incubated
with primary antibodies against STAT3, p-STAT3, PTEN, PDCD4,
TIMP-3, Ki67, MMP2 and Bcl-2 (Santa Cruz, USA), followed by
incubation with HRP-conjugated secondary antibody (Zymed, USA).
GAPDH (Santa Cruz) was used as internal control. The specific
protein was detected using a SuperSignal protein detection kit
(Pierce, USA).
TSCCA xenograft tumor assay
Nude mouse subcutaneous TSCCA xenograft model was
performed as described before (14). The mice were randomly divided into
three groups when tumors reached ~10 mm in length. Two groups of
mice were treated with DMSO (local injection, once every 3 days for
21 days) or WP1066 (40 mg/kg, local injection) alone. One group of
mice was taken as control. The tumor volume was measured with a
caliper every 3 days using the formula: Volume = length ×
width2/2. At the end of the observation period, the mice
bearing xenograft tumors were sacrificed and the tumor tissues were
removed for formalin fixation and preparation of paraffin-embedded
sections.
IHC staining
The paraffin-embedded tissue sections were used for
examination of STAT3, p-STAT3, PTEN, PDCD4, TIMP-3, Ki67, MMP2 and
Bcl-2 expressions, as well as HE staining. For IHC assay, sections
were incubated with primary antibodies (1:200 dilutions) overnight
at 4°C, followed by a biotin-labeled secondary antibody (1:100
dilutions) for 1 h at room temperature. Sections were incubated
with ABC-peroxidase and diaminobenzidine (DAB), counterstained with
hematoxylin and visualized using light microscopy.
TUNEL assay
Apoptotic cell death in the tumor specimens of mouse
models was examined by the TUNEL method using an in situ
cell death kit (Roche, USA). Positive cells were visualized using
fluorescence microscopy. The reaction mixture was incubated without
enzyme in a control coverslip to detect non-specific staining.
Nuclei were counterstained with DAPI karyotyping kit and visualized
using FV-1000 laser scanning confocal biological microscopes and
analyzed using IPP5.1 (Olympus).
Statistical analysis
Results are presented as averages of at least three
experiments performed in triplicate with respective standard
errors. Statistical evaluation for data analysis was determined by
t-test and χ2 test; differences with P<0.05 were
considered statistically significant.
Results
STAT3 expression correlates with miR-21
in OSCC tissue samples
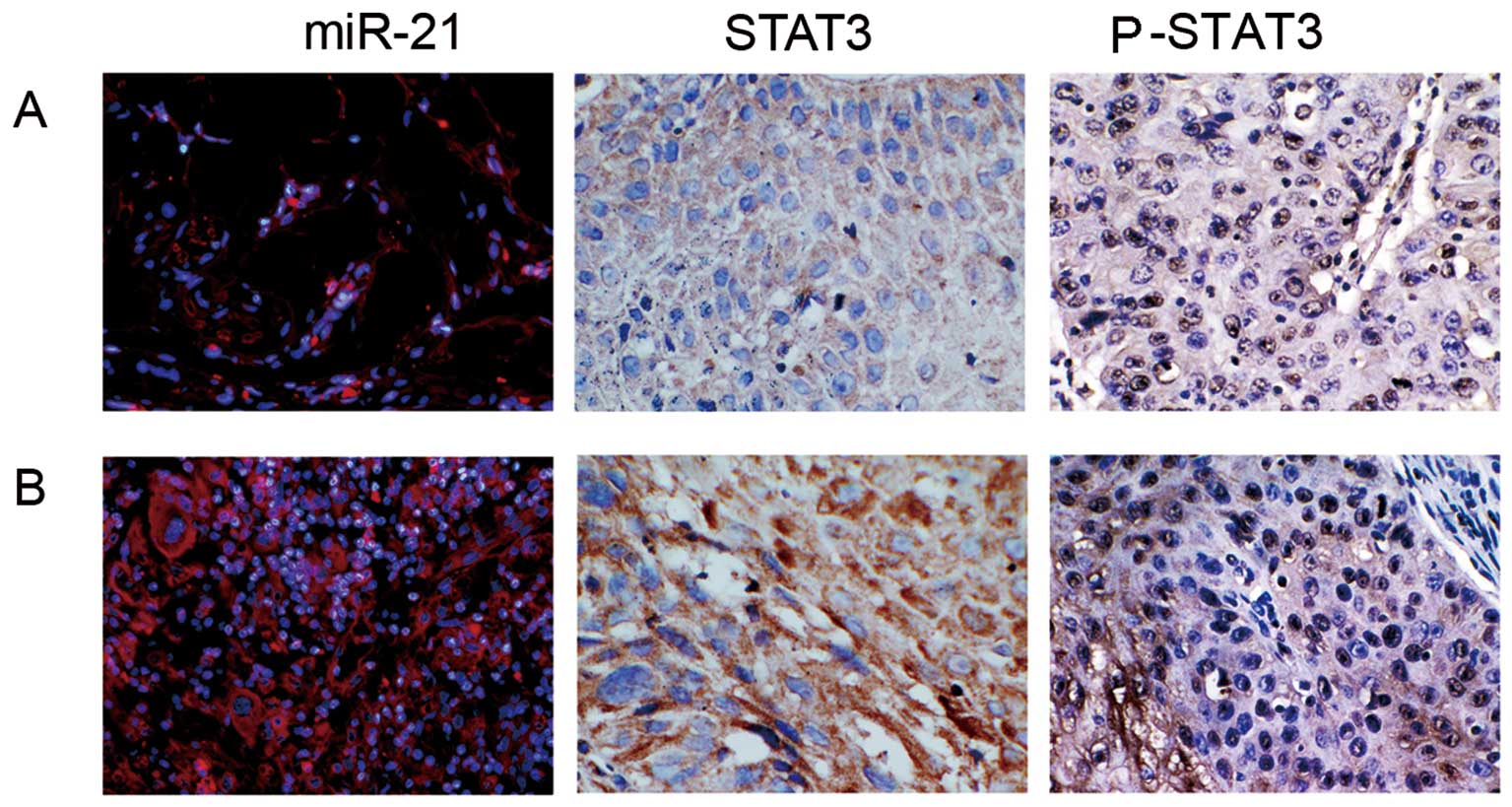
miR-21 is overexpressed in OSCC, including cultured
cell lines and tumor tissue samples (16,17).
In the present study, we assessed miR-21 expression in 90 OSCC
tissue samples with distinct differentiation status to study the
regulatory mechanism of miR-21 in OSCC oncogenesis. Using an
LNA-modified antisense miR-21 probe, miR-21 expressions in
well-differentiated (positive rate, 45.0±15.2), moderately
differentiated (positive rate, 71.7±17.1) and poorly differentiated
(positive rate, 91.8±9.20) OSCC samples were shown (Fig. 1). IHC staining indicated that
positive STAT3 signal resided in both nucleus and cytoplasm. The
STAT3 expression rates for poorly differentiated, moderately
differentiated, and well-differentiated OSCC samples were:
89.2±14.5, 68.1±20.2 and 46.3±12.5, respectively (Fig. 1). Pearson’s rank correlation
analysis showed a high, positive correlation (R=0.902, P<0.05)
between STAT3 and miR-21 expressions among different OSCC
samples.
Reduction of STAT3 by WP1066 inhibits
OSCC growth in vitro
We employed WP1066, a specific STAT3 small-molecule
inhibitor, to suppress STAT3 activity in OSCC cancer cells.
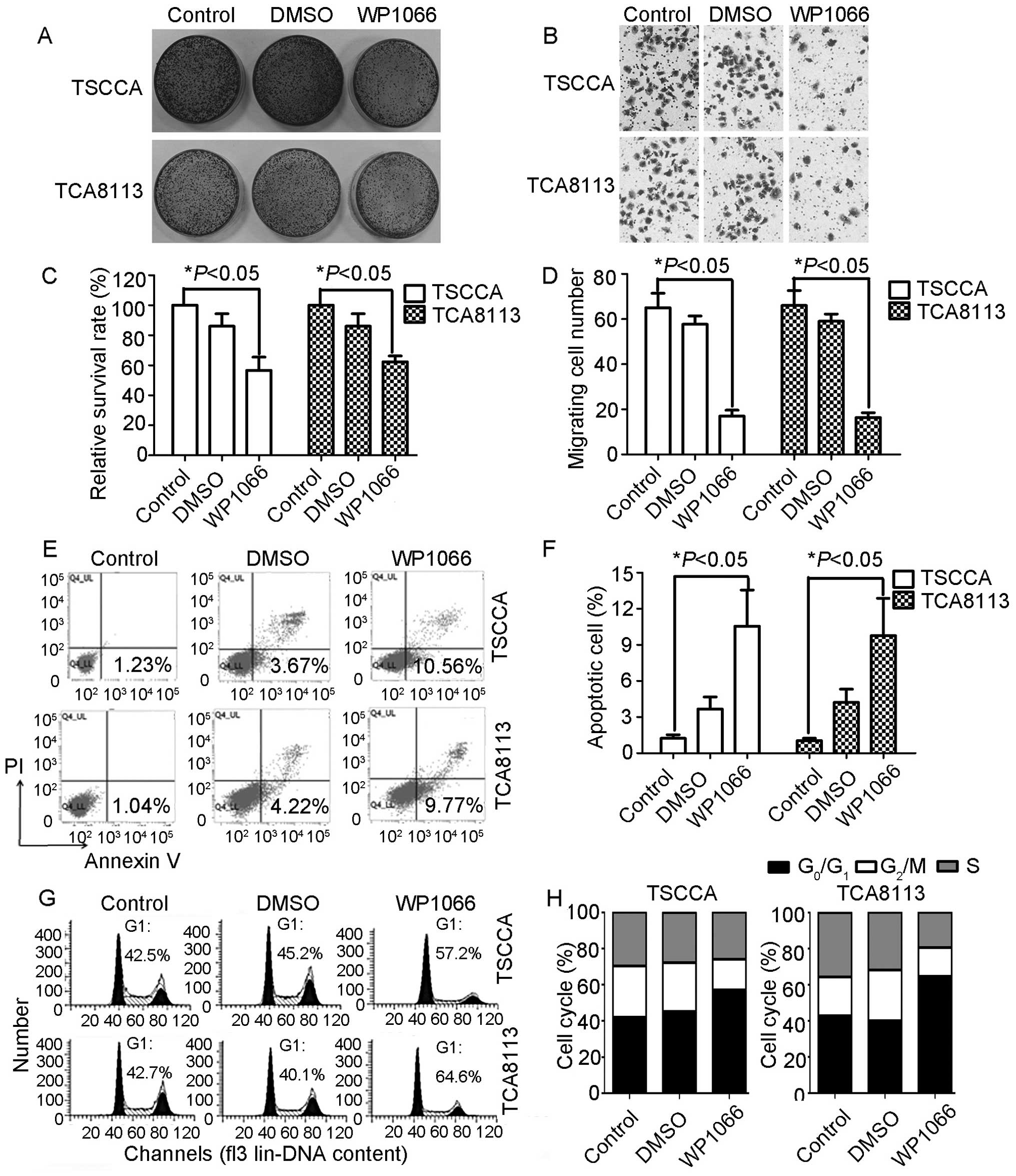
Calculation of cell clone numbers revealed that TSCCA and TCA8113
cells exhibited more decreased proliferation ability than control
and DMSO-treated groups following treatment with WP1066. The
survival rate of WP1066-treated TSCCA cells was 56.67% (F=19.99,
P=0.002; Fig. 2A and C), and the
survival rate of WP1066-treated TCA8113 cells was 62.33% (F=61.81,
P=0.000). Transwell assay was employed to measure STAT3’s effects
on OSCC in vitro cell invasiveness. The number of invasive
cells in WP1066-treated cultures was reduced relative to the
control cells. Fig. 2B and D show
reduction from 65.00±11.14 to 17.00±4.58 in TSCCA cells and
66.00±11.53 to 16.33±3.79 in TCA8113 cells. Transwell assay
indicated that the invasiveness of cancer cells and the migration
ability were significantly attenuated through inhibition of STAT3
activity. The effect of decreased STAT3 on apoptosis was analyzed
through Annexin V and PI double staining. The Annexin V-positive
early-phase apoptotic cells significantly increased in
WP1066-treated cells (10.56% for TSCCA cells and 9.77% for TCA8113
cells) when compared with parental and DMSO-treated cells (P=0.01;
Fig. 2E and F). The early-phase
apoptosis data were consistent with the cell cycle blockage in both
cell lines. G1 phase percentage of TSCCA cells was 57.2%, a value
that was 14.7% higher than that in the control group, 72 h
following WP1066 treatment. Meanwhile, G1 phase percentage of
TCA8113 cells was 64.6%, a value that was 21.9% higher than that in
the control group. Both TSCCA and TCA8113 cells displayed G1 phase
blockage (Fig. 2G and H). Western
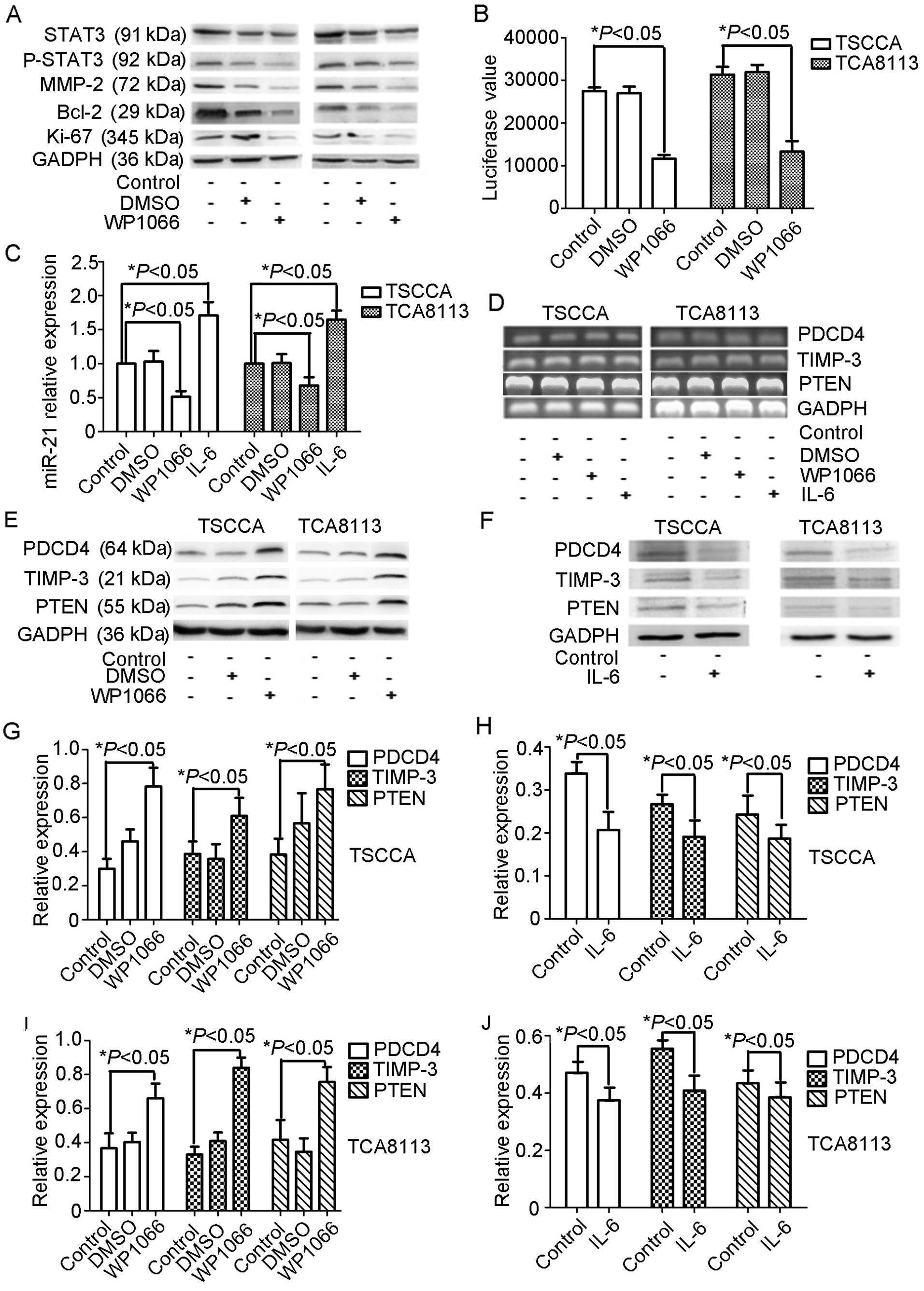
blot analysis indicated that MMP-2 expression was inhibited
effectively in WP1066-treated TSCCA and TCA8113 cells, Ki-67 and
Bcl-2 (Fig. 4A).
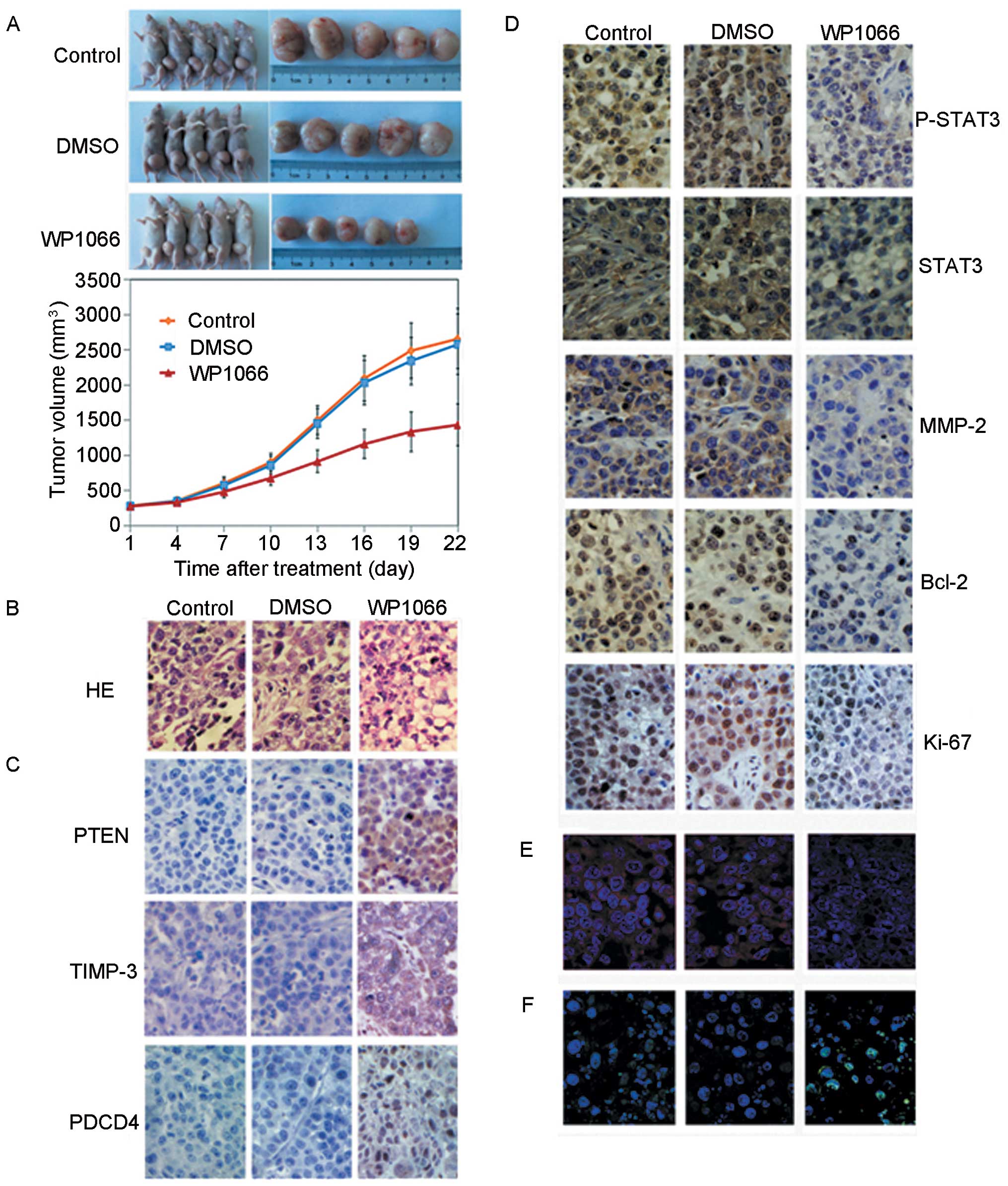
WP1066 suppresses TSCCA growth in a
xenograft model
In vitro experiments indicated that STAT3 was
a potential target in OSCC therapy. We performed a
proof-of-principle experiment using a TSCCA cell xenograft model
and a DMSO-diluted WP1066 therapy approach for confirmatory
purposes. Ten mice were challenged by in situ injection of
DMSO as a negative control group, and another 10 mice were treated
with PBS as a control group. The mean tumor volume of the mice in
the control, the DMSO and the WP1066-treated groups was ~280
mm3, with no statistically significant difference at the
onset of the treatment. The mice were monitored and treated every 3
days for 3 weeks, and the tumor volume of the mice in each group
was measured. Fig. 3A shows that a
significant decrease in tumor volume was only observed in the
WP1066-treated group (F=15.390, P=0.000).
HE stain was revealed in WP1066-treated tumor
samples. Atypia tumor cells and necrosis cells reduced in number
and in size. Tumor tissue derived from WP1066-treated groups showed
decreased microvessel density and chromatin staining compared with
those from the control groups (Fig.
3B). TUNEL staining assay data showed that more apoptotic
nuclei were found in the WP1066-treated group, revealing that STAT3
inhibition caused tumor cell apoptosis and contributed to tumor
growth suppression of the xenografts (Fig. 3F). STAT3 and p-STAT3 expressions
were inhibited significantly (Fig.
3D), and miR-21 expression was decreased in xenograft tumors
(Fig. 3E) for the WP1066-treated
group. Thus, STAT3-miR-21 adhered to an in vivo regulatory
mechanism. We observed decreased Ki-67, MMP-2 and Bcl-2
expressions, consistent with in vitro results, in the
WP1066-treated tumors (Fig. 3D). We
measured the expression status of miR-21 in the animal model.
Fig. 3C shows an increase of PTEN,
PDCD4 and TIMP-3 from the IHC staining data.
WP1066 inhibits miR-21 expression in
OSCC
We generated a pMIR-21-Report-Luc reporter plasmid
containing the putative STAT3 binding sites to miR-21 promoter in
an attempt to address the stable relationship between STAT3 and
miR-21 in oral cancer cell lines. The pMIR-21-Report-Luc reporter
construct was separately co-transfected with WP1066 or DMSO into
TSCCA and TCA8113 cells, followed by measurement of luciferase
activity in the transfected cells 3 days after transfection.
Results showed that WP1066 caused 50–60% decrease in luciferase
activity (Fig. 4B), indicating that
STAT3 promotes miR-21 gene transcription.
We examined the expression changes in the biological
direct targets of miR-21 (PDCD4, PTEN and TIMP-3) at mRNA and
protein levels. RT-PCR results indicated no significant difference
in PDCD4, PTEN and TIMP-3 mRNA expression levels among the control,
DMSO-treated and WP1066-treated TSCCA and TCA8113 cell groups
(Fig. 4D). The protein levels of
PDCD4, PTEN and TIMP-3 increased following treatment with WP1066 in
both cell lines (Fig. 4E, G and I)
compared with the control and the DMSO-treated groups.
To further illustrate the hypothesized STAT3-miR-21
regulatory axis, we enrolled IL-6 to upregulate STAT3 activity and
to examine miR-21 and biological target expressions. miR-21
expression increased in IL-6-treated TSCCA and TCA8113 cells
(Fig. 4C). PDCD4, PTEN and TIMP-3
expressions exhibited decreased protein levels, but no significant
changes were found in the mRNA levels (Fig. 4D and E). These data provide
additional evidence of the influence of STAT3 on miR-21 expression
at the transcription level.
Discussion
Previous studies have reported on the activation of
STAT family and the control of aberrantly expressed miRNAs in the
most basic mechanisms of oral squamous cell carcinoma (OSCC)
(19,20). In the present study, we showed that
miR-21 and STAT3 were more co-overexpressed in human OSCC tissue
samples compared with tumor-adjacent tissues. miR-21 is one of the
most overexpressed miRNAs in a number of medium-scale and
large-scale profiling experiments designed to detect dysregulated
miRNAs in human cancers (21).
Overexpression of miR-21 could increase cell proliferation,
migration, invasion and survival, as confirmed in breast cancer,
glioma, hepatocellular carcinoma, OSCC, myeloma and colon cancer
(4). Systemic review and
meta-analysis have shown that miR-21 detection has a prognostic
value in cancer patients, especially in HNSCC and gastrointestinal
cancers. Higher expression of miR-21 has been correlated with
overall survival in HNSCC and the combined HR is 1.46 (22). Data suggested that miR-21 is a
candidate biomarker for cancer treatment; thus, the mechanism of
elevated expression of miR-21 in human cancers should be studied in
the future.
Several studies have established that miR-21 is
induced by diverse mechanisms, such as genetic alterations,
mutations, single nucleotide polymorphisms, Drosha and Dicer
activity, methyltransferases and histone deacetylases (23). For instance, Toll ligand receptor
activation by LPS upregulates miR-21 in numerous cells types,
including macrophages, fibroblasts and peripheral blood mononuclear
cells (24). IL-6 promotes the
survival of multiple myeloma cells through induction of miR-21
expression by STAT3 activation (25). In situ hybridization and IHC
data indicated a linear correlation expression mechanism of STAT3
and miR-21 gene putative promoter region. We employed WP1066, a
specific STAT3 inhibitor, to suppress STAT3 activation in TSCCA and
TCA8113 cells (26). In the OSCC
cell lines and animal model, miR-21 expression downregulated
linearly upon treatment with WP1066. Tumor suppressor genes, such
as PTEN, TIMP-3 and PDCD4, are functional targets of miR-21
(5–7). STAT3 inhibition triggered upregulation
of protein expression levels but not mRNA levels, and IL-6 induced
STAT3 activation could inhibit these gene expressions. Luciferase
assay suggested that the miR-21 promoter region contained a STAT3
putative regulatory sequence. Data suggested the existence of a
STAT3→miR-21 transcriptional regulatory axis in OSCC.
Early studies indicated that HNSCC and the derived
cell lines exhibit markedly elevated levels of phosphorylated STAT3
(29) among the STAT protein family
members, whereas constitutive activation of STAT3 has been
demonstrated in many types of cancer, including breast, lung and
thyroid cancer (27,28). Increased STAT3 levels alter cell
cycle progression and promote the proliferation and survival of
tumor cells (30). Both in
vitro and in vivo treatment of WP1066 in OSCC cells
showed significant reduction of cancer cell clone formation
ability, increased apoptotic nuclei, cancer cell migration,
invasion inhibition and cell cycle blockage at G1 phase. We
inferred that miR-21 reduction brought by WP1066 treatment could
partially explain the mechanism of the results. PTEN expression
increased and the AKT signaling pathway was suppressed due to the
downregulation of miR-21 in OSCC cells. The AKT pathway is
responsible for cancer survival and metastasis by regulating Bcl-2
and MMP2/9 (31). TIMP3, the
ECM-bound protease regulator, are key inhibitors of several MMPs
and the expression is prognostic for cancer (32). Thus, the recovery of TIMP-3 protein
is responsible for the migration and reduction of invasion in OSCC
cells. PDCD4 reportedly inhibits AP-1-mediated trans-activation and
induces expression of the cyclin-dependent kinase inhibitor p21
(33). Thus, the OSCC cell cycle
blockage in G1 phase occurred due to the upregulation of PDCD4.
Our results provide evidence of the inhibition of
STAT3 by WP1066, resulting in upregulation of PTEN, PDCD4 and
TIMP-3 by suppression of miR-21, as well as the contribution of
STAT3/miR-21 pathway in in vitro and in vivo OSCC
growth. In addition, WP1066 could be a novel candidate drug to
treat OSCC by inhibiting STAT3/miR-21 axis.
Acknowledgements
This study was supported by the China National
Natural Scientific Fund (grant numbers 81172573, 81101916 and
51103107), and the Tianjin Science and Technology Committee (grant
numbers 11JCYBJC10800 and 12JCYBJC33800).
References
|
1
|
Posner M and Vermorken JB: Induction
therapy in the modern era of combined-modality therapy for locally
advanced head and neck cancer. Semin Oncol. 35:221–228. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cohen EE, Lingen MW and Vokes EE: The
expanding role of systemic therapy in head and neck cancer. J Clin
Oncol. 9:1743–1752. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big roll in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan X, Wang ZX and Wang R: MicroRNA-21: a
novel therapeutic target in human cancer. Cancer Biol Ther.
12:1224–1232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Down-regulation of Pdcd4 by mir-21 facilitates glioblastoma
pro-liferation in vivo. Neuro Oncol. 13:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng F, Henson R, Wehbe-Janek H, et al:
MicroRNA-21 regulates expression of the PTEN tumor suppressor gene
in human hepatocellular cancer. Gastroenterology. 133:647–658.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rawlings JS, Rosler KM and Harrison DA:
The JAK/STAT signaling pathway. J Cell Sci. 117:1281–1283. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He G and Karin M: NF-κB and STAT3 - key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011.
|
|
10
|
Wang YY, Sun G, Luo H, et al: MiR-21
modulates hTERT through a STAT3-dependent manner on glioblastoma
cell growth. CNS Neurosci Ther. 18:722–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han L, Yue X, Zhou X, et al: MicroRNA-21
expression is regulated by β-catenin/STAT3 pathway and promotes
glioma cell invasion by direct targeting RECK. CNS Neurosci Ther.
18:573–583. 2012.
|
|
12
|
Yang CH, Yue J, Pfeffer SR, et al:
MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma
cells. J Biol Chem. 286:39172–39178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lederle W, Depner S, Schnur S, et al: IL-6
promotes malignant growth of skin SCCs by regulating a network of
autocrine and paracrine cytokines. Int J Cancer. 128:2803–2814.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Ren Y, Moore L, et al:
Downregulation of miR-21 inhibits EGFR pathway and suppresses the
growth of human glioblastoma cells independent of PTEN status. Lab
Invest. 90:144–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han L, Zhang A, Wang H, et al:
Tat-BMPs-PAMAM conjugates enhance therapeutic effect of small
interference RNA on U251 glioma cells in vitro and in vivo. Hum
Gene Ther. 4:417–426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Huang H, Sun L, et al: MiR-21
indicates poor prognosis in tongue squamous cell carcinomas as an
apoptosis inhibitor. Clin Cancer Res. 15:3998–4008. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu ZW, Zhong LP, Ji T, et al: MicroRNAs
contribute to the chemoresistance of cisplatin in tongue squamous
cell carcinoma lines. Oral Oncol. 46:317–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Parkin DM, Bray F, Ferlay J, et al: Global
cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
19
|
Grandis JR, Drenning SD, Zeng Q, et al:
Constitutive activation of Stat3 signaling abrogates apoptosis in
squamous cell carcinogenesis in vivo. Proc Natl Acad Sci.
97:4227–4232. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bourguignon LY, Earle C, Wong G, et al:
Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21
expression and chemoresistance in hyaluronan/CD44-activated head
and neck squamous cell carcinoma cells. Oncogene. 31:149–160. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volinia S, Calin GA, Liu CG, et al: A
micro-RNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu X, Han Y, Wu Y, et al: Prognostic role
of microRNA-21 in various carcinomas: a systematic review and
meta-analysis. Eur J Clin Invest. 41:1245–1253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sheedy FJ, Palsson-McDermott E, Hennessy
EJ, et al: Negative regulation of TLR4 via targeting of the
proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat
Immunol. 11:141–147. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Löffler D, Brocke-Heidrich K, Pfeifer G,
et al: Interleukin-6 dependent survival of multiple myeloma cells
involves the Stat3-mediated induction of microRNA-21 through a
highly conserved enhancer. Blood. 110:1330–1333. 2007.PubMed/NCBI
|
|
26
|
Horiguchi A, Asano T, Kuroda K, et al:
STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell
carcinoma. Br J Cancer. 102:1592–1599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Darnell JE: Validating Stat3 in cancer
therapy. Nat Med. 11:595–596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bromberg J: Stat proteins and oncogenesis.
J Clin Invest. 109:1139–1142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grandis JR, Drenning SD, Chakraborty A, et
al: Requirement of Stat3 but not Stat1 activation for epidermal
growth factor receptor-mediated cell growth in vitro. J Clin
Invest. 102:1385–1392. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johnson DE: Targeting proliferation and
survival pathways in head and neck cancer for therapeutic benefit.
Chin J Cancer. 31:319–326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kotzsch M, Farthmann J, Meye A, et al:
Prognostic relevance of uPAR-del4/5 and TIMP-3 mRNA expression
levels in breast cancer. Eur J Cancer. 41:2760–2768. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Göke R, Barth P, Schmidt A, et al:
Programmed cell death protein 4 suppresses CDK1/cdc2 via induction
of p21Waf1/Cip1. Am J Physiol Cell Physiol.
287:C1541–C1546. 2004.PubMed/NCBI
|