|
1
|
Wang X, Tang S, Le SY, et al: Aberrant
expression of oncogenic and tumor-suppressive microRNAs in cervical
cancer is required for cancer cell growth. PLoS One. 3:e25572008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilting SM, van Boerdonk RA, Henken FE, et
al: Methylation-mediated silencing and tumour suppressive function
of hsa-miR-124 in cervical cancer. Mol Cancer. 9:1672010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang HJ: Aberrant DNA methylation in
cervical carcinogenesis. Chin J Cancer. 32:42–48. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Faridi R, Zahra A, Khan K and Idrees M:
Oncogenic potential of human papillomavirus (HPV) and its relation
with cervical cancer. Virol J. 8:2692011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Whiteside MA, Siegel EM and Unger ER:
Human papillomavirus and molecular considerations for cancer risk.
Cancer. 113(Supp 10): 2981–2994. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saavedra KP, Brebi PM and Roa JC:
Epigenetic alterations in preneoplastic and neoplastic lesions of
the cervix. Clin Epigenetics. 4:132012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lechner M, Fenton T, West J, et al:
Identification and functional validation of HPV-mediated
hypermethylation in head and neck squamous cell carcinoma. Genome
Med. 5:152013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chaiwongkot A, Vinokurova S, Pientong C,
et al: Differential methylation of E2 binding sites in episomal and
integrated HPV 16 genomes in preinvasive and invasive cervical
lesions. Int J Cancer. 132:2087–2094. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Das P, Thomas A, Mahantshetty U,
Shrivastava SK, Deodhar K and Mulherkar R: HPV genotyping and site
of viral integration in cervical cancers in Indian women. PLoS One.
7:e410122012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-218 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng ZM and Wang X: Regulation of
cellular miRNA expression by human papillomaviruses. Biochim
Biophys Acta. 1809.668–677. 2011.PubMed/NCBI
|
|
13
|
Rao Q, Shen Q, Zhou H, Peng Y, Li J and
Lin Z: Aberrant microRNA expression in human cervical carcinomas.
Med Oncol. 29:1242–1248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Liu J, Yuan C, Cui B, Zou X and Qiao
Y: High-risk human papillomavirus reduces the expression of
microRNA-218 in women with cervical intraepithelial neoplasia. J
Int Med Res. 38:1730–1736. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dreher A, Rossing M, Kaczkowski B, Nielsen
FC and Norrild B: Differential expression of cellular microRNAs in
HPV-11 transfected cells. An analysis by three different array
platforms and qRT-PCR. Biochem Biophys Res Commun. 403:357–362.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sato F, Tsuchiya S, Meltzer SJ and Shimizu
K: MicroRNAs and epigenetics. FEBS J. 278:1598–1609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Colón-López V, Ortiz AP and Palefsky J:
Burden of human papillomavirus infection and related comorbidities
in men: implications for research, disease prevention and health
promotion among Hispanic men. P R Health Sci J. 29:232–240.
2010.PubMed/NCBI
|
|
18
|
Chaturvedi AK: Beyond cervical cancer:
burden of other HPV-related cancers among men and women. J Adolesc
Health. 46:S20–S26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu Q, Ma D and Zhao S: DNA methylation
changes in cervical cancers. Methods Mol Biol. 863:155–176. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Botezatu A, Goia-Rusanu CD, Iancu IV, et
al: Quantitative analysis of the relationship between
microRNA-124a, -34b and -203 gene methylation and cervical
oncogenesis. Mol Med Rep. 4:121–128. 2011.PubMed/NCBI
|
|
21
|
Sasagawa T, Takagi H and Makinoda S:
Immune responses against human papillomavirus (HPV) infection and
evasion of host defense in cervical cancer. J Infect Chemother.
18:807–815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Correa de Adjounian MF, Adjounian H and
Adjounian SH: Silenciamiento de genes mediante RNA interferencia:
consideraciones sobre el mecanismo y diseño de los sistemas
efectores. AVFT. 27:22–25. 2008.
|
|
23
|
Rouhi A, Mager DL, Humphries RK and
Kuchenbauer F: MiRNAs, epigenetics, and cancer. Mamm Genome.
19:517–525. 2008. View Article : Google Scholar
|
|
24
|
Bock C: Epigenetic biomarker development.
Epigenomics. 1:99–110. 2009. View Article : Google Scholar
|
|
25
|
Yang N, Coukos G and Zhang L: MicroRNA
epigenetic alterations in human cancer: one step forward in
diagnosis and treatment. Int J Cancer. 122:963–968. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valeri N, Vannini I, Fanini F, Calore F,
Adair B and Fabbri M: Epigenetics, miRNAs, and human cancer: a new
chapter in human gene regulation. Mamm Genome. 20:573–580. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar
|
|
28
|
Henken FE, Wilting SM, Overmeer RM, et al:
Sequential gene promoter methylation during HPV-induced cervical
carcinogenesis. Br J Cancer. 97:1457–1464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berdasco M and Esteller M: Aberrant
epigenetic landscape in cancer: how cellular identity goes awry.
Dev Cell. 19:698–711. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saito Y and Jones PA: Epigenetic
activation of tumor suppressor microRNAs in human cancer cells.
Cell Cycle. 5:2220–2222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lujambio A and Esteller M: CpG island
hypermethylation of tumor suppressor microRNAs in human cancer.
Cell Cycle. 6:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Toyota M, Suzuki H, Sasaki Y, et al:
Epigenetic silencing of microRNA-34b/c and B-cell
translocation gene 4 is associated with CpG island methylation
in colorectal cancer. Cancer Res. 68:4123–4132. 2008.PubMed/NCBI
|
|
34
|
Huang YW, Liu JC, Deatherage DE, et al:
Epigenetic repression of microRNA-129-2 leads to
overexpression of SOX4 oncogene in endometrial cancer.
Cancer Res. 69:9038–9046. 2009.PubMed/NCBI
|
|
35
|
Leonard SM, Wei W, Collins SI, et al:
Oncogenic human papillomavirus imposes an instructive pattern of
DNA methylation changes which parallel the natural history of
cervical HPV infection in young women. Carcinogenesis.
33:1286–1293. 2012. View Article : Google Scholar
|
|
36
|
Missaoui N, Hmissa S, Dante R and Frappart
L: Global DNA methylation in precancerous and cancerous lesions of
the uterine cervix. Asian Pac J Cancer Prev. 11:1741–1744.
2010.PubMed/NCBI
|
|
37
|
Kalantari M, Calleja-Macias IE, Tewari D,
et al: Conserved methylation patterns of human papillomavirus type
16 DNA in asymptomatic infection and cervical neoplasia. J Virol.
78:12762–12772. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burgers WA, Blanchon L, Pradhan S, et al:
Viral oncoproteins target the DNA methyltransferases. Oncogene.
26:1650–1655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
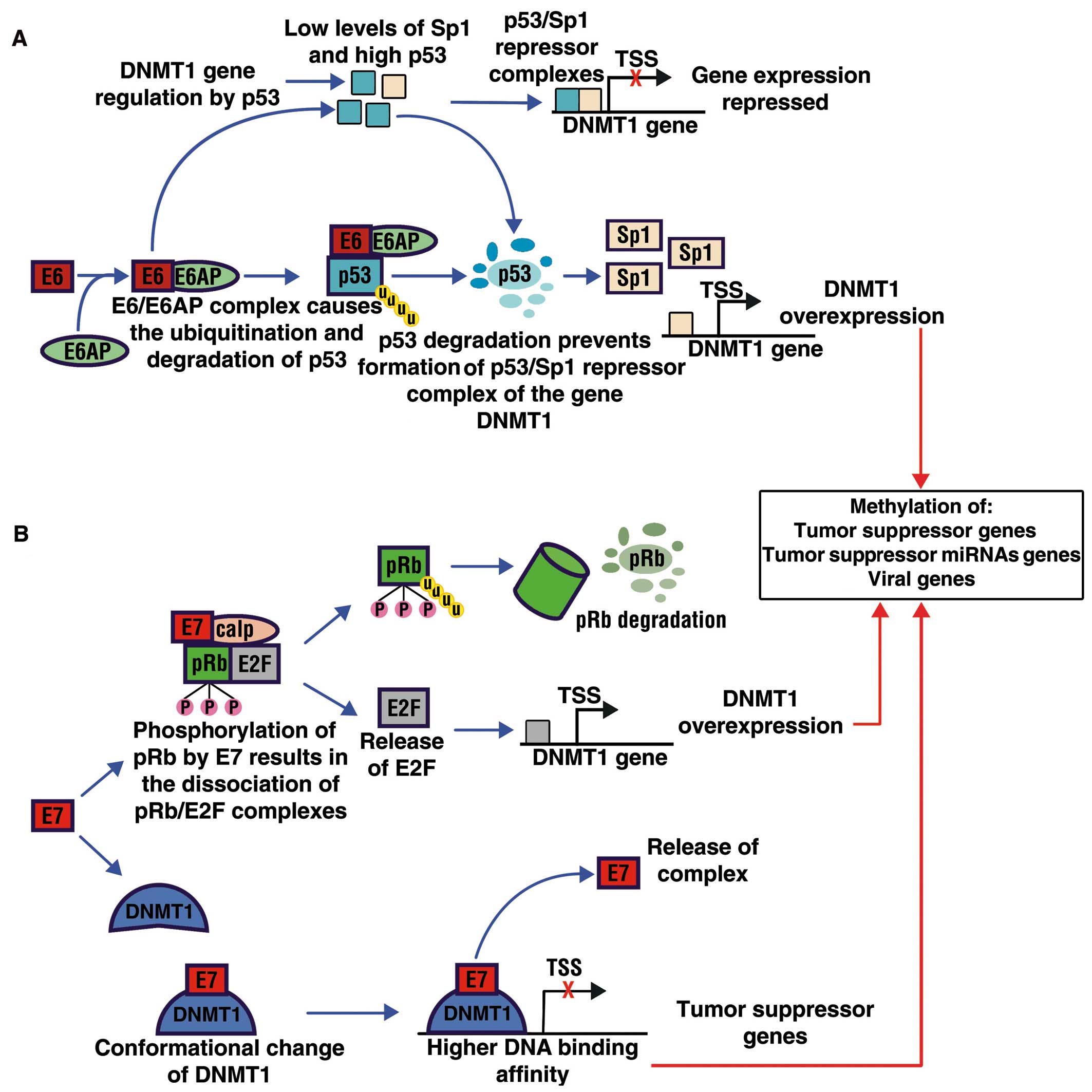
Au Yeung CL, Tsang WP, Tsang TY, Co NN,
Yau PL and Kwok TT: HPV-16 E6 upregulation of DNMT1 through
repression of tumor suppressor p53. Oncol Rep. 24:1599–1604.
2010.PubMed/NCBI
|
|
40
|
McCabe MT, Davis JN and Day ML: Regulation
of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res.
65:3624–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Richards KL, Zhang B, Baggerly KA, et al:
Genome-wide hypomethylation in head and neck cancer is more
pronounced in HPV-negative tumors and is associated with genomic
instability. PLoS One. 4:e49412009. View Article : Google Scholar
|
|
42
|
Lin RK, Wu CY, Chang JW, et al:
Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1
overexpression in lung cancer. Cancer Res. 70:5807–5817. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin-Tao W, Ling D, Shi-Wen J, et al:
Folate deficiency and aberrant expression of DNA methyltransferase
1 were associated with cervical cancerization. Curr Pharm Des. Jul
19–2013.(Epub ahead of print).
|
|
44
|
Nambaru L, Meenakumari B, Swaminathan R
and Rajkumar T: Prognostic significance of HPV physical status and
integration sites in cervical cancer. Asian Pac J Cancer Prev.
10:355–360. 2009.PubMed/NCBI
|
|
45
|
Turan T, Kalantari M, Cuschieri K, Cubie
HA, Skomedal H and Bernard HU: High-throughput detection of human
papillomavirus-18 L1 gene methylation, a candidate biomarker for
the progression of cervical neoplasia. Virology. 361:185–193. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Davis-Dusenbery BN and Hata A: MicroRNA in
cancer: the involvement of aberrant microRNA biogenesis regulatory
pathways. Genes Cancer. 1:1100–1114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shivdasani RA: MicroRNAs: regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pavicic W, Perkiö E, Kaur S, et al:
Altered methylation at microRNA-associated CpG islands in
hereditary and sporadic carcinomas: a methylation-specific
multiplex ligation-dependent probe amplification (MS-MLPA)-based
approach. Mol Med. 17:726–735. 2011. View Article : Google Scholar
|
|
50
|
Shen Y, Li Y, Ye F, et al: Identification
of miR-23a as a novel microRNA normalizer for relative
quantification in human uterine cervical tissues. Exp Mol Med.
43:358–366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Greco D, Kivi N, Qian K, et al: Human
papillomavirus 16 E5 modulates the expression of host microRNAs.
PLoS One. 6:e216462011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
So AY, Jung JW, Lee S, Kim HS and Kang KS:
DNA methyltransferase controls stem cell aging by regulating BMI1
and EZH2 through microRNAs. PLoS One. 6:e195032011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ferreira HJ, Heyn H, Moutinho C, et al:
CpG island hypermethylation-associated silencing of small nucleolar
RNAs in human cancer. RNA Biol. 9:881–890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hata A and Davis BN: Control of microRNA
biogenesis by TGFβ signaling pathway - a novel role of Smads in the
nucleus. Cytokine Growth Factor Rev. 20:517–521. 2009.
|
|
55
|
Fazi F and Nervi C: MicroRNA: basic
mechanisms and transcriptional regulatory networks for cell fate
determination. Cardiovasc Res. 79:553–561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang G, Wang Y, Teng M, Zhang D, Li L and
Liu Y: Signal transducers and activators of transcription-1 (STAT1)
regulates microRNA transcription in interferon γ-stimulated HeLa
cells. PLoS One. 5:e117942010.PubMed/NCBI
|
|
57
|
Bandres E, Agirre X, Bitarte N, et al:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Siomi H and Siomi MC: Posttranscriptional
regulation of microRNA biogenesis in animals. Mol Cell. 38:323–332.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ and Kerin MJ: MicroRNAs as novel biomarkers for breast cancer. J
Oncol. 2009:9502012009.PubMed/NCBI
|
|
60
|
Chen J, Yao D, Li Y, et al: Serum microRNA
expression levels can predict lymph node metastasis in patients
with early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013.PubMed/NCBI
|
|
61
|
Gocze K, Gombos K, Juhasz K, Kovacs K,
Kajtar B, Benczik M, et al: Unique microRNA expression profiles in
cervical cancer. Anticancer Res. 33:2561–2567. 2013.PubMed/NCBI
|
|
62
|
Wilting SM, Snijders PJ, Verlaat W, et al:
Altered microRNA expression associated with chromosomal changes
contributes to cervical carcinogenesis. Oncogene. 32:106–116. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huang L, Lin JX, Yu YH, Zhang MY, Wang HY
and Zheng M: Downregulation of six microRNAs is associated with
advanced stage, lymph node metastasis and poor prognosis in small
cell carcinoma of the cervix. PLoS One. 7:e337622012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cheung TH, Man KN, Yu MY, et al:
Dysregulated microRNAs in the pathogenesis and progression of
cervical neoplasm. Cell Cycle. 11:2876–2884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ma D, Zhang YY, Guo YL, Li ZJ and Geng L:
Profiling of microRNA-mRNA reveals roles of microRNAs in cervical
cancer. Chin Med J. 125:4270–4276. 2012.PubMed/NCBI
|
|
66
|
Pereira PM, Marques JP, Soares AR, Carreto
L and Santos MA: MicroRNA expression variability in human cervical
tissues. PLoS One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hu X, Schwarz JK, Lewis JS Jr, et al: A
microRNA expression signature for cervical cancer prognosis. Cancer
Res. 70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lee JW, Choi CH, Choi JJ, et al: Altered
microRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yao T, Rao Q, Liu L, et al: Exploration of
tumor-suppressive microRNAs silenced by DNA hypermethylation in
cervical cancer. Virol J. 10:1752013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wilting SM, Verlaat W, Jaspers A, et al:
Methylation-mediated transcriptional repression of microRNAs during
cervical carcinogenesis. Epigenetics. 8:220–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kalimutho M, Di Cecilia S, Del Vecchio
Blanco G, et al: Epigenetically silenced miR-34b/c as a novel
faecal-based screening marker for colorectal cancer. Br J Cancer.
104:1770–1778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Vogt M, Munding J, Grüner M, et al:
Frequent concomitant inactivation of miR-34a and
miR-34b/c by CpG methylation in colorectal, pancreatic,
mammary, ovarian, urothelial, and renal cell carcinomas and soft
tissue sarcomas. Virchows Arch. 458:313–322. 2011.PubMed/NCBI
|
|
73
|
Zhang Y, Wang X, Xu B, et al: Epigenetic
silencing of miR-126 contributes to tumor invasion and angiogenesis
in colorectal cancer. Oncol Rep. 30:1976–1984. 2013.PubMed/NCBI
|
|
74
|
Zhu A, Xia J, Zuo J, et al: MicroRNA-148a
is silenced by hypermethylation and interacts with DNA
methyltransferase 1 in gastric cancer. Med Oncol. 29:2701–2709.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Tsai KW, Wu CW, Hu LY, et al: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tsai KW, Liao YL, Wu CW, et al: Aberrant
hypermethylation of miR-9 genes in gastric cancer.
Epigenetics. 6:1189–1197. 2011.PubMed/NCBI
|
|
77
|
Kim K, Lee HC, Park JL, et al: Epigenetic
regulation of microRNA-10b and targeting of oncogenic
MAPRE1 in gastric cancer. Epigenetics. 6:740–751. 2011.
|
|
78
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Deng H, Guo Y, Song H, et al: MicroRNA-195
and microRNA-378 mediate tumor growth suppression by epigenetical
regulation in gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yan-Fang T, Jian N, Jun L, et al: The
promoter of miR-663 is hypermethylated in Chinese pediatric acute
myeloid leukemia (AML). BMC Med Genet. 14:742013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Köhler CU, Bryk O, Meier S, et al:
Analyses in human urothelial cells identify methylation of miR-152,
miR-200b and miR-10a genes as candidate bladder cancer biomarkers.
Biochem Biophys Res Commun. 438:48–53. 2013.PubMed/NCBI
|
|
82
|
Datta J, Kutay H, Nasser MW, et al:
Methylation mediated silencing of microRNA-1 gene and its role in
hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jung CJ, Iyengar S, Blahnik KR, et al:
Epigenetic modulation of miR-122 facilitates human embryonic stem
cell self-renewal and hepatocellular carcinoma proliferation. PLoS
One. 6:e277402011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
He Y, Cui Y, Wang W, et al:
Hypomethylation of the hsa-miR-191 locus causes high expression of
hsa-mir-191 and promotes the epithelial-to-mesenchymal transition
in hepatocellular carcinoma. Neoplasia. 13:841–853. 2011.PubMed/NCBI
|
|
85
|
Zhang Y, Yan LX, Wu QN, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Vrba L, Muñoz-Rodríguez JL, Stampfer MR
and Futscher BW: miRNA gene promoters are frequent targets of
aberrant DNA methylation in human breast cancer. PLoS One.
8:e543982013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Rauhala HE, Jalava SE, Isotalo J, et al:
miR-193b is an epigenetically regulated putative tumor suppressor
in prostate cancer. Int J Cancer. 127:1363–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hulf T, Sibbritt T, Wiklund ED, et al:
Epigenetic-induced repression of microRNA-205 is associated with
MED1 activation and a poorer prognosis in localized prostate
cancer. Oncogene. 32:2891–2899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Formosa A, Lena AM, Markert EK, et al: DNA
methylation silences miR-132 in prostate cancer. Oncogene.
32:127–134. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Heller G, Weinzierl M, Noll C, et al:
Genome-wide miRNA expression profiling identifies miR-9-3
and miR-193a as targets for DNA methylation in non-small
cell lung cancers. Clin Cancer Res. 18:1619–1629. 2012.PubMed/NCBI
|
|
92
|
Incoronato M, Urso L, Portela A, et al:
Epigenetic regulation of miR-212 expression in lung cancer. PLoS
One. 6:e277222011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kitano K, Watanabe K, Emoto N, et al: CpG
island methylation of microRNAs is associated with tumor size and
recurrence of non-small-cell lung cancer. Cancer Sci.
102:2126–2131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang Z, Chen Z, Gao Y, et al: DNA
hypermethylation of microRNA-34b/c has prognostic value for stage I
non-small cell lung cancer. Cancer Biol Ther. 11:490–496. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wong KY1, Liang R, So CC, Jin DY, Costello
JF and Chim CS: Epigenetic silencing of MIR203 in multiple
myeloma. Br J Haematol. 154:569–578. 2011.PubMed/NCBI
|