Introduction
Esophageal cancer (EC) is the third most prevalent
gastrointestinal malignancy in the world. It is a deadly disease
with 5-year survival rates ranging from 10 to 16% (1). Esophageal squamous cell carcinoma
(ESCC) is the major histologic subtype of EC. Annually, there are
462,000 reported new cases and 386,000 deaths; 75% of the patients
die within one year of diagnosis (1). The incidence of EC shows great
geographical variation. Its high prevalence is of particular
importance in Northern China. Worldwide, the south side of the
Taihang Mountains in North-central China along the northern borders
of Hebei, Henan and Shanxi Provinces forms one of the highest risk
regions of EC (2–4). In Linzhou County of Henan Province in
1993–1997, the age-standardized incidence rate of EC in males and
females was 121/100,000 and 80/100,000, respectively (5). Epidemiological studies suggest that
environmental factors such as alcohol abuse, tobacco smoke and
dietary carcinogens may play a substantial role in the etiology of
EC (6,7). However, strong evidence from familial
aggregation of EC in Yangcheng County and segregation analysis of
EC in Yangquan indicate the existence of genetic susceptibility
loci for EC predisposition (3,8).
Historically, the Chaoshan people in Nanao, which is another
high-risk region of EC in Southern China (9), originally migrated from the Taihang
Mountain region. The distinctly high EC incidence in the Chaoshan
population further strengthens the etiologic role played by genetic
components in familial EC. A third line of evidence supporting a
causative role of genetic factors in familial EC comes from
molecular genetic studies of loss of heterozygosity (LOH), SNP
association, microarray profiling and BRCA2 germline
mutations (10–14).
Recent technological advances allow the
identification of long regions of homozygosity in genomic DNAs
using high-density SNP arrays. These homozygous regions, which may
dictate the level of consanguinity of an individual, are referred
to as identity-by-descent (IBD) segments, also known as
autozygosity (15,16). The IBD segments represent the
sharing of common sequences from an ancestor in those regions.
Previous studies suggest that a high rate of consanguinity, which
produces germline genomic homozygosity, is associated with cancers
(17–20). A recent study using SNP array
analysis showed that the signatures of autozygosity correlate with
colorectal cancer (CRC) incidence, and the IBD regions may help
localize the genes that contribute to CRC heritability (15). However, IBD regions are difficult to
locate without pedigree information. We hypothesized that the IBD
approach is useful for mapping the common susceptibility locus with
low-penetrance SNPs in hereditary ESCC patients in Henan, one of
the highest ESCC risk regions in the world. Screening programs
based on knowledge of the candidate cancer susceptibility locus may
reduce cancer mortality by prevention. Thus, the long-term aim is
to better understand the genetic basis for ESCC in Henan to achieve
the ultimate goal of improving ESCC patient survival.
Materials and methods
Samples for SNP analysis
Samples for IBD analysis consisted of 50 blood
samples from 18 healthy unrelated individuals and 32 FH+
ESCC patients from the high-risk region of Henan Province and
nearby counties from the Linzhou Center Hospital and Yaocun
Esophageal Cancer Hospital collected from 2001 to 2008. Table I summarizes the clinical information
of the 32 cases and 18 controls and details of the family history
of the ESCC patients. Thirty-one of the 32 FH+ patients
analyzed involved at least two generations having a family history
of ESCC. Approval for use of human blood and/or information was
obtained from the Committee for Ethical Review of Research
Involving Human Subjects at Zhengzhou University. The study was
conducted according to Declaration of Helsinki principles. Informed
written consent was obtained from each individual.
 | Table IClinical information and details of
the family history of the ESCC patients. |
Table I
Clinical information and details of
the family history of the ESCC patients.
| Sample name | Gender | Age at
diagnosis | No. of
generations | No. of ESCC
cases/family members | Relationship with
index case |
|---|
| FEDTM001002 | Female | 64 | 3 | 8/21 | Grandfather,
mother, aunt, 5 siblings, aSibling with dysplasia |
|
FEDTM001004a | Male | 50 | | |
| FELJB007001 | Male | 82 | 3 | 14/7 | 4 Mother, 6
siblings, 2 sibling’s wives, 2 nieces, 2 nephews, nephew’s
wife |
| FELYS010002 | Male | 57 | 2 | 8/35 | Father, 2 uncles,
uncle’s wife, aunt, 2 siblings, cousin |
| FELPK021001 | Male | 74 | 2 | 6/52 | Mother and father,
uncle, 3 siblings |
| FE016CZH022 | Male | 56 | 4 | 14/49 | Grandfather,
father, 5 uncles, 3 cousins, wife, brother’s wife, niece |
| FE016CZH023 | Female | 56 | 3 | 11/27 | Grandfather,
mother, 2 uncles, husband, sibling’s wife, 4 cousins |
| FE016CZH010 | Female | 82 | | |
| FE08YHM012 | Male | 71 | 1 | 4/67 | 3 Cousins |
| FH(+)B3 | Female | 48 | 2 | Unk | Father |
| FH(+)B4 | Male | 55 | 2 | Unk | Mother |
| FH(+)B5 | Female | 62 | 2 | Unk | Mother |
| FH(+)-3/B18 | Male | 56 | 2 | 3/8 | Mother and
father |
| FH(+)-3/B19 | Male | 56 | 2 | 2/12 | Mother |
| FH(+)-3/B20 | Male | 48 | 2 | 3/11 | Mother and
father |
| FH(+)-3/B22 | Male | 58 | 2 | 2/8 | Mother |
| FH(+)-3/B23 | Male | 55 | 2 | 2/7 | Mother |
| FH(+)-3/B24 | Male | 63 | 2 | 2/7 | Mother |
| FH(+)-3/B25 | Male | 55 | 2 | 2/7 | Father |
| FH(+)-3/B26 | Male | 48 | 2 | Unk | Father |
| FH(+)-3/B27 | Male | 51 | 2 | 2/8 | Mother |
| FH(+)-3/B28 | Male | 62 | 2 | Unk | Father |
| FH(+)-3/B29 | Male | 50 | 2 | 2/8 | Mother |
| FH(+)-3/B30 | Male | 55 | 2 | 2/8 | Father |
| FH(+)-3/B31 | Female | 51 | 2 | 2/13 | Father |
| FH(+)-3/B33 | Male | 64 | 2 | 3/9 | Father and
sister |
| FH(+)-3/B34 | Female | 58 | 2 | 2/12 | Mother |
| FH(+)-3/B36 | Female | 58 | 2 | 2/7 | Mother |
| FH(+)-3/B37 | Male | 62 | 2 | 3/6 | Father and
brother |
| FH(+)-3/B38 | Male | 58 | 2 | 2/6 | Mother |
| FH(+)-3/B39 | Female | 63 | 2 | 2/9 | Father |
| FH(+)-3/B40 | Male | 47 | 2 | Unk | Mother and
father |
| Samples | Sex ratio
(M:F) | Average |
|---|
| 32 FH+
ESCC | 2.6:1.0 | 58.6 |
| 18 Healthy
unrelated individuals | 1.0:1.0 | 45.9 |
DNA extraction from clotted blood
Between 0.4 and 2 ml of clotted blood was used for
DNA extractions. DNA extraction was performed as described by Kanai
et al (21). Briefly, 250 μl
of lysis solution was mixed with each 200 μl of clotted blood and
incubated overnight at 55–65°C. Phase separation was performed by
centrifugation, and DNA was precipitated in ethanol.
SNP array and genotyping procedure
The SNP genotyping was performed at the UCLA
microarray center with Affymetrix GeneChip Human mapping (~248K
SNPs) Sty I SNP Array (Affymetrix). Genotypes were called by the
GeneChip DNA Analysis Software (v2.0, Affymetrix).
CHB HapMap genotype data
Genotype data of Affymetrix GeneChip Human mapping
(~238K SNPs) Sty I SNP Array of 45 unrelated individuals from Han
Chinese in Beijing, China (CHB), which is publicly accessible, were
downloaded from the Affymetrix technical documentation (http://www.affymetrix.com/support/technical/sample_data/500k_hapmap_genotype_data.affx).
Quality control
A total of 238,032 SNPs were filtered with a SNPs
call rate ≥0.90, individual call rate ≥0.94, and a minor allele
frequency (MAF) >0.05. SNPs were selected to have Hardy-Weinberg
equilibrium (HWE), P-values of ≥0.001 in controls and the CHB
HapMap dataset. After applying these stringent quality control
measures for the inherited ESCC IBD study, genotypes for 150,940
SNPs were available for 28 FH+ cases, 16 controls and a
45 CHB HapMap dataset.
Statistical and bioinformatic
analysis
PLink (22) (v1.06)
software (http://pngu.mgh.harvard.edu/~purcell/plink/contact.shtml#cite)
was used to detect IBD segments of different combinations with a
threshold limit of IBD segments ranging from 1 to 4 Mb in length
and comprised of 20–50 SNPs in each individual’s genome. The
threshold limit of IBD segments was defined as a stretch of runs of
at least e.g. 50 consecutive homozygous SNPs encompassing a minimum
of e.g. 2 Mb. Remaining options were set to default values. BEAGLE
software was used to detect the sharing of IBD regions in paired
individuals (23). A fast IBD score
<10−10 and the shared haplotype length of ≥1 Mb were
used as thresholds to indicate strong evidence of the shared IBD
haplotypes among the genomes of pairs of individuals (case/case vs.
control/control pairing). Comparisons of the difference in the
average lengths of IBDs between cases, controls and CHB HapMap
dataset were performed by ANOVA. Statistical analysis for
comparison of the distribution of categorical variables was
performed by Chi-square test and Fisher’s exact test in the
pair-IBD approach.
ESCC tissue specimens and real-time
quantitative PCR analysis
Total RNAs from 31 pairs of matched esophageal
epithelial non-tumor and tumor specimens from ESCC patients were
extracted by TRIzol reagent as previously described (24). These included 11 Henan
FH+, 10 Henan family FH−, and 10 Hong Kong
ESCC specimens. Henan and Hong Kong ESCC specimens were collected
from Yaocun Esophageal Cancer Hospital, Linzhou, Henan, China from
2005 to 2007 and Queen Mary Hospital from 2004 to 2008,
respectively. Approval for the present study was obtained from the
Hospital Institutional Review Board at the University of Hong Kong,
Hong Kong. Two micrograms of total RNA was reverse transcribed into
cDNAs by SuperScript III reverse transcriptase (Invitrogen), as
described in the manufacturer’s manual. Real-time qPCR was
performed in a StepOnePlus machine (Applied Biosystems) using
FastStart Universal Probe Master Mix (Roche Diagnostics) and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) TaqMan
probes (Applied Biosystems). The universal probe library (UPL)
(Roche Diagnostics) was used to validate the expression of
PLCE1 (NM_016341.3), CYP2-C18 (NM_000772.2),
SIAH1 (NM_001006610.1) and GPT2 (NM_001142466.1), as
previously described (25). Primer
sequences and UPL probe number used are provided upon request.
Results
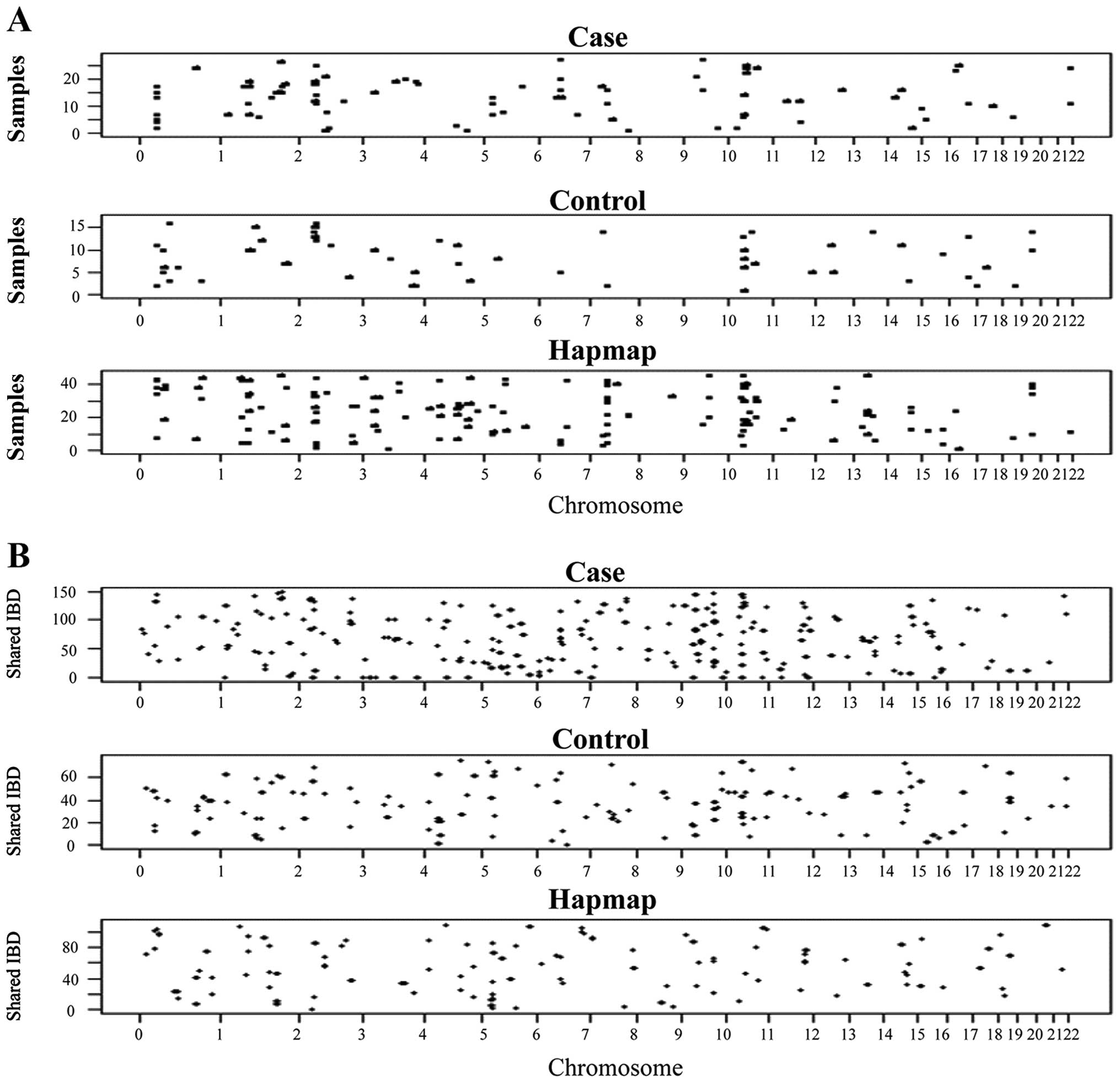
IBD segments: individual germline genomic
homozygosity with exceptionally long stretches of SNPs
The germline homozygosity in 28 Henan FH+
ESCC patients was explored by analysis with Affymetrix GeneChip
Human mapping SNP array (~238K SNPs, Sty I). The IBD segment
approach scores the individual germline homozygosity (IBD segments)
by detection of long stretches of at least 2 Mb containing a
minimum of 50 consecutive homozygous SNPs in the blood DNAs. These
IBD segments were distributed across the genomes of the 28
FH+ ESCC cases, 16 Henan normal controls and 45
individuals from the CHB HapMap datasets (Fig. 1A). The threshold was set to a
minimum of 2 Mb in length containing at least 50 consecutive
homozygous SNPs. Longer average IBD segment length and higher
average number of SNPs in IBD segments were also detected in the
cases compared to the controls and CHB HapMap controls, when the
threshold was set ranging from 20 to 50 consecutive homozygous SNPs
(data not shown). A total of 26 IBD segments in the FH+
ESCC samples having no overlap with the control and CHB HapMap were
detected (data not shown). The physical map is based on the Human
Feb. 2009 (GRCh37/hg19) assembly (http://genome.ucsc.edu/cgi-bin/hgGateway). Among the
28 FH+ ESCC samples, patient FH3B31 had the longest
total length of 29.3 Mb IBD segments distributed in 6 regions.
Patient FH3B24 had the longest IBD segment of 21.1 Mb encompassing
799 SNPs at chromosome 2q31.1-q32.2. When the IBD segment was
defined as a minimum of 2 Mb in length containing at least 50
consecutive homozygous SNPs, 22/28, 14/16 and 38/45 (78.6, 87.5 and
84.4%) of FH+ ESCC patients, Henan controls and CHB
HapMap controls were observed as having more than one IBD segment,
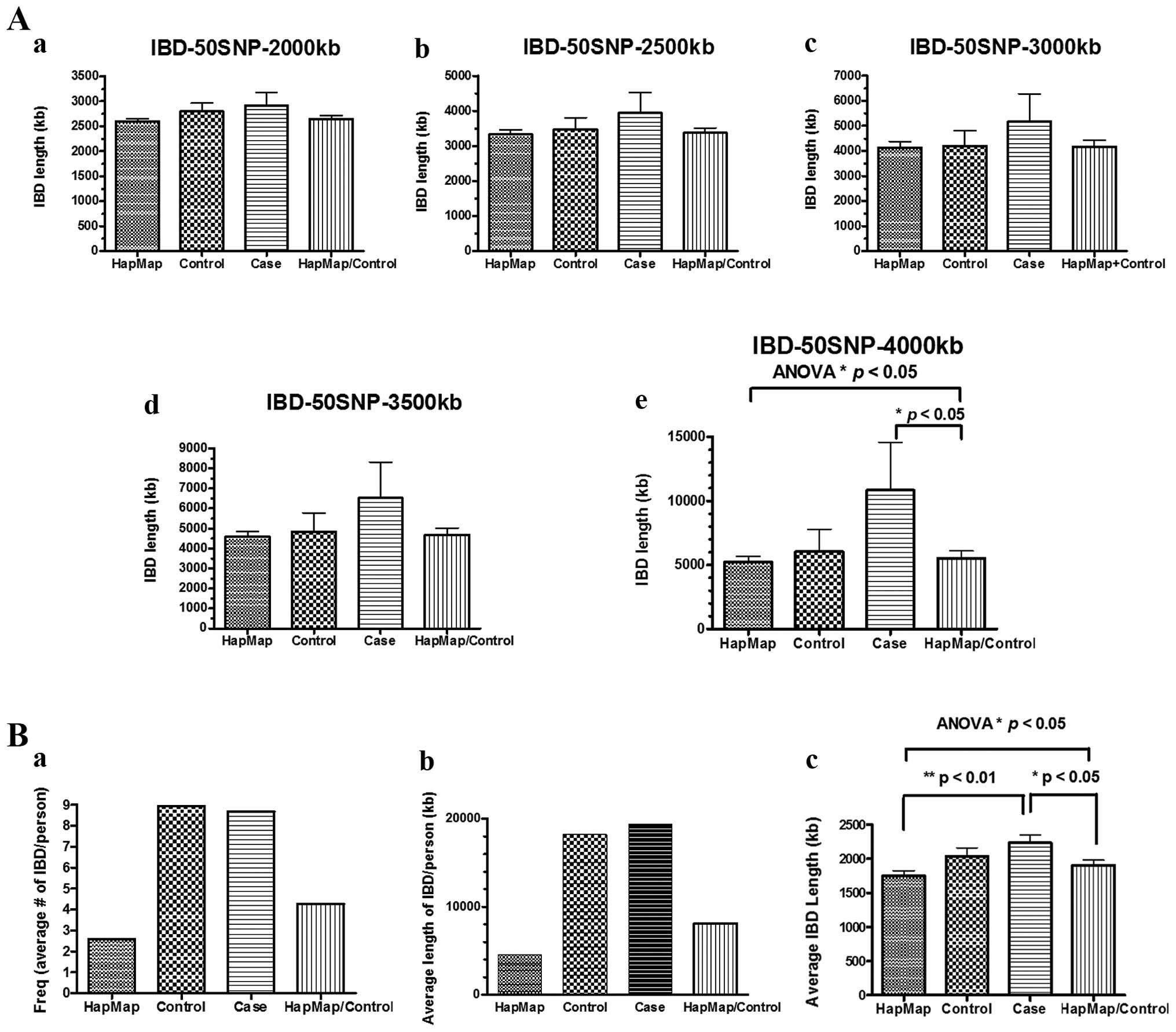
respectively. However, a trend of longer average IBD segments, but
not a higher frequency of IBD segments, was observed in
FH+ ESCC patients compared to the Henan controls and
Henan control/CHB HapMap, when the threshold was set ranging from 2
to 3.5 Mb in length containing at least 50 consecutive homozygous
SNPs, although the difference did not reach statistical
significance (Fig. 2A-a–d).
FH+ ESCC patients had statistically significant longer
average IBD segments compared to the Henan control/CHB HapMap, when
the threshold was set to 4 Mb in length containing at least 50
consecutive homozygous SNPs (Fig.
2A-e).
Pairing analysis for sharing of IBD
segment approach: eight critical regions with significantly higher
frequencies of sharing of IBD segments among the FH+
ESCC patients compared to the CHB HapMap/Henan normal datasets
The pairing analysis approach, performed with the
BEAGLE software, was used for identification of the shared IBD
segments distributed across the genomes of the FH+ ESCC
cases, Henan normal controls, and individuals from the CHB HapMap
datasets (Fig. 1B). When the
bioinformatic analysis was carried out based on the homozygosity
mapping approach of case/case and control/control pairing, the
frequency (number of IBD segments per person) and average length of
IBD per person were higher in the cases compared to the CHB HapMap
dataset, but similar in the cases and controls from Henan (Fig. 2B-a and -b). The FH+ ESCC
patients had statistically significant longer average IBD lengths
compared to the CHB HapMap alone and the CHB HapMap/Control. A
longer length of IBD segments was also observed, when a comparison
was made between the cases and controls, although the difference
did not reach statistical significance (Fig. 2B-c).
Among the 28, 16 and 45 individuals in the case,
control, and CHB HapMap groups, the numbers of pairs for
calculation of sharing events were 378, 120 and 990, respectively.
Table II summarizes the 8 critical
regions that are shared at a statistically significant higher
number of events in the FH+ ESCC cases compared to the
CHB HapMap controls. The 8 critical regions include 2q32.1-q32.2,
3p22.3-p22.2, 4q21.1-q21.21, 7p22.2, 8q23.2-q23.3, 10q23.33-q24.1,
14q24.3 and 16q11.2-q12.1. The genes involved are also listed in
Table II. Table III summarizes the statistical
analysis of the eight critical IBD regions in Henan FH+
ESCC identified by excessive sharing of homozygous segments in the
case/case and control/control pairing. These regions include 7p22.2
with four and zero sharing events of IBD segments; six regions at
2q32.1-q32.2, 3p22.3-p22.2, 4q21.1-q21.21, 8q23.2-q23.3, 14q24.3
and 16q11.2-q12.1 were detected to have three and zero sharing of
IBD segments and a critical region at 10q23.33-q24.1 was detected
to have six and two sharing events of IBD segments out of 378 and
990 pairing events in the cases and CHB HapMap dataset,
respectively (Table III).
 | Table IIPairing analysis of sharing of the
IBD segment approach: eight critical regions with significantly
higher frequency of sharing of IBD segments among the
FH+ ESCC patients compared to the control and CHB HapMap
dataset.a |
Table II
Pairing analysis of sharing of the
IBD segment approach: eight critical regions with significantly
higher frequency of sharing of IBD segments among the
FH+ ESCC patients compared to the control and CHB HapMap
dataset.a
| Individual ID1 | Individual ID2 | Start SNP | End SNP | Chromosome | Start Pos | End Pos | Length (kb) | Critical
cytoBand | Genes |
|---|
| FHB5 | FH3B39 | rs10931294 | rs4667249 | 2 | 188440696 | 189700120 | 1,259.4 |
2q32.1-q32.2b | MIR561,
GULP1, DIRC1 |
| FHB3 | FHB5 | rs10931294 | rs4667249 | 2 | 188440696 | 189700120 | 1,259.4 |
2q32.1-q32.2b | |
| FHB3 | FH3B39 | rs935806 | rs16830983 | 2 | 187836723 | 189855427 | 2,018.7 |
2q32.1-q32.2b | |
| HB6 | HB9 | rs850889 | rs4278898 | 2 | 186208248 | 188707452 | 2,499.2 | 2q32.1b | |
| FH3B27 | FH3B39 | rs6775328 | rs197729 | 3 | 36405852 | 37495846 | 1,090.0 |
3p22.3-p22.2c | TRANK1,
EPM2AIP1, MLH1, LRRFIP2, GOLGA4,
C3orf35, ITGA9 |
| FH3B39 | FH3B40 | rs3749279 | rs17036845 | 3 | 36484823 | 37775974 | 1,291.2 |
3p22.3-p22.2c |
| FHB3 | FH3B39 | rs4328757 | rs9311168 | 3 | 36938180 | 37977417 | 1,039.2 | 3p22.2c | |
| NA18524 | NA18563 | rs10510690 | rs4364115 | 3 | 37525566 | 38895359 | 1,369.8 | 3p22.2c | |
| FH3B25 | FH3B30 | rs4075858 | rs6837860 | 4 | 97991815 | 99559661 | 1,567.8 | 4q22.3-q23 | C4orf37,
RAP1GDS1 |
| FH3B30 | FH3B34 | rs4075858 | rs6837860 | 4 | 97991815 | 99559661 | 1,567.8 | 4q22.3-q23 | |
| FH3B25 | FH3B34 | rs6835122 | rs1154435 | 4 | 96733630 | 100285148 | 3,551.5 | 4q22.3-q23 | |
| FH2B18 | FH3B22 | rs10807829 | rs17134637 | 7 | 3204223 | 4266506 | 1,062.3 | 7p22.2 | SDK1 |
| FH3B20 | FH3B22 | rs2644270 | rs2189973 | 7 | 2921582 | 4333275 | 1,411.7 | 7p22.2 | |
| FH2B18 | FH3B20 | rs1525558 | rs10239020 | 7 | 3323181 | 4376641 | 1,053.5 | 7p22.2 | |
| FH2B18 | FH3B33 | rs17133874 | rs7782329 | 7 | 3848379 | 4925012 | 1,076.6 | 7p22.2-p22.1 | |
| FH3B38 | FH3B39 | rs9297452 | rs1513524 | 8 | 112743439 | 114450518 | 1,707.1 | 8q23.3 | CSMD3 |
| FH3B28 | FH3B39 | rs2350728 | rs10107411 | 8 | 111184071 | 114509172 | 3,325.1 | 8q23.2-q23.3 | |
| FH3B40 | FEDTM1 | rs1492660 | rs10505248 | 8 | 113470071 | 116156852 | 2,686.8 | 8q23.2-q23.3 | |
| FHB5 | FH3B31 | rs4919587 | rs11188222 | 10 | 95607549 | 96936657 | 1,329.1 | 10q23.33 | LGI1,
SLC35G1, PLCE1, TBC1D12, HELLS,
CYP2C18, CYP2C19, CYP2C9, CYP2C8,
C10orf129, PDLIM1, SORBS1,
ALDH18A1 |
| FH3B23 | FH3B30 | rs6583904 | rs3737015 | 10 | 95488351 | 97028413 | 1,540.1 | 10q23.33-q24.1 |
| FH3B30 | FH3B31 | rs1925246 | rs7084645 | 10 | 95868120 | 97069762 | 1,201.6 | 10q23.33-q24.2 |
| FH3B23 | FH3B31 | rs1925246 | rs7084645 | 10 | 95868120 | 97069762 | 1,201.6 | 10q23.33-q24.1 |
| FH3B19 | FH3B28 | rs10509671 | rs10882652 | 10 | 96069054 | 97441326 | 1,372.3 | 10q23.33-q24.1 | |
| FH3B33 | FE016CZH010 | rs7899038 | rs10748673 | 10 | 96772412 | 98122352 | 1,349.9 | 10q23.33-q24.1 | |
| NA18563 | NA18573 | rs10786148 | rs7083399 | 10 | 95655566 | 97105133 | 1,449.6 | 10q23.33-q24.1 | |
| NA18564 | NA18632 | rs4918070 | rs501603 | 10 | 95828553 | 97333879 | 1,505.3 | 10q23.33-q24.1 | |
| FH3B25 | FH3B39 | rs12586708 | rs177213 | 14 | 76840497 | 78533453 | 1,693.0 | 14q24.3d | ISM2,
SPTLC2, ALKBH1, SLIRP, SNW1,
C14orf178, ADCK1 |
| FH3B22 | FH3B39 | rs17103928 | rs177213 | 14 | 76507299 | 78533453 | 2,026.2 | 14q24.3d |
| FH3B22 | FH3B25 | rs12586708 | rs177170 | 14 | 76840497 | 78573143 | 1,732.6 | 14q24.3d | |
| HB19 | HB21 | rs760233 | rs11622359 | 14 | 76393968 | 77939449 | 1,545.5 | 14q24.3d | |
| FH3B26 | FE08 | rs34738934 | rs8046716 | 16 | 46710869 | 48560941 | 1,850.1 | 16q11.2-q12.1 | VPS35,
ORC6, MYLK3, GPT2, DNAJA2,
NETO2, PHKB, ABCC12, ABCC11,
SIAH1, C16orf87, ITFG1, LONP2,
LOC100507577, MIR548AE2 |
| FH3B40 | FE08 | rs34738934 | rs8046716 | 16 | 46710869 | 48560941 | 1,850.1 | 16q11.2-q12.1 |
| FH3B26 | FH3B40 | rs17839567 | rs893174 | 16 | 31057945 | 49303349 | 18,245.4 | 16q11.2-q12.1 |
 | Table IIIStatistical analysis of the eight
critical IBD regions in Henan FH+ ESCC cases identified
by excessive sharing of homozygous segments in case/case and
control/control pairing. |
Table III
Statistical analysis of the eight
critical IBD regions in Henan FH+ ESCC cases identified
by excessive sharing of homozygous segments in case/case and
control/control pairing.
| No. | Critical IBD
region | Frequency of IBD
sharing events in cases, (n=28) | Frequency of IBD
sharing events in controls (n=16) | P-value (Fisher
exact test, 2-tailed) | Frequency of IBD
sharing events in CHB HapMap (n=45) | P-value (Fisher
exact test, 2-tailed) |
|---|
| 1 | 2q32.1-q32.2 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
| 2 | 3p22.3-p22.2 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
| 3 | 4q21.1-q21.21 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
| 4 | 7p22.2 | 4/378 | 0/120 | 0.5768 | 0/990 | 0.0058 |
| 5 | 8q23.2-q23.3 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
| 6 | 10q23.33-q24.1 | 6/378 | 0/120 | 0.3436 | 2/990 | 0.0071 |
| 7 | 14q24.3 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
| 8 | 16q11.2-q12.1 | 3/378 | 0/120 | 1.0000 | 0/990 | 0.0210 |
Validation of target gene expression
identified by pairing IBD analysis by quantitative RT-PCR
To examine the differential gene expression
profiling among familial ESCC cases from high-risk Mainland China
and cases from moderate-risk Hong Kong, an oligonucleotide
microarray analysis with the 28K gene chip prepared at the Genome
Institute of Singapore, as previously described (26), was performed using 31 pairs of ESCC
specimens from Henan (11 FH+, 10 FH−) and
Hong Kong (10 cases) (data not shown). Four critical target genes
at 10q23.33-q24.1 and 16q11.2-q12.1 (Table II) were chosen for further study to
demonstrate their clinical and biological relevance after
comparison for overlap between the two lists of candidate genes
identified by pairing IBD analysis and microarray differential
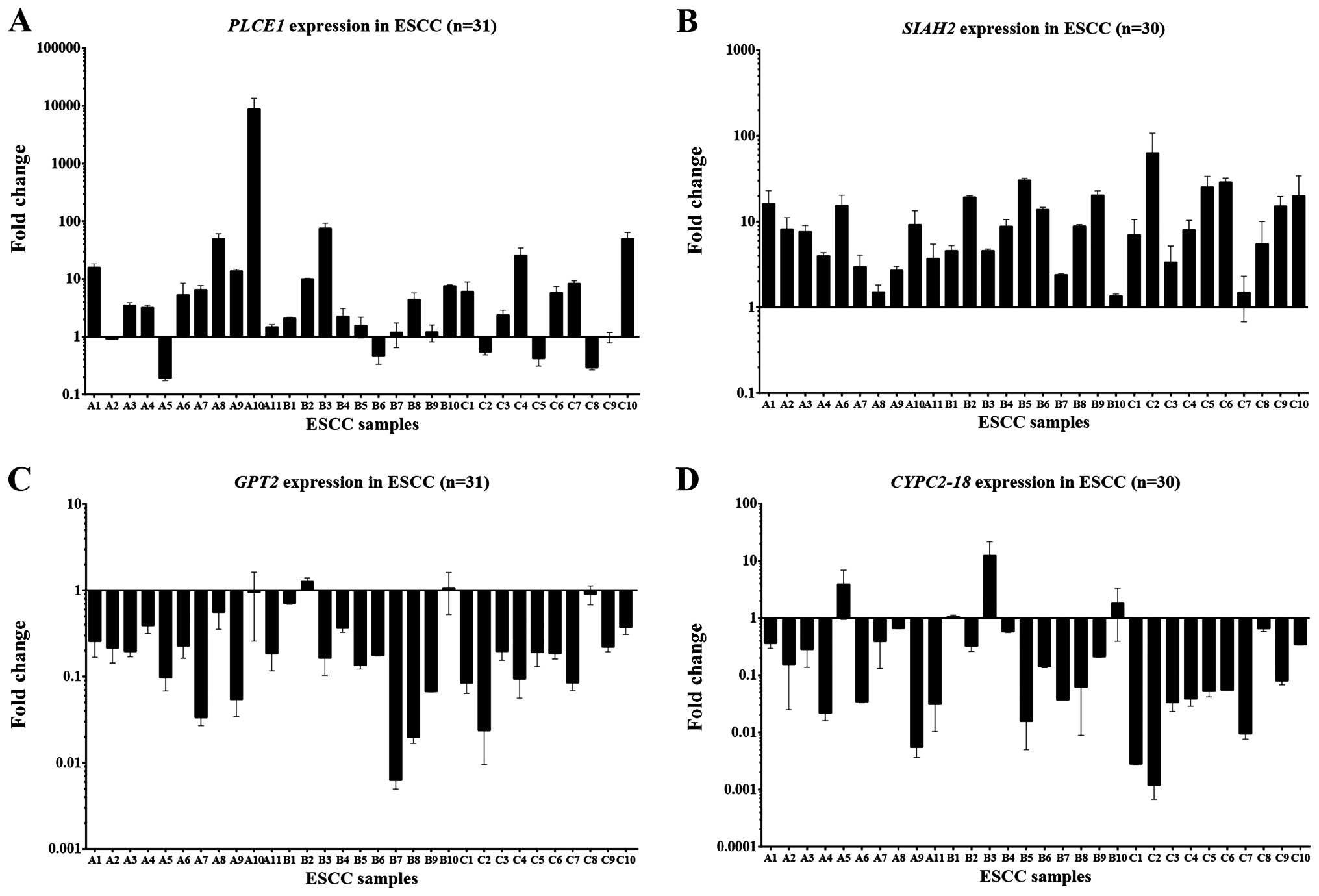
expression. Validation of target gene expression by quantitative
RT-PCR demonstrated significant overexpression of PLCE1 and
SIAH1, as well as downregulation of CYP2-C18 and
GPT2 expression using GAPDH expression for
normalization (Fig. 3). Among the
31 pairs of ESCC cases, the frequencies of PLCE1 and
SIAH1 overexpression were 64.5 (20/31) and 90.0% (27/30),
respectively, while the frequencies of GPT2 and
CYP2-C18 downregulation were 80.6 (25/31) and 76.7% (23/30)
in ESCC primary tumor tissues, respectively, when the threshold was
set at a 2-fold difference.
Discussion
Increased risk of cancer incidence has been observed
in inbred populations (18,20,27).
An inbred population is characterized by the sharing of longer IBD
segments. These observations raise the question as to whether
germline homozygosity is involved in cancer predisposition,
particularly in inherited cancer. Technological advances in SNP
array provide the opportunity to address this issue by the
identification of these long stretches of IBD segments among cancer
patients. Homozygosity mapping was reported as an efficient
strategy to map human recessive traits with the DNA of affected
children from consanguineous marriages (28,29).
The information obtained from a single affected child of a first
cousin marriage is the same as those using linkage mapping with a
nuclear family with three affected children. By studying DNA of
less than a dozen unrelated, affected inbred individuals,
homozygosity mapping makes it possible to map recessive diseases
(28). An example of the
application of homozygosity mapping of five unrelated
consanguineous families with autosomal recessive cutis laxa (ARCL)
identified a candidate region on chromosome 17q25, and
disease-causing mutations in PYCR1 were detected by
high-throughput sequencing of the candidate region (29).
China is one of the highest risk regions of EC
worldwide; the reported age-standardized incidence rate of EC in
males and females was 37.9/100,000 and 12.0/100,000, respectively,
in 2002 (1). The
Taihang-Hebei-Henan-Shanxi area in North-central China forms an
extraordinarily high risk region for this type of cancer (2–4). The
age-standardized incidence rate of EC in males and females was
~3-fold and 6.5-fold higher, respectively, in Linzhou County of
Henan Province in 1993–1997 compared to that in other parts of
China (5). There was an ~10-fold
increase in esophageal carcinoma incidence in these highest risk
regions of the Taihang-Hebei-Henan-Shanxi area compared to a
moderate-risk region such as Hong Kong. The present study used the
homozygosity mapping approach of affected ESCC patients from those
villages with exceptionally high incidence of ESCC. Consanguineous
marriages in these villages were highly suspected as longer average
IBD segments were observed in Henan villages among both the
controls and cases (Fig. 2B-a and
-b; Table IV). The presence of
homozygous segments in an individual’s genome can be explained by
tracing one’s parental lineage to a common ancestor. The first
approach identified 26 non-overlapping IBD segments that fulfilled
a threshold criteria set (50 SNPs, 2 Mb) in the present study of
FH+ ESCC cases from Henan. Two individuals are identical
by descent at a locus, when they share the same genetic materials
from a common ancestor. The second approach in the present study
detected excessive sharing of eight critical IBD regions in the
genomes between two affected individuals with FH+ ESCC.
Table IV summarizes the results of
IBD regions identified by individual homozygosity mapping and
shared IBD segments in the case/case and control/control pairing
approaches. Both approaches detected longer average IBD lengths in
FH+ ESCC cases, when compared to the CHB HapMap dataset
(Table IV). In the second
approach, compared to the CHB HapMap dataset, there was a ~4.3-fold
increase in the average IBD length per person in the Henan controls
and FH+ ESCC cases. There was an ~3.3-fold increase in
the frequency of IBD segments in the Henan controls and
FH+ ESCC cases compared to the CHB HapMap dataset. The
data suggested that populations from Henan villages (both
FH+ ESCC and controls) showed a higher rate of
consanguinity compared to the CHB HapMap dataset. The critical
regions detected by the pair-wise comparison provided additional
information on the potential cancer susceptibility loci for ESCC
development and should be further studied with a few affected
inbred individuals or a higher number of FH+ ESCC
patients.
 | Table IVSummary of the results of the IBD
regions identified by individual homozygosity mapping and sharing
of homozygous segments in case/case and control/control pairing
approaches. |
Table IV
Summary of the results of the IBD
regions identified by individual homozygosity mapping and sharing
of homozygous segments in case/case and control/control pairing
approaches.
| Approaches |
|---|
|
|
|---|
| Homozygosity
mapping (50 SNPs, 2 Mb) | Pairing of IBD
segments |
|---|
|
|
|
|---|
| Data set | Total IBD length
(Mb)/no. of IBD segments | Average IBD length
(Mb)a | Average IBD length
(Mb)/person | Frequency of IBD
segments | Total IBD length
(Mb)/no. of IBD segments | Average IBD length
(Mb)a | Average IBD length
(Mb)/personb | Frequency of IBD
segmentsc |
|---|
| CHB HapMap,
n=45 | 378.7/146 | 2.59 | 8.42 | 3.2 | 204.3/117 | 1.75 | 4.54 | 2.6 |
| Henan control,
n=16 | 156.5/56 | 2.79 | 9.78 | 3.5 | 291.1/143 | 2.04 | 18.20 | 8.9 |
| FH+
ESCC, n=28 | 239.4/82 | 2.92 | 8.55 | 2.9 | 542.4/243 | 2.23 | 19.37 | 8.7 |
The only overlapping region, which may predispose an
individual to develop ESCC, identified by both approaches was at
2q31.1-q32.2, in which a region of 21.1 Mb (chr 2:
170,290,090–191,418,198) identified by IBD detected in each
individual alone was further narrowed down to 1.26 Mb containing
one microRNA, MIR561, and two candidate genes Homo
sapiens GULP, engulfment adaptor PTB domain containing 1
(GULP1), and Homo sapiens disrupted in renal
carcinoma 1 (DIRC1) (chr 2: 188,440,696–189,700,120) by
considering pairing information in the latter approach.
GULP1 is an adapter protein necessary for the engulfment of
apoptotic cells by phagocytes. Disruption of DIRC1 by
translocation t(2;3)(q33;q21) is associated with familial clear
cell renal cancer. The gene expression of GULP1 and
DIRC1 in ESCC tumor tissues at 2q32.1-q32.2 was 0.89-fold
and 1.27-fold, respectively, in our unpublished microarray dataset.
Further study of candidate genes in this locus is necessary to
verify their role in ESCC susceptibility. Another potential genetic
susceptibility locus for ESCC at 10q23.33-q24.1 worthy of attention
harbors one of the candidate genes located within the region,
PLCE1 (phospholipase C, epsilon 1). Notably, PLCE1
was recently reported independently to be associated with ESCC in
the high-risk region of Northern China by a genome-wide association
study (GWAS) (30–32). The same locus at 10q23 was
identified independently by three different groups as risk factors
for high ESCC incidence areas in China with a combined large sample
size of 16,499 ESCC and 21,998 controls. It has been hypothesized
that germline IBD regions represent low-penetrance factors for
cancer predisposition (17). The
potential cancer susceptibility loci identified in our data did not
localize to loci of high penetrance cancer susceptibility genes
such as BRCA1 at 17q12, BRCA2 at 13q14, or
TP53 at 17p13-p15, but reside at regions similar to
PLCE1 at 10q23 with low-penetrance susceptibility for cancer
development. Combining the data from shared IBD segments by pairing
analysis and the microarray profiling allowed us to narrow down the
candidate genes for validation. These studies support the
biological relevance of at least four target genes residing within
the IBD segments at 10q23.33-q24.1 and 16q11.2-q12.1 having
significant differential expression in primary ESCC tumors.
PLCE1 is a phospholipase responsible for hydrolysis of
phosphatidyl-inositol 4,5-bisphosphate regulating
1,2-diacylglycerol downstream signaling and an effector of GTPases,
such as, Ras (33).
CYP2-C18, (cytochrome P450, family 2, subfamily C,
polypeptide 18) located within a cluster of cytochrome P450 genes
at 10q24, encodes one of the cytochrome P450 superfamily of
enzymes, which are monooxygenases involved in drug metabolism and
synthesis of cholesterol, steroids and other lipids. SIAH1
(siah E3 ubiquitin protein ligase 1) is an E3 ligase. It may play a
functional role in Parkinson’s disease development, regulation of
the cellular response to hypoxia and apoptosis. GPT2
[glutamic pyruvate transaminase (alanine aminotransferase 2)]
encodes an enzyme for amino acid metabolism and gluconeogenesis.
Importantly, PLCE1, GPT2, SIAH1 and
CYP2-C18, appear to play an important role in ESCC
tumorigenesis. Further in-depth genomic studies on these genes and
others residing in these critical IBD regions are needed to
elucidate their roles in ESCC.
The increased length of germline genomic
homozygosity associated with hereditary ESCC in Henan was observed.
To strategically increase the power of the present study, we
focused on high ESCC incidence region samples with ESCC
FH+ cases and with multiple ESCC cases within one
family. The ancestries of Henan cases and control samples were also
carefully matched to avoid false signals introduced by small
differences in ancestry. However, the results were still limited by
the small sample size. The importance of these IBD segments in the
etiology and development of ESCC in high-risk areas requires
further investigation with an expanded sample size for validation,
and capture region targeted sequencing of the eight potential
cancer susceptibility loci identified in this study is required to
search and validate the importance of the disease-causing genetic
variants responsible for familial ESCC development. Importantly,
using a genetic IBD approach for the study of inherited ESCC, it is
clear that host genetic susceptibility does indeed contribute to
ESCC development in high-risk Henan.
Acknowledgements
We acknowledge the Research Grant Council of Hong
Kong SAR, P.R. China for the funding to M.L.L.
Abbreviations:
|
ARCL
|
autosomal recessive cutis laxa
|
|
CHB
|
Han Chinese in Beijing, China
|
|
CRC
|
colorectal cancer
|
|
CYP2C18
|
cytochrome P450, family 2, subfamily
C, polypeptide 18
|
|
DIRC1
|
Homo sapiens disrupted in renal
carcinoma 1
|
|
EC
|
esophageal cancer
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
FH−
|
family history-negative
|
|
FH+
|
family history-positive
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
GPT2
|
glutamic pyruvate transaminase
(alanine aminotransferase 2)
|
|
GULP1
|
Homo sapiens GULP, engulfment
adaptor PTB domain containing 1
|
|
GWAS
|
genome-wide association study
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
IBD
|
identity-by-descent
|
|
LOH
|
loss of heterozygosity
|
|
MAF
|
minor allele frequency
|
|
NT
|
non-tumor
|
|
PLCE1
|
phospholipase C, epsilon 1
|
|
SIAH1
|
Siah E3 ubiquitin protein ligase 1
|
|
T
|
tumor
|
|
UPL
|
universal probe library
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Tran GD, Sun XD, Abnet CC, et al:
Prospective study of risk factors for esophageal and gastric
cancers in the Linxian general population trial cohort in China.
Int J Cancer. 113:456–463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang W, Bailey-Wilson JE, Li W, et al:
Segregation analysis of esophageal cancer in a moderately
high-incidence area of northern China. Am J Hum Genet. 67:110–119.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiao YL, Hou J, Yang L, et al: The trends
and preventive strategies of esophageal cancer in high-risk areas
of Taihang Mountains, China. Zhongguo Yi Xue Ke Xue Yuan Xue Bao.
23:10–14. 2001.(In Chinese).
|
|
5
|
Xibib S, Meilan H, Moller H, et al: Risk
factors for oesophageal cancer in Linzhou, China: a case-control
study. Asian Pac J Cancer Prev. 4:119–124. 2003.PubMed/NCBI
|
|
6
|
Yokokawa Y, Ohta S, Hou J, et al:
Ecological study on the risks of esophageal cancer in Ci-Xian,
China: the importance of nutritional status and the use of well
water. Int J Cancer. 83:620–624. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang CS: Research on esophageal cancer in
China: a review. Cancer Res. 40:2633–2644. 1980.PubMed/NCBI
|
|
8
|
Hu N, Dawsey SM, Wu M, et al: Familial
aggregation of oesophageal cancer in Yangcheng County, Shanxi
Province, China. Int J Epidemiol. 21:877–882. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang G, Su M, Wang D, et al: Genetic
heterogeneity of oesophageal cancer in high-incidence areas of
Southern and Northern China. PLoS One. 5:e96682010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su H, Hu N, Shih J, et al: Gene expression
analysis of esophageal squamous cell carcinoma reveals consistent
molecular profiles related to a family history of upper
gastrointestinal cancer. Cancer Res. 63:3872–3876. 2003.
|
|
11
|
Hu N, Roth MJ, Polymeropolous M, et al:
Identification of novel regions of allelic loss from a genomewide
scan of esophageal squamous-cell carcinoma in a high-risk Chinese
population. Genes Chromosomes Cancer. 27:217–228. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu N, Wang C, Su H, et al: High frequency
of CDKN2A alterations in esophageal squamous cell carcinoma
from a high-risk Chinese population. Genes Chromosomes Cancer.
39:205–216. 2004.
|
|
13
|
Hu N, Wang C, Ng D, et al: Genomic
characterization of esophageal squamous cell carcinoma from a
high-risk population in China. Cancer Res. 69:5908–5917. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu N, Su H, Li WJ, et al: Allelotyping of
esophageal squamous-cell carcinoma on chromosome 13 defines
deletions related to family history. Genes Chromosomes Cancer.
44:271–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bacolod MD, Schemmann GS, Wang S, et al:
The signatures of autozygosity among patients with colorectal
cancer. Cancer Res. 68:2610–2621. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Broman KW and Weber JL: Long homozygous
chromosomal segments in reference families from the centre d’Etude
du polymorphisme humain. Am J Hum Genet. 65:1493–1500.
1999.PubMed/NCBI
|
|
17
|
Assie G, LaFramboise T, Platzer P and Eng
C: Frequency of germline genomic homozygosity associated with
cancer cases. JAMA. 299:1437–1445. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rudan I, Rudan D, Campbell H, et al:
Inbreeding and risk of late onset complex disease. J Med Genet.
40:925–932. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lebel RR and Gallagher WB: Wisconsin
consanguinity studies. II: Familial adenocarcinomatosis. Am J Med
Genet. 33:1–6. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shami SA, Qaisar R and Bittles AH:
Consanguinity and adult morbidity in Pakistan. Lancet. 338:9541991.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanai N, Fujii T, Saito K and Tokoyama T:
Rapid and simple method for preparation of genomic DNA from easily
obtainable clotted blood. J Clin Pathol. 47:1043–1044. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Purcell S, Neale B, Todd-Brown K, et al:
PLINK: a tool set for whole-genome association and population-based
linkage analyses. Am J Hum Genet. 81:559–575. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Browning SR and Browning BL:
High-resolution detection of identity by descent in unrelated
individuals. Am J Hum Genet. 86:526–539. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chan SH, Yee Ko JM, Chan KW, et al: The
ECM protein LTBP-2 is a suppressor of esophageal squamous cell
carcinoma tumor formation but higher tumor expression associates
with poor patient outcome. Int J Cancer. 129:565–573. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheung AK, Ko JM, Lung HL, et al:
Cysteine-rich intestinal protein 2 (CRIP2) acts as a
repressor of NF-κB-mediated proangiogenic cytokine transcription to
suppress tumorigenesis and angiogenesis. Proc Natl Acad Sci USA.
108:8390–8395. 2011.PubMed/NCBI
|
|
26
|
Cheung AK, Lung HL, Hung SC, et al:
Functional analysis of a cell cycle-associated, tumor-suppressive
gene, protein tyrosine phosphatase receptor type G, in
nasopharyngeal carcinoma. Cancer Res. 68:8137–8145. 2008.
View Article : Google Scholar
|
|
27
|
Rudan I: Inbreeding and cancer incidence
in human isolates. Hum Biol. 71:173–187. 1999.PubMed/NCBI
|
|
28
|
Lander ES and Botstein D: Homozygosity
mapping: a way to map human recessive traits with the DNA of inbred
children. Science. 236:1567–1570. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reversade B, Escande-Beillard N,
Dimopoulou A, et al: Mutations in PYCR1 cause cutis laxa
with progeroid features. Nat Genet. 41:1016–1021. 2009.
|
|
30
|
Wang LD, Zhou FY, Li XM, et al:
Genome-wide association study of esophageal squamous cell carcinoma
in Chinese subjects identifies susceptibility loci at PLCE1
and C20orf54. Nat Genet. 42:759–763. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abnet CC, Freedman ND, Hu N, et al: A
shared susceptibility locus in PLCE1 at 10q23 for gastric
adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet.
42:764–767. 2010.
|
|
32
|
Wu C, Hu Z, He Z, et al: Genome-wide
association study identifies three new susceptibility loci for
esophageal squamous-cell carcinoma in Chinese populations. Nat
Genet. 43:679–684. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bunney TD and Katan M: Phospholipase C
epsilon: linking second messengers and small GTPases. Trends Cell
Biol Biol. 16:640–648. 2006. View Article : Google Scholar : PubMed/NCBI
|