Introduction
Gastric cancer is the fourth most common malignancy
worldwide, with an estimated 738,000 deaths in 2008 (1). In China, gastric cancer is the most
commonly diagnosed carcinoma and the leading cause of
cancer-related mortality. Notably, Gansu Province is one of the
high-risk areas, particularly in Wuwei City where the mortality
rate is nearly 5 times as high as the average level (2). To date, surgical resection is the
primary treatment for gastric cancer (3), but both local and distant recurrence
are high due to delayed treatment because of diagnostic limitations
in early non-invasive detection.
microRNAs (miRNAs) are an abundant class of
non-coding RNAs, ~19–25 nucleotides in length. miRNAs regulate
various processes involved in cell progression such as cellular
division and differentiation (4),
development (5), cell cycle
regulation (6) and apoptosis
(7) by binding to 3′-UTR of the
target gene mRNA, either by inducing mRNA degradation or by
inhibiting mRNA translation. It is widely recognized that miRNAs
play important roles in tumorigenesis acting as potent oncogenes or
tumor-suppressor genes (8,9) and may be novel biomarkers for cancer
diagnosis and prognosis (10).
According to recent experimental evidence, miRNA expression is
dysregulated in gastric cancer (11), and miRNA clusters such as
miR-106b-25 (12) and miR-222-221
(13) take part in cellular growth
or apoptosis. Thus, it is hopeful that miRNAs may become efficient
targets for the diagnosis and prognosis of gastric cancer.
Radiotherapy, one widely used treatment for most
malignant tumors, has become an integral part of gastric cancer
treatment, particularly in the early and intermediate stages of
gastric cancer (14–16). Cells present DNA damage, cell cycle
arrest or apoptosis in response to ionizing radiation (IR)
(17). Recently, it has been
reported that radiation modulates the miRNA expression profile in
human endothelial cells (18),
prostate cancer (19,20), brain cancer (21) and lung cancer (21,22).
Moreover, miRNAs play a critical role in multidrug resistance in
human gastric cancer (23–25). However, no research concerning the
change in expression profile in response to radiation and the roles
of miRNAs in DNA damage responses have been reported in gastric
cancer to date. In the present study, we aimed to identify the
dysregulation of miRNAs responding to IR and their potential
influences on cellular responses in gastric cancer.
Materials and methods
Cell culture
Human gastric cancer cell lines BGC823 and MGC803
were purchased from the Institutes of Biochemistry and Cell
Biology, Chinese Academy of Sciences (Shanghai, China). Cells were
maintained at 37°C in a 5% CO2 atmosphere in RPMI-1640
medium (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum
(Hyclone, Logan, UT, USA) and 100 U/ml penicillin and 100 mg/ml
streptomycin.
Clinical samples
Ten human gastric cancer tissue samples were
obtained from the Department of Oncology, The First People’s
Hospital of Lanzhou City. The corresponding adjacent normal gastric
tissues were also obtained. Human gastric cancer tissue samples
from patients were obtained with informed consent under
institutional review board approved protocols. Samples were
immersed in RNAlater (Ambion, Foster City, CA, USA) 2 h after
surgery and were then stored at −20°C.
Irradiation
Clinical tissue samples were irradiated immediately
after surgical resection at Lanzhou General Hospital, Lanzhou
Command with 4 Gy of X-rays (6 MV, 0.8 Gy/min). Cells were
irradiated with a laboratory X-ray source (RX-650; Faxitron
Bioptics, USA) at a dose rate of 0.8 Gy/min (100 keV, 5 mA). Cells
and tissue samples were split with TRIzol reagent (Invitrogen,
Eugene, OR, USA) 2 h after irradiation.
RNA preparation
Total RNAs of cells and tissue samples were
extracted using TRIzol reagent according to the manufacturer’s
instructions. The quality and quantity of total RNAs were tested by
BioPhotometer Plus (Eppendorf, Hamburg, Germany) and denaturing
agarose gel electrophoresis.
microRNA microarray analysis
Two different clinical tissue samples were used for
microarray analysis. Total RNAs were isolated with TRIzol reagent 2
h after irradiation. Then the miRNAs were labeled with Hy3 using
the miRCURY™ Array Power Labeling kit (Exiqon, Vedbaek, Denmark)
and hybridized respectively on a miRCURY™ LNA microRNA array
(version 10.0, Exiqon) as previously described (26). Microarray images were obtained by
using a Genepix 4000B scanner (Axon Instruments, Union City, CA,
USA) and processed and analyzed with Genepix Pro 6.0 (Axon
Instruments) and Microsoft Excel software (Microsoft Campus,
Redmond, WA, USA).
Quantitative RT-PCR for miRNA
Total RNA was extracted using TRIzol reagent
according to the manufacturer’s protocol. Reverse transcription was
performed using the All-in-One™ First-Strand cDNA Synthesis kit
(GeneCopoeia, Guangzhou, China). To quantify hsa-miR-300 and
hsa-miR-642 expression, real-time PCR was performed using
All-in-One™ miRNA qPCR Detection kit (GeneCopoeia) based on
SYBR-Green according to the manufacturer’s protocol. U6 snRNA was
used to normalize the relative amount of miRNA. The fold-change of
miRNA was calculated using the 2−ΔΔCT method (27).
miRNA target prediction and gene
ontology
Target prediction of candidate miRNAs was carried
out with TargetScanHuman (release 5.1) (http://www.targetscan.org/). The functions of the
miRNA target genes were analyzed on the Gene Ontology website
(http://www.geneontology.org/).
Cell cycle assay
MGC803 or BGC823 cells (1.5×105) were
seeded into 12-well plates. Transfection of miRNA duplexes (Ambion)
including miR-300 precusor (pre-miR-300) and negative control
(pre-neg) was performed 24 h later using Lipofectamine 2000
(Invitrogen). Cells were exposed to 1 Gy of X-rays after 5 h and
the medium was then replaced with fresh culture medium. Cells were
fixed at −20°C and then stained with 50 μg/ml propidium iodide 16 h
after irradiation. A FACSCalibur flow cytometer (Becton Dickinson,
Franklin Lakes, NJ, USA) and FlowJo software (version 6.0, Tree
Star, Ashland, OR, USA) were used to analyze the cell cycle for
each sample.
Immunofluorescence
Immunofluorescence assay was conducted according to
a previous report (28). In brief,
cells were fixed in 4% paraformaldehyde for 10 min and in methanol
at −20°C for 20 min, permeabilized with 0.5% Triton X-100 for 10
min, blocked with 5% skim milk for 2 h and stained with the primary
antibody [rabbit anti-53BP1 antibody purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA) for 2 h. The bound antibody was
visualized using Alexa Fluor® 594 anti-rabbit antibody
(Molecular Probes, Eugene, OR, USA), and cell nuclei were
counterstained with DAPI solution (Invitrogen). Images were
captured with a laser confocal microscope (Carl Zeiss LSM 700;
Jena, Germany). 53BP1 foci were counted using a fluorescence
microscope (Leica DMI6000B; Wetzlar, Germany), and a minimum of 100
cells were scored for each sample.
Statistical analysis
Results are expressed as means ± SD. The statistical
significance of the results was determined by Student’s t-tests
using Microsoft Excel software (Microsoft Campus).
Results
Alteration of the miRNA profile in
gastric cancer exposed to IR
To identify the miRNA expression changed by ionizing
radiation, we exposed two independent clinical gastric cancer
tissues to 4 Gy of X-rays. The miRNA profiles of unirradiated and
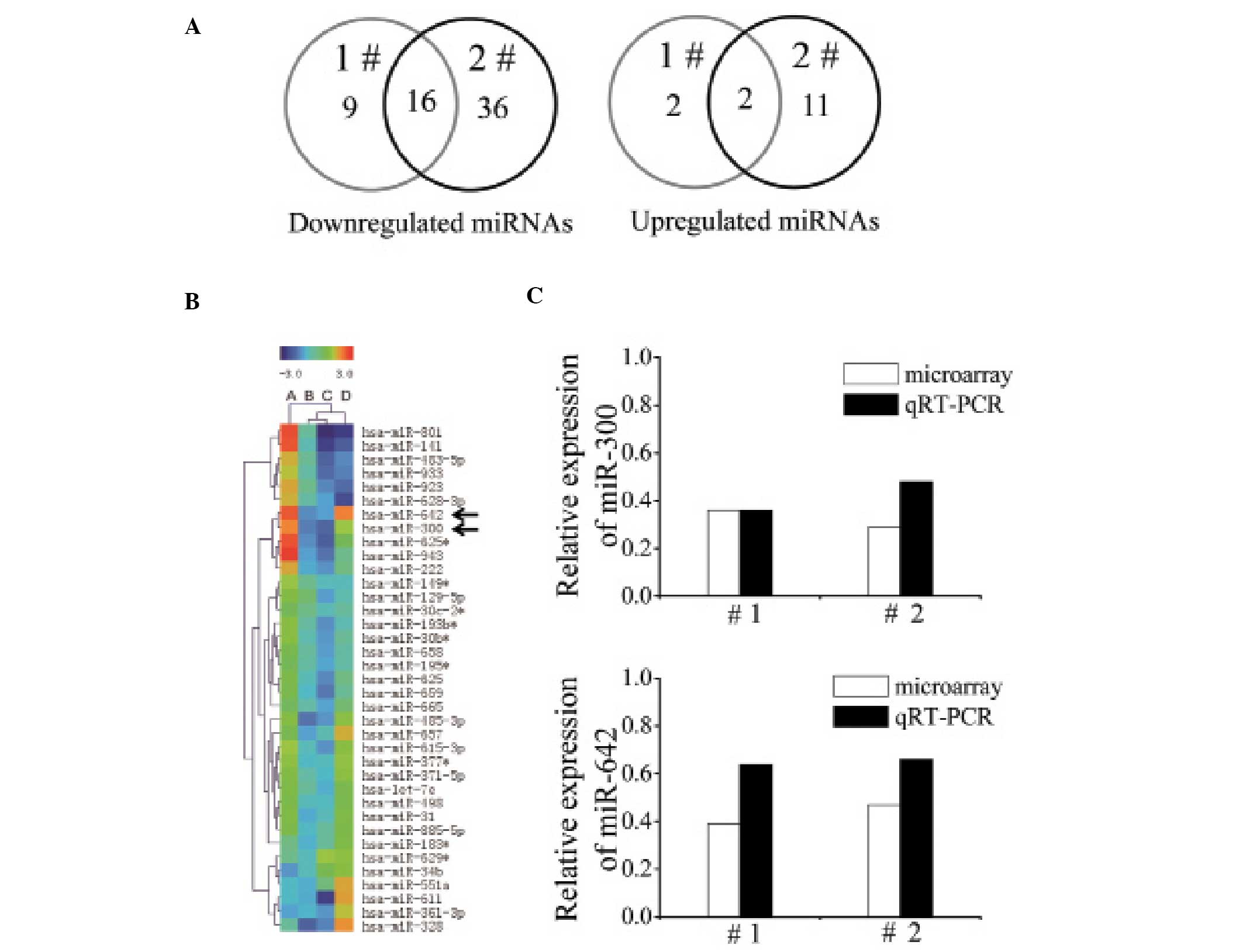
irradiated samples were obtained by miRNA microarray. The Venn
diagram presenting the number of downregulated and upregulated
miRNAs after 4 Gy of X-ray exposure is shown in Fig. 1A. In both irradiated samples, we
identified that expression levels of 18 miRNAs were altered
>1.5-fold, among which 16 miRNAs were downregulated and 2 miRNAs
were upregulated. These miRNAs and the fold change are shown in
Table I. Notably, hierarchical
clustering results showed that miR-300 and miR-642 expression
levels were upregulated in gastric cancer while IR reduced their
expression levels (Fig. 1B). Next,
we validated the decreased expression of miR-300 and miR-642 which
were downregulated >2-fold in the miRNA microarray results using
quantitative RT-PCR (qRT-PCR) in the two gastric cancer samples
exposed to X-rays (Fig. 1C).
 | Table ILevels of miRNA altered more than
1.5-fold following irradiation in two clinical samples. |
Table I
Levels of miRNA altered more than
1.5-fold following irradiation in two clinical samples.
| Fold changea |
|---|
|
|
|---|
| miRNA | # 1b | # 2c |
|---|
| hsa-let-7a | −2.00 | −1.85 |
| hsa-miR-138-1* | 1.58 | 5.17 |
| hsa-miR-141 | −1.52 | −5.26 |
| hsa-miR-24-1* | −1.85 | −2.94 |
| hsa-miR-300 | −2.78 | −3.45 |
| hsa-miR-377* | −1.75 | −1.67 |
| hsa-miR-423-3p | −1.54 | −2.00 |
| hsa-miR-485-3P | −3.03 | −2.33 |
| hsa-miR-490-5p | −1.92 | −1.72 |
| hsa-miR-498 | −1.61 | −1.72 |
| hsa-miR-615-3p | −1.61 | −2.38 |
| hsa-miR-625 | −1.54 | −2.56 |
| hsa-miR-625* | −2.50 | −3.85 |
| hsa-miR-637 | 1.80 | 2.32 |
| hsa-miR-642 | −2.56 | −2.13 |
| hsa-miR-657 | −2.17 | −1.52 |
| hsa-miR-659 | −1.79 | −2.94 |
| hsa-miR-943 | −2.17 | −3.03 |
Verification of the downregulation and
putative target prediction of miR-300 and miR-642
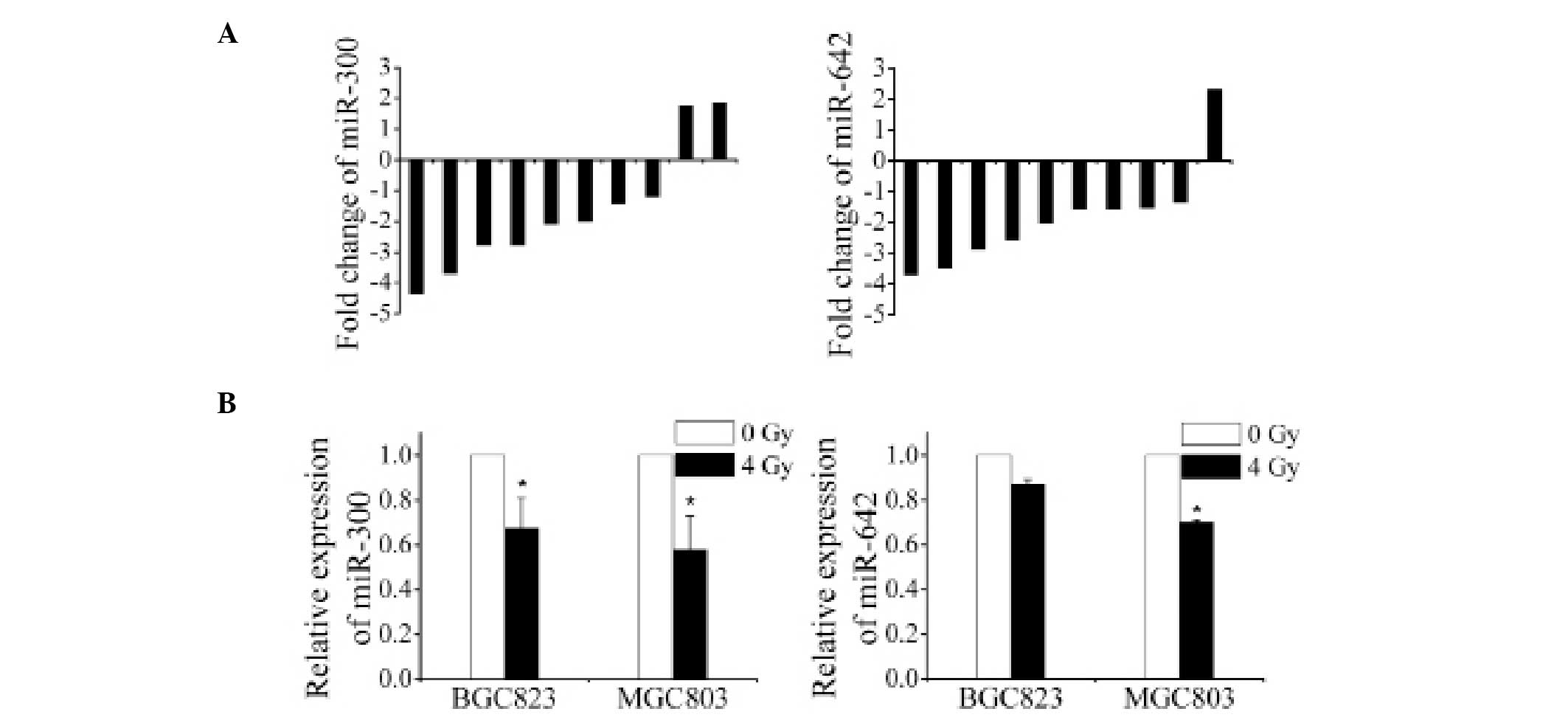
Since miR-300 and miR-642 expression levels were
decreased in the two gastric cancer samples, we verified their
downregulation using additionalclinical gastric cancer tissues
(n=10) and two cultured gastric cancer cell lines (BGC823 and
MGC803) exposed to 4 Gy of X-rays by qRT-PCR method. The results
showed that miR-300 was significantly downregulated in 8 of the 10
clinical gastric cancer tissues and in both cell lines while
miR-642 was downregulated in 9 of the 10 clinical gastric cancer
tissues and in both cell lines (Fig. 2A
and B), implying the general downregulation of miR-300 and
miR-642 in gastric cancer after irradiation.
We employed TargetScanHuman (release 5.1) to predict
the potential target genes of miR-300 and miR-642. Hundreds of
candidate targets were obtained for each miRNA through the 2–8
nucleotides as seed sequence. Then, we classified all the candidate
targets with Gene Ontology (GO). Based on the cellular response to
ionizing radiation damage (17),
the targets related to apoptosis (GO: 0006915), cell cycle
regulation (GO: 0000074) or DNA damage and repair (GO: 0006281) are
summarized in Table II. The
results suggest that both miR-300 and miR-642 may play important
roles in cellular radiation response by modulating related
pathways.
 | Table IIPredicted targets of miR-300 and
miR-642. |
Table II
Predicted targets of miR-300 and
miR-642.
| Function of the
target genes |
|---|
|
|
|---|
| miRNA | Apoptosis (GO:
0006915) | Cell cycle (GO:
0000074) | DNA damage repair
(GO: 0006281) |
|---|
| miR-300 | BCL2L11 | CCNK | ATAD5 |
| GAS2 | KLF4 | TP53BP1 |
| CASP8AP2 | NASP | |
| APAF1 | TP53 | |
| DLC1 | | |
| miR-642 | TP53 | CDK10 | XPA |
| CASPS2 | CHFR | MRE11A |
| CASPS7 | ING4 | PRELD1 |
| CASPS9 | GRB2 | |
| CASPS10 | | |
| BCL2L11 | | |
miR-300 rescues IR-induced G2 cell cycle
arrest and promotes DNA damage repair
To validate the target prediction that miR-300 and
miR-642 may be involved in cell cycle regulation and DNA damage
repair, we tested the cell cycle and the 53BP1 focus kinetics in
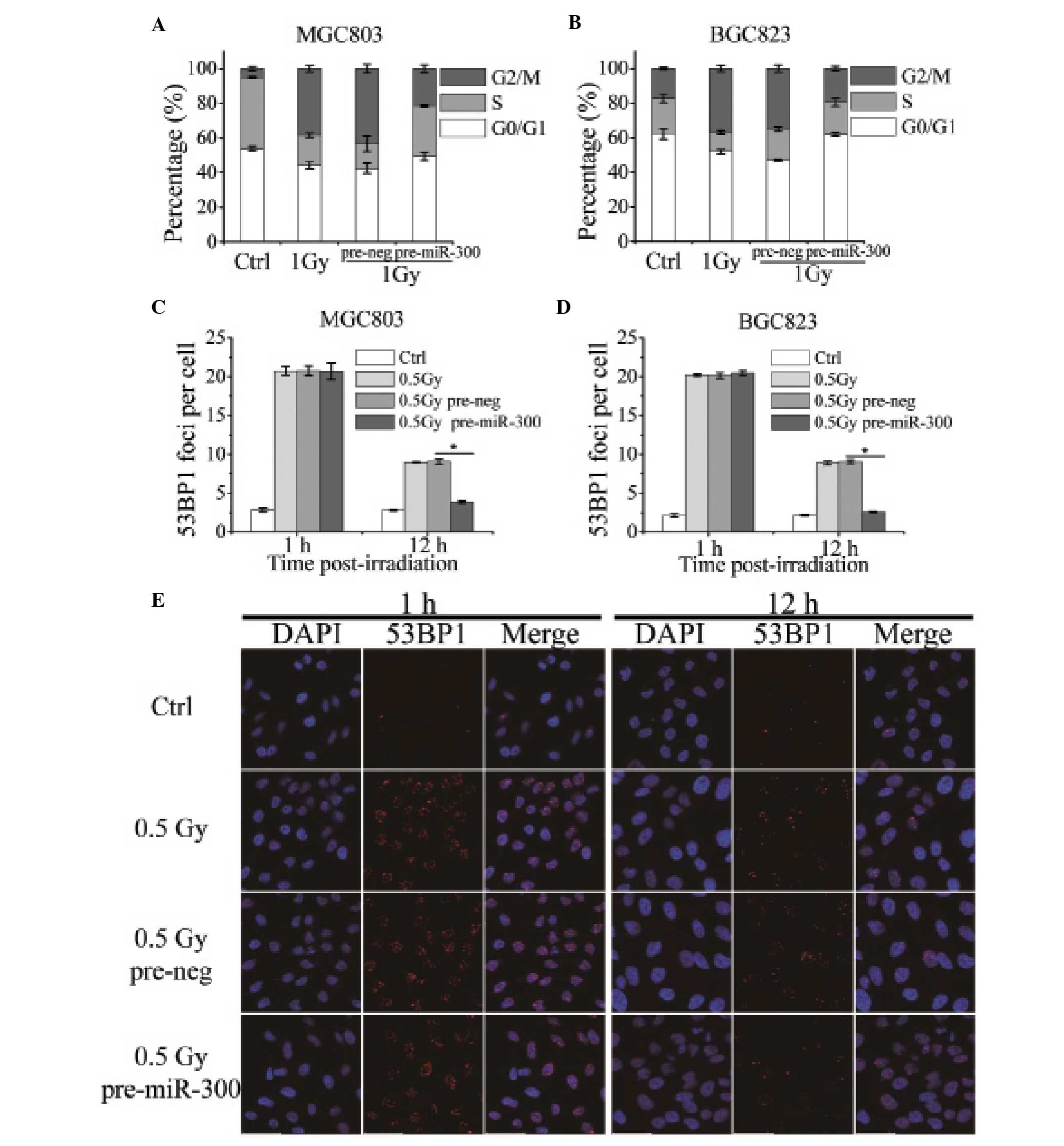
MGC803 and BGC823 cells exposed to X-rays.
Results of the flow cytometry showed that 1 Gy of
X-rays induced marked G2 cell cycle blockage, while cells in the G2
phase were decreased from 43.33±2.45 to 21.48±2.17% (p=0.039) in
the MGC803 cells transfected with pre-miR-300 (Fig. 3A). In addition, the effects were
more evident in the BGC823 cells (p=0.0027); miR-300 overexpression
released G2 cell arrest induced by 1 Gy of X-rays nearly to the
unirradiated level (Fig. 3B). These
results indicate that IR-induced miR-300 suppression impairs the
cellular cell cycle recover.
The formation of 53BP1 foci is widely used as a
marker of DNA double-strand breaks induced by ionizing radiation.
To investigate the effect of miR-300 on DNA damage and repair
ability, we tested 53BP1 focus kinetics in MGC803 and BGC823 cells
transfected with pre-miR-300 or pre-neg. The measurement of 53BP1
foci at 1 h and 12 h post-irradiation with 0.5 Gy of X-rays showed
that no difference was observed 1 h after irradiation between the
two groups. However, 12 h after irradiation the number of 53BP1
foci in the group transfected with pre-miR-300 was significantly
less than that in the MGC803 cells transfected with pre-neg
(Fig. 3C and E). BGC823 cells
demonstrated similar results (Fig.
3D). These data imply that miR-300 does not influence the
cellular response to DNA damage but elevates the cellular DNA
damage repair ability.
Discussion
Radiotherapy is widely used for the treatment of
cancer. Since miRNAs are often located in fragile sites of the
genome (29), it has been suggested
that IR can trigger or disrupt the expression of these fragile
sites consequently resulting in a dysregulated miRNA profile. To
date, there are vast amounts of data concerning miRNA alterations
and their functions in response to ionizing radiation in human
normal or tumor cells (30–34); however, few data are available in
regards to gastric cancer. Moreover, further investigation must be
carried out to clarify whether IR or chemo-induced miRNA
alterations in tumor cells are a product of disruption in
epigenetics or an initiating mutational event.
In the present study, we tested the miRNA profile of
clinical gastric cancer tissues irradiated with X-rays and found
that 18 miRNAs were markedly dysregulated in gastric cancer in
response to IR, among which 16 miRNAs were significantly
downregulated and 2 miRNAs were upregulated. The changes of miRNA
expression profile may provide a new method for the diagnosis and
prognosis of gastric cancer.
An miRNA can modulate the cellular response to
IR-induced DNA damage through inhibition of its target genes. Lal
et al showed that overexpression of miR-24 enhanced
radiosensitivity by targeting H2AX (35). miR-421 (36) and miR-101 (37) were found to efficiently target ATM
and sensitize tumor cells to IR. Wang et al found that IR
downregulated miR-185 expression, while elevation of miR-185
sensitized renal carcinoma cells to IR and enhanced
radiation-induced apoptosis and proliferation inhibition by
repressing ATR (38). miR-182
directly downregulates BRCA1 expression and impairs homologous
recombination-mediated repair and renders cells hypersensitive to
IR (39). On the other hand, DNA
damage response factors, ATM (40)
and BRCA1 (41) can regulate miRNA
biogenesis via KSRP or DROSHA microprocessor complex, which are key
players of miRNA biogenesis, suggesting that miRNA expression may
be regulated by IR indirectly. In gastric cancer, it has been
reported that inhibition of miR-221/222 enhanced radiosensitivity
by targeting PTEN (42). We further
identified that miR-300, one of the downregulated miRNAs post
irradiation, can affect the cellular responses to radiation by
relieving IR-induced G2 cell cycle arrest and promoting the
cellular DNA damage repair ability.
The abundance of miR-300 and miR-642 is not high in
various human tissues and cell lines (43,44) so
that little has been reported concerning their expression patterns
and functions until recently. Recently, studies indicate that
miR-300 and miR-642 are involved in the cisplatin-based
chemo-response in cancer cells. Kumar et al identified that
miR-300 and miR-642 were upregulated in a human cisplatin-resistant
ovarian cancer cell line and that miR-300 may be an important
regulator for ovarian cancer chemotherapy by targeting TGF-β and
apoptotic pathways (45). In
addition, Nordentoft et al found that downregulation of
miR-642 increased cisplatin sensitivity in human bladder cancer
cell lines (46). Our data
demonstrated that expression of miR-300 and miR-642 was upregulated
in gastric cancer compared with normal gastric tissues and both
miRNAs were downregulated after exposure to X-rays. Although
increased expression of miR-300 and miR-642 in gastric cancer
suggests their oncogenicity and decreased expression in response to
chemotherapy or radiation exposure provides evidence for cancer
therapy, the direct target genes and regulation pathways of miR-300
and miR-642 need to be further investigated. In conclusion, our
study provides evidence that miRNAs are involved in the regulation
of radiation responses in gastric cancer and changes in expression
of specific miRNAs such as miR-300 have the potential to be used in
the treatment, diagnosis and prognosis of gastric cancer.
Acknowledgements
The authors are grateful to Dr Sha Li of the Lanzhou
General Hospital, Lanzhou Command, Lanzhou, China, for her
assistance with clinical sample radiation; Hailong Pei, Qingxiang
Gao and Liang Peng for assistance with flow cytometry; and Xurui
Zhang for assistance with confocal microscopy. This study was
supported by grants from the National Natural Science Foundation of
China (nos. 31270895, 81272454 and U1232125).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006.
|
|
3
|
Zhu X and Li J: Gastric carcinoma in
China: current status and future perspectives (Review). Oncol Lett.
1:407–412. 2010.PubMed/NCBI
|
|
4
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
9
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: the implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu WK, Lee CW, Cho CH, et al: MicroRNA
dysregulation in gastric cancer: a new player enters the game.
Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Petrocca F, Visone R, Onelli MR, et al:
E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest
and apoptosis in gastric cancer. Cancer Cell. 13:272–286. 2008.
|
|
13
|
Kim YK, Yu J, Han TS, et al: Functional
links between clustered microRNAs: suppression of cell-cycle
inhibitors by microRNA clusters in gastric cancer. Nucleic Acids
Res. 37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hazard L, O’Connor J and Scaife C: Role of
radiation therapy in gastric adenocarcinoma. World J Gastroenterol.
12:1511–1520. 2006.PubMed/NCBI
|
|
15
|
McCloskey SA and Yang GY: Benefits and
challenges of radiation therapy in gastric cancer: techniques for
improving outcomes. Gastrointest Cancer Res. 3:15–19.
2009.PubMed/NCBI
|
|
16
|
Buergy D, Lohr F, Baack T, et al:
Radiotherapy for tumors of the stomach and gastroesophageal
junction - a review of its role in multimodal therapy. Radiat
Oncol. 7:1922012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li L, Story M and Legerski RJ: Cellular
responses to ionizing radiation damage. Int J Radiat Oncol Biol
Phys. 49:1157–1162. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wagner-Ecker M, Schwager C, Wirkner U,
Abdollahi A and Huber PE: MicroRNA expression after ionizing
radiation in human endothelial cells. Radiat Oncol. 5:252010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Josson S, Sung SY, Lao K, Chung LW and
Johnstone PA: Radiation modulation of microRNA in prostate cancer
cell lines. Prostate. 68:1599–1606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
O’Kelly F, Marignol L, Meunier A, Lynch
TH, Perry AS and Hollywood D: MicroRNAs as putative mediators of
treatment response in prostate cancer. Nat Rev Urol. 9:397–407.
2012.PubMed/NCBI
|
|
21
|
Niemoeller OM, Niyazi M, Corradini S, et
al: MicroRNA expression profiles in human cancer cells after
ionizing radiation. Radiat Oncol. 6:292011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shin S, Cha HJ, Lee EM, et al: Alteration
of miRNA profiles by ionizing radiation in A549 human non-small
cell lung cancer cells. Int J Oncol. 35:81–86. 2009.PubMed/NCBI
|
|
23
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu W, Xu H, Zhu D, et al: miR-200bc/429
cluster modulates multidrug resistance of human cancer cell lines
by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 69:723–731.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Li T, Zhang B, et al:
MicroRNA-19a/b regulates multidrug resistance in human gastric
cancer cells by targeting PTEN. Biochem Biophys Res Commun.
434:688–694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Castoldi M, Schmidt S, Benes V, et al: A
sensitive array for microRNA expression profiling (miChip) based on
locked nucleic acids (LNA). RNA. 12:913–920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔC(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He J, Li J, Ye C, et al: Cell cycle
suspension: a novel process lurking in G2 arrest. Cell
Cycle. 10:1468–1476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Simone NL, Soule BP, Ly D, et al: Ionizing
radiation-induced oxidative stress alters miRNA expression. PLoS
One. 4:e63772009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kraemer A, Anastasov N, Angermeier M,
Winkler K, Atkinson MJ and Moertl S: MicroRNA-mediated processes
are essential for the cellular radiation response. Radiat Res.
176:575–586. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chaudhry MA, Sachdeva H and Omaruddin RA:
Radiation-induced micro-RNA modulation in glioblastoma cells
differing in DNA-repair pathways. DNA Cell Biol. 29:553–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwon JE, Kim BY, Kwak SY, Bae IH and Han
YH: Ionizing radiation-inducible microRNA miR-193a-3p induces
apoptosis by directly targeting Mcl-1. Apoptosis. 18:896–909. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao L, Lu X and Cao Y: MicroRNA and
signal transduction pathways in tumor radiation response. Cell
Signal. 25:1625–1634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lal A, Pan Y, Navarro F, et al:
miR-24-mediated downregulation of H2AX suppresses DNA repair in
terminally differentiated blood cells. Nat Struct Mol Biol.
16:492–498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu H, Du L, Nagabayashi G, Seeger RC and
Gatti RA: ATM is downregulated by N-Myc-regulated microRNA-421.
Proc Natl Acad Sci USA. 107:1506–1511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan D, Ng WL, Zhang X, et al: Targeting
DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS
One. 5:e113972010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, He J, Su F, et al: Repression of
ATR pathway by miR-185 enhances radiation-induced apoptosis and
proliferation inhibition. Cell Death Dis. 4:e6992013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Moskwa P, Buffa FM, Pan Y, et al:
miR-182-mediated downregulation of BRCA1 impacts DNA repair and
sensitivity to PARP inhibitors. Mol Cell. 41:210–220. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang X, Wan G, Berger FG, He X and Lu X:
The ATM kinase induces microRNA biogenesis in the DNA damage
response. Mol Cell. 41:371–383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawai S and Amano A: BRCA1 regulates
microRNA biogenesis via the DROSHA microprocessor complex. J Cell
Biol. 197:201–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang C-Z, Lei H, Zhang A-L, et al:
MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Landgraf P, Rusu M, Sheridan R, et al: A
mammalian microRNA expression atlas based on small RNA library
sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008.PubMed/NCBI
|
|
45
|
Kumar S, Kumar A, Shah PP, Rai SN,
Panguluri SK and Kakar SS: MicroRNA signature of cisplatin
resistant vs. cisplatin sensitive ovarian cancer cell lines. J
Ovarian Res. 4:172011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nordentoft I, Birkenkamp-Demtroder K,
Agerbaek M, et al: miRNAs associated with chemo-sensitivity in cell
lines and in advanced bladder cancer. BMC Med Genomics. 5:402012.
View Article : Google Scholar : PubMed/NCBI
|