Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer worldwide and the third most common cause of
cancer-related mortality, with >600,000 deaths/year (1). The heavy burden of HCC has generated
extensive studies of molecular mechanisms underlying this disease
in the hope of elucidating its development and determining better
methods for treatment.
MicroRNAs (miRNAs) are evolutionarily conserved,
small non-coding RNAs that are considered to play fundamental roles
in various biological processes through regulation of gene
expression at the level of post-transcription. Several studies have
reported some important aberrant miRNA expression profiles in HCC
and found multiple specific dysregulated miRNAs associated with the
development of liver cancer through regulating oncogenes or tumor
suppressors (2–8). Transcription factors (TFs) are
essential for the regulation of gene expression by binding to
specific DNA sequences on the promoter of target genes. Many
studies have proved that TFs are key regulators in HCC, in
particular, the protooncogenes c-Myc, c-fos and c-jun are
frequently overexpressed in HCC and play critical roles in
modulating cellular growth, differentiation and apoptosis by
regulating a number of genes (9–12).
Both the TFs and miRNAs are key regulators of gene expression, and
they may mutually regulate each other to form feedback loops, or
they regulate the same target gene to form a feed-forward loop
(FFL) (13). For example, the
cyclin D1 and miRNA-17/20 feedback loop in breast cancer and
TP53/miR-106b/E2F FFL in cell proliferation have been
experimentally verified (14,15).
However, the combined regulation of miRNAs and TFs in HCC remains
elusive.
Previously, we identified a specific aberrant miRNA
expression profiling in HCC by comparison of miRNA expression
profiles in cancerous hepatocytes with normal primary human
hepatocytes and found that miR-221 was the most overexpressed miRNA
in HCC (16). Several other studies
also demonstrated that miR-221 was one of the most upregulated
miRNAs in liver cancer cell lines and tissues (6,8,17).
Consistently, upregulation of miR-221 in glioblastoma, lung, liver,
stomach, colon, pancreatic, kidney, bladder, prostate and thyroid
cancer strengthened its importance in tumorigenesis (8,17–24).
Recently, Callegari et al developed a liver-specific
overexpression of miR-221 transgenic mouse model and found that
these miR-221 transgenic animals exhibited a strong predisposition
to the development of liver tumors (25). miR-221 can promote cell
proliferation and increase the cell number in S-phase by
suppressing p27 (18), p57
(26) and PTEN (17); inhibit cell apoptosis by modulating
BMF (27); accelerate angiogenesis
by regulating TIMP3 (17); and
regulate DNA damage and repair by targeting DDIT4 (8). These studies strongly suggested an
important role of miR-221 upregulation in hepatocarcinogenesis;
however, little is known regarding its upstream regulatory
mechanisms and there is a lack of a comprehensive understanding of
miR-221 in the HCC signaling pathway.
In the present study, bioinformatics analysis was
used to reveal dysregulated miRNAs in modulating the signaling
network of HCC. We focused on upregulated miR-221 in HCC, and
studied the functions of miR-221 silencing in liver cancer in
vitro and in vivo. In particular, the therapeutic value
of stable miR-221 silencing by lentivirus-mediated-anti-miR-221 was
evaluated in nude mice bearing hepatoma xenografts.
Materials and methods
Bioinformatics analysis
We previously identified a specific miRNA expression
profiling in liver cancer and the microarray data were deposited in
NCBI’s Gene Expression Omnibus (GEO) public database (http://www.ncbi.nlm.nih.gov/geo/, GEO accession
no. GSE20077) (16). Here, the most
differentially expressed miRNAs were used for further
bioinformatics analysis. We obtained the candidate miRNA targets
using the combinatorial utilization of two different databases
[TargetScan (28) and miRanda
(29)] as our previous study
(13) and chose the overlapped
predicted targets based on evolutionary conservation among
mammalian. Then, we merged the overlap targets with the
experimentally reported miRNA targets from TarBase and miR2Disease
resource (30,31). Finally, we chose the candidate genes
involved in HCC from the predicted targets as our interested miRNA
targets. To characterize the FFLs among miRNA, TF and HCC genes, we
parsed the ENCODE ChIP-Seq data from the UCSC genome browser
database (http://genome.ucsc.edu/) to obtained the
TF targets (32). For those >100
TFs in ENCODE data, we identified 29 TFs, which were reported to be
related with HCC through literature review. Using the strategy we
previously worked, we obtained the FFLs among miRNA, TF and HCC
genes (13). The miRNA regulatory
network was constructed by merging the FFLs and miRNA target pairs
through our inner Perl scripts. The presented network images were
drawn using the Cytoscape software (version 2.6.1) (33).
Cell lines, miR-221 precursor/inhibitor
and cell transfection
The hepatoma cell lines SK-HEP-1, HepG2, SMMC-7721
and cervical cancer cell line HeLa were cultured and maintained as
previously described (16).
Stability-enhanced miR-221 precursor (pre-miR-221), miR-221
inhibitor (anti-miR-221) and their matched negative controls
(pre-miR-NC and anti-miR-NC) were from Ambion (Austin, TX, USA).
Cell transfections were performed using Lipofectamine™ 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions.
RNA and protein extraction
The mirVana™ PARIS™ kit (Ambion) was applied to
isolate total RNA and protein from the same experimental samples
according to the manufacturer’s instructions.
TaqMan qRT-PCR
The expression of mature miRNAs was assayed using
the TaqMan MicroRNA Assays specific for hsa-miR-221 and RUN6B (both
from Applied Biosystems, Foster City, CA, USA) according to the
manufacturer’s instructions.
Cell proliferation, cell cycle/apoptosis,
cell migration/invasion and clonogenicity assay
These assays were detected as we previously
described (16).
SYBR-Green qRT-PCR
SK-HEP-1 cells were harvested for RNA extraction 48
h after transfection. The First Strand cDNA Synthesis kit
(Fermentas, Burlington, VT, USA) was applied to synthesize the
first strand cDNAs from 1 μg of total tested RNA. A 20 μl PCR
reaction/well in 384-well reaction plates was prepared by using the
Platinum® SYBR-Green qPCR SuperMix UDG reagent
(Invitrogen). The PCR reaction was carried out on Applied
Biosystems 7900HT Fast Real-Time PCR System. GAPDH RNA was used as
a control. The primer sequences are listed in Table I.
 | Table IPrimers used in SYBR-Green
qRT-PCR. |
Table I
Primers used in SYBR-Green
qRT-PCR.
| Gene name | Primers |
|---|
| BMF | F:
5′-CCAGCCTCCCAGCTAAAG-3′ |
| BMF | R:
5′-CCTGGGGATGAACAAAATG-3′ |
| BBC3 | F:
5′-TGGGACTCCTGCCCTTAC-3′ |
| BBC3 | R:
5′-GGCTGGGAGTCCAGTATG-3′ |
| ANGPTL2 | F:
5′-GTTTGGTACTGTCCATGTCTG-3′ |
| ANGPTL2 | R:
5′-GCAGATTCGTGTCATTACAAG-3′ |
Luciferase assay
To verify the targets of miR-221, the pMIR-REPORT™
system (Applied Biosystems) was applied. Briefly, 55-mer
double-stranded oligonucleotides containing the predicted miR-221
binding sites were synthesized with the single-stranded overhangs
of the restriction sites SpeI and HindIII, and
inserted into the multiple cloning site of the pMIR-REPORT™
luciferase vector to establish the pLUC-targets i.e. pLUC-BMF,
pLUC-BBC3 and pLUC-ANGPTL2 vectors. As a control, the
pLUC-muttargets, i.e. pLUC-mutBMF, pLUC-mutBBC3, pLUC-mutANGPTL2
plasmids, were also prepared by the same method, except for the
synthesized oligonucleotides containing corresponding mutated
nucleotides in the seed-match sequence. The mutated scrambling
sequences were prepared by the online tool on the web site
https://www.genscript.com/ssl-bin/app/scramble.
Synthesized sequences are listed in Table II. In 96-well plates, SK-HEP-1
cells were cotransfected with 0.1 μg of each pLUC-target or
pLUC-muttarget, 0.01 μg of pMIR-REPORT™ β-gal plasmid served as an
internal transfection efficiency control, and pre-miR-221 or
pre-miR-NC with a 50 nM final concentration. At 24 h
post-transfection, luciferase and β-galactosidase activities were
measured using the Dual-Light Assay System (Applied Biosystems)
according to the manufacturer’s instructions.
 | Table IIThe sequences of 55-mer
double-stranded oligonucleotides containing the predicted miRNA
binding sites. |
Table II
The sequences of 55-mer
double-stranded oligonucleotides containing the predicted miRNA
binding sites.
| Targeted gene | Sequence |
|---|
| BMF | F:
5′-CTAGTCAGGGGCTATCGAGGAGACCCAGTGAGAATGTAGCATTTTGTTCATCCCA-3′
R:
5′-AGCTTGGGATGAACAAAATGCTACATTCTCACTGGGTCTCCTCGATAGCCCCTGA-3′ |
| Mutated | BMF F:
5′-CTAGTCAGGGGCTATCGAGGAGACCCAGTGAGATGTATCGATTTTGTTCATCCCA-3′
R:
5′-AGCTTGGGATGAACAAAATCGATACATCTCACTGGGTCTCCTCGATAGCCCCTGA-3′ |
| BBC3 | F:
5′-CTAGTGGCCAGCGCGGGGGACTTTCTCTGCACCATGTAGCATACTGGACTCCCAA-3′
R:
5′-AGCTTTGGGAGTCCAGTATGCTACATGGTGCAGAGAAAGTCCCCCGCGCTGGCCA-3′ |
| Mutated | BBC3 F:
5′-CTAGTGGCCAGCGCGGGGGACTTTCTCTGCACCTGTATCGATACTGGACTCCCAA-3′
R:
5′-AGCTTTGGGAGTCCAGTATCGATACAGGTGCAGAGAAAGTCCCCCGCGCTGGCCA-3′ |
| ANGPTL2 | F:
5′-CTAGTTACCTCAGCATTTCTCACAAAGTGTACCATGTAGCATGTT
TTGTGTATAA-3′
R:
5′-AGCTTTATACACAAAACATGCTACATGGTACACTTTGTGAGAAATGCTGAGGTAA-3′ |
| Mutated | ANGPTL2 F:
5′-CTAGTTACCTCAGCATTTCTCACAAAGTGTACCTGTATCGATGTTTTGTGTATAA-3′
R:
5′-AGCTTTATACACAAAACATCGATACAGGTACACTTTGTGAGAAATGCTGAGGTAA-3′ |
Western blot analysis
SK-HEP-1 cells were harvested for protein extraction
48 h after transfection. Cell protein lysates were used for western
blot analysis as previously described (16). The following antibodies were used:
anti-BMF (ab9655), anti-BBC3 (ab9643), anti-ANGPTL2 (ab35574,) (all
from Abcam), anti-β-actin (sc-47778), goat anti-mouse IgG-HRP
(sc-2005) and goat anti-rabbit IgG-HRP (sc-2004) (all from Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Construction of miR-221 silencing
lentivirus
In order to elucidate the role of miR-221 in
vivo, we constructed a recombinant lentivirus termed
miR-221(D)-LV to generate stable loss-of-function of miR-221 in
hepatoma cells. We first prepared a recombinant lentivirus vector
named as pmiR-221(D)-LV as previously described (34). Briefly, two oligonucleotides
(forward sequence, 5′-CCGGCGAAACCCAGCAGACAAT
GTAGCTTTTTTTTGGAAG-3′ and reverse
sequence, 5′-AATTCTTCCAAAAAAAAGCTACATTGTCTGCTGGG TTTC-3′)
were chemically synthesized and annealed to form a double-stranded
nucleotide with the overhangs (Italic letters in the sequences) of
AgeI and EcoRI in 5′ and 3′ flanking ends,
respectively. The bold letters in the forward sequence refer to the
complementary sequence to miR-221. This double-stranded nucleotide
was directionally cloned into the AgeI/EcoRI-digested
lentivirus vector pGCSIL-GFP (GeneChem Co., Ltd., Shanghai, China)
to form the recombinant lentivirus vector pmiR-221(D)-LV.
pGCSIL-GFP has a polymerase III promoter U6 which can promote the
transcription of small non-coding RNAs. The underlined polyT in the
3′ flanking region of forward sequence is the transcription
termination signal for promoter U6. A control recombinant
lentivirus vector, pNC-GFP-LV, containing a scrambling sequence was
also constructed and used as an internal control. All recombinant
vectors were verified by sequencing. To obtain high titer
recombinant lentivirus, a lentivirus package system (GeneChem Co.,
Ltd.) was applied, containing three vectors: i) our recombinant
lentivirus vector [pmiR-221(D)-LV or pNC-GFP-LV], ii) pHelper1.0,
and iii) pHelper2.0. pHelper1.0 contains gag gene and pol gene of
human immunodeficiency virus (HIV), coding the major structural
protein Gag and the virus-specific enzyme Pol of HIV, respectively.
pHelper 2.0 contains VSV-G gene of herpes simplex virus (HSV),
coding envelope protein needed by virus package. These three
vectors were cotransfected into the packaging cell line 293T to
produce VSV.G-pseudo-typed lentiviral particles. At 8-h
post-transfection, the culture media was replaced by complete
media. The supernatant of transfected 293T was collected at 48 h
post-transfection, and was further concentrated into high titer
lentivirus as previously described (35). The titer of lentivirus was evaluated
by limiting dilution assay (36).
Animal experiments
BALB/c athymic nude mice (male, 4–6 weeks old, 16–20
g) were purchased from Hubei Research Center of Laboratory Animal
(Wuhan, China) and bred in pathogen-free conditions in the Animal
Centre of Tongji Medical College. All animals received humane care
and all animal experiments were carried out in accordance with the
Guide for the Care and Use of Laboratory Animals of Tongji Medical
College (Permit no. 130321u). SK-HEP-1 cells were incubated with
lentivirus stocks diluted at an MOI of 50 in Opti-MEM supplemented
with 5 μg/ml polybrene for 10 h. Transduction efficiency was
examined by fluorescence microscopy 72 h later. SK-HEP-1 cells
(2×106) infected with miR-221(D)-LV or NC-GFP-LV were
suspended in 100 μl DMEM and injected subcutaneously into the back
of mice. Tumor growth was examined every 4 days for >1 month. To
establish hepatoma xenograft model, 2×106 SK-HEP-1 cells
were inoculated subcutaneously on the right flank of nude mice.
After 8 days, the transplanted nude mice were randomly divided into
2 groups (n=6 each). TU (2×108) miR-221(D)-LV or
NC-GFP-LV/animal was administered a single intratumor injection.
Tumor volume (V) was monitored by measuring the length
(L) and width (W) with vernier caliper and calculated
with the formula V = (LxW2)
× 0.5.
Immunohistochemistry
Resected tumor tissues were fixed in 4%
paraformaldehyde, embedded in paraffin, cut into 4 μm pieces and
mounted on polylysine-coated slides. Haematoxylin and eosin
(H&E) and Ki67 immunohistochemistry assay were detected as
previously described (16).
Statistical analysis
All data are presented as means ± standard error
from 3 separate experiments performed in triplicate except
otherwise noted. The differences between groups were analyzed by
Student’s t-test and P<0.05 was considered to indicate a
statistically significant result.
Results
miR-221 plays a central role in HCC
regulatory network
To reveal the regulation of dysregulated miRNAs
involved in HCC, bioinformatics analysis was used to predict
regulatory targets and TFs-related to hepatocarcinogenesis and
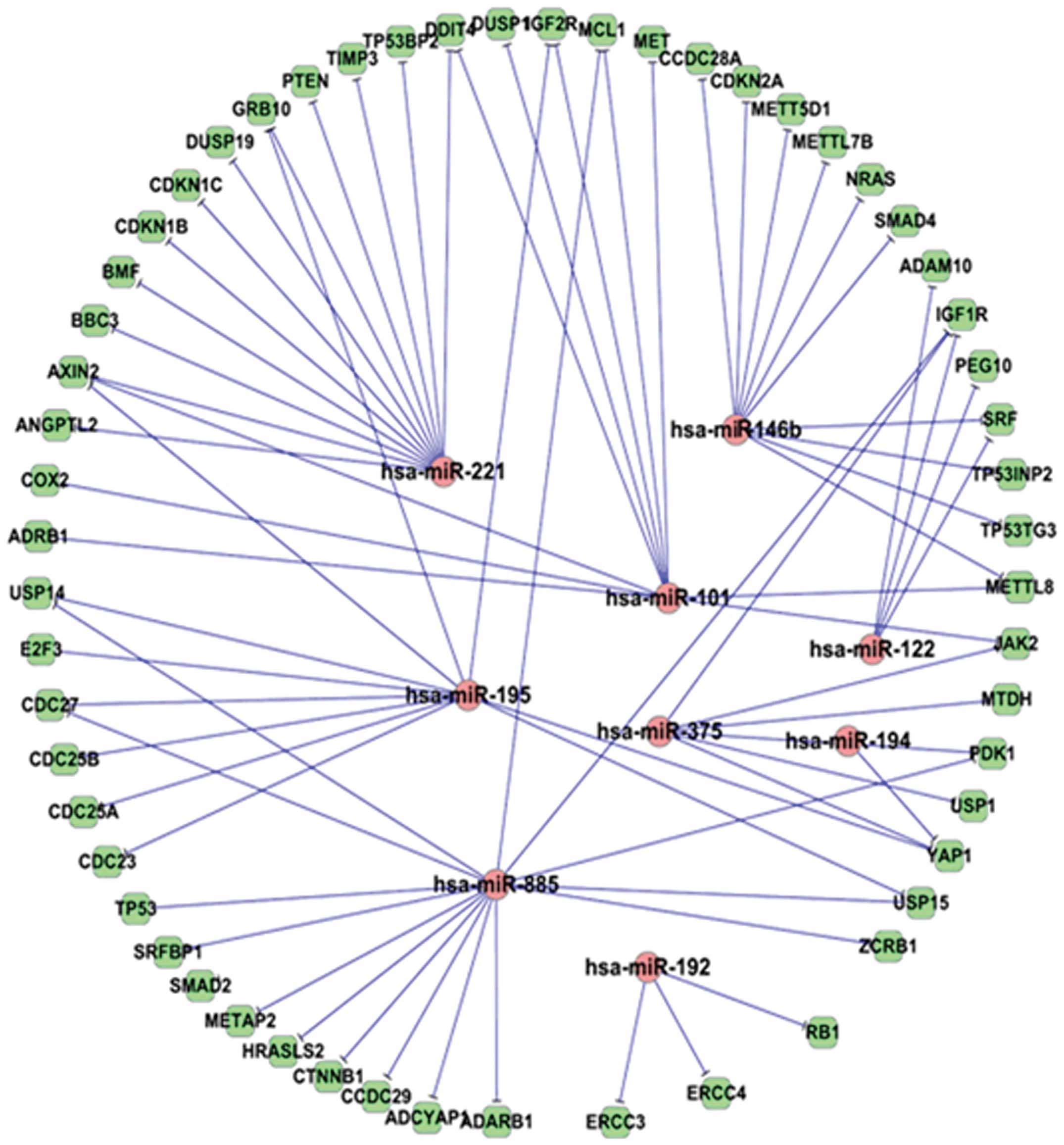
construct regulatory networks for dysregulated miRNAs. As shown in
Fig. 1, the 9 mostly differentially
expressed miRNAs in our miRNA microarray assay (16) were predicted to target 56
HCC-related genes with 73 edges in the HCC miRNA target network. In
this network, miR-221 is one of the miRNAs having the most targets
of HCC-related genes. Furthermore, since TF is another type of key
regulator of gene expression, we also investigated the TFs involved
in the HCC regulatory network. Using the ENCODE ChIP-Seq data in
UCSC genome browser, we obtained the TF targets in the HCC genes
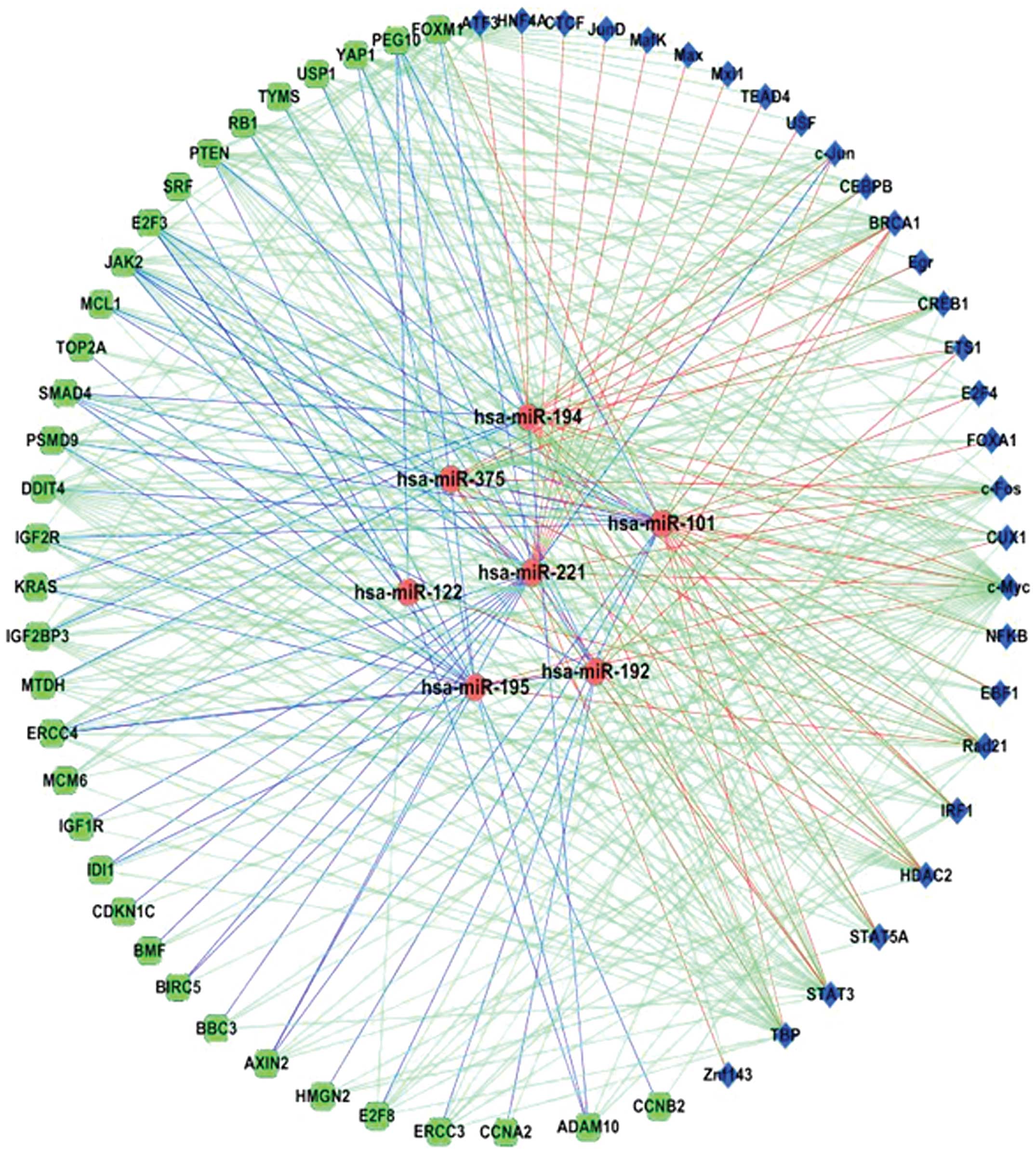
and miRNAs. Choosing those TFs involved in HCC and using the same
strategy as described in our previous study (13), we constructed the co-regulatory
network among the HCC-related TFs, genes and miRNAs (Fig. 2). Fig.
2 shows that there are many FFLs among TFs, miRNAs and HCC
genes. In an FFL, the TF regulates miRNA and HCC gene, while the
miRNA also regulates the same HCC gene. This network contains 332
regulatory relations (edges) among 70 molecules (nodes), including
7 miRNAs, 29 TFs and 34 HCC-related genes. As shown in this
network, miR-221 is a core miRNA regulating many HCC genes and
being regulated by many TFs, while c-Myc, c-Jun and STAT are core
TFs. Notably, FOXM1 is an HCC-related gene and also a TF. Thus, in
the network, it regulates other HCC-related genes or miRNAs and
also as a target of other TFs or miRNAs. Since miR-221 is the most
highly expressed miRNA in our microarray results and it is also a
core miRNA in our regulatory networks, we then focused on miR-221.
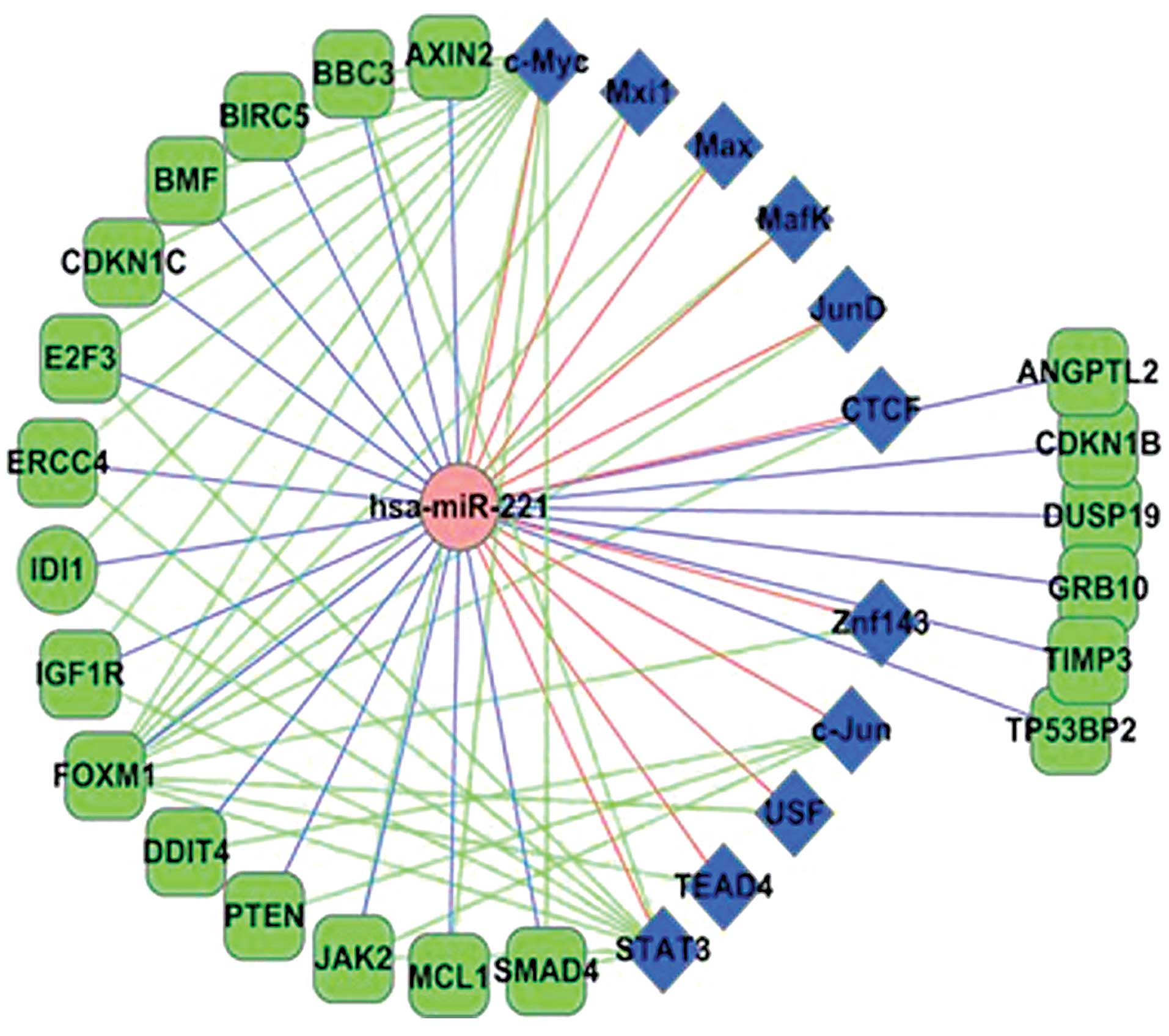
Extracting miR-221-related targets and TFs in Fig. 2 co-regulatory network and adding
some other known miR-221 targets, we obtained the miR-221
regulatory network (Fig. 3). As
shown in Fig. 3, there are many FFL
regulations particularly among the c-Myc, miR-221 and HCC-related
genes. These may indicate that c-Myc plays an important role in the
miR-221 functional pathway in HCC development. These analyses
indicate that miR-221 is a critical modulator of HCC and could be
considered as a potential therapeutic target.
miR-221 silencing inhibits tumorigenic
properties of liver cancer cells in vitro
To assess the role of miR-221 in
hepatocarcinogenesis, we used gain- and loss-of-function methods.
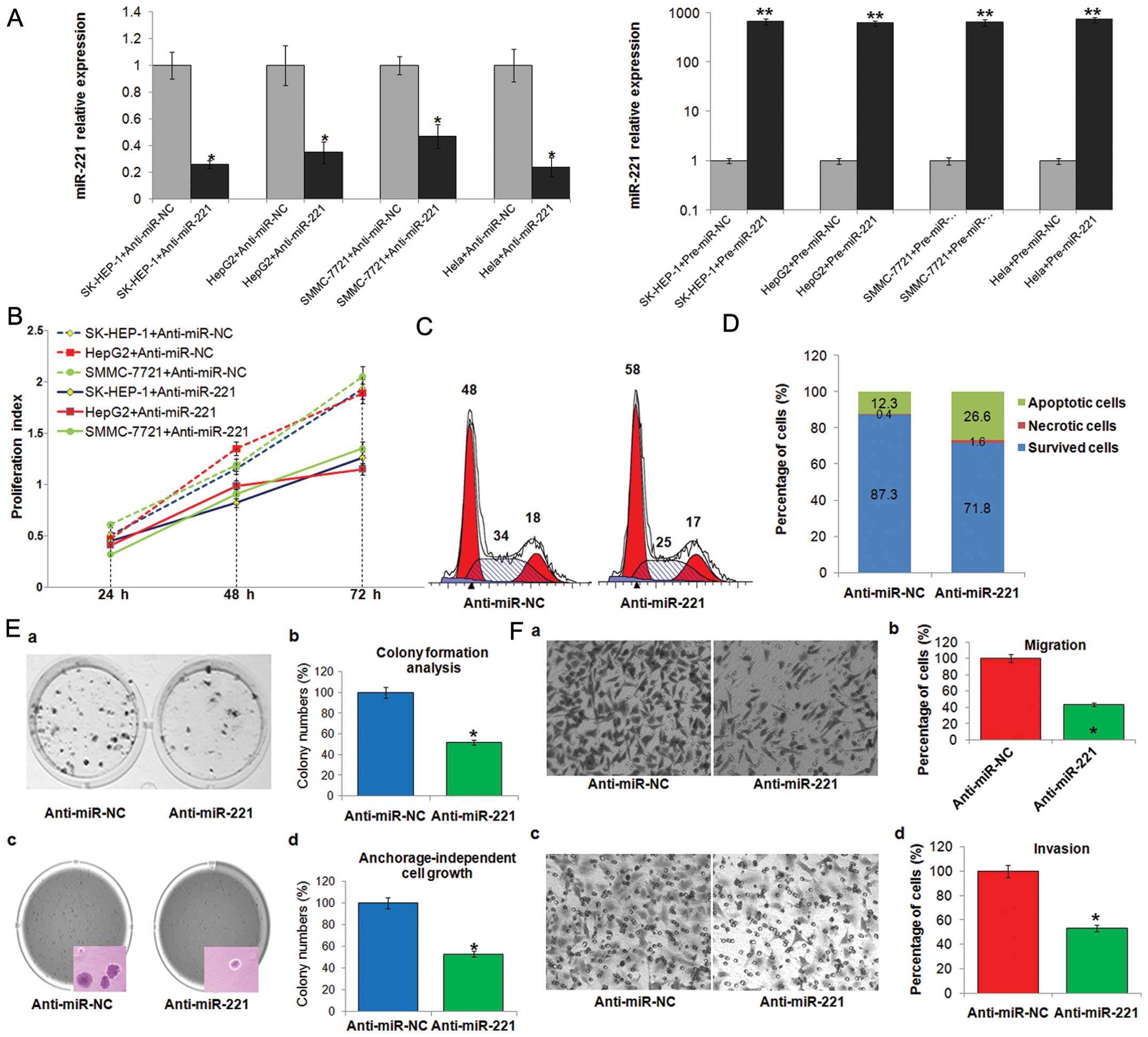
As shown in Fig. 4A, the increase
of miR-221 or inhibition of miR-221 expression were verified by
TaqMan qRT-PCR in SK-HEP-1, HepG2, SMMC-7721 or HeLa cells
transfected with pre-miR-221 or anti-miR-221.
Sustained cell proliferation is the most distinctive
property of cancer. Inhibiting miR-221 markedly suppressed the
proliferation of liver cancer cells (Fig. 4B). To examine whether compromised
cell proliferation could be attributed to the cell cycle arrest, we
further analyzed the status of cell cycle. As shown in Fig. 4C, miR-221 silencing significantly
increased the percentages of cells in G1 phase, which indicated the
occurrence of G1 arrests in treated SK-HEP-1 cells, while the cell
percentages in S phase were obviously shrunk by nearly 10%.
Escaping from cell apoptosis is another advantage of tumor cells
for overwhelming growth. The result from Annexin V/PI combined
labeling flow cytometry analysis showed that the numbers of
apoptotic cells were clearly increased in SK-HEP-1 cells treated
with anti-miR-221 (Fig. 4D). To
assess the functional role of miR-221 in tumor formation, the
capacity of colony formation and anchorage-independent growth were
measured in SK-HEP-1 transfected with anti-miR-221. Notably,
anti-miR-221-transfected cells displayed much fewer and smaller
colonies than control transfectants (Fig. 4E). Metastasis is another hallmark of
cancer. As shown in Fig. 4F,
miR-221 inhibition effectively suppressed the ability of SK-HEP-1
cell migration and invasion.
These data indicate that miR-221 functions as an
oncogene and its silencing inhibits tumorigenic properties of liver
cancer cells in vitro.
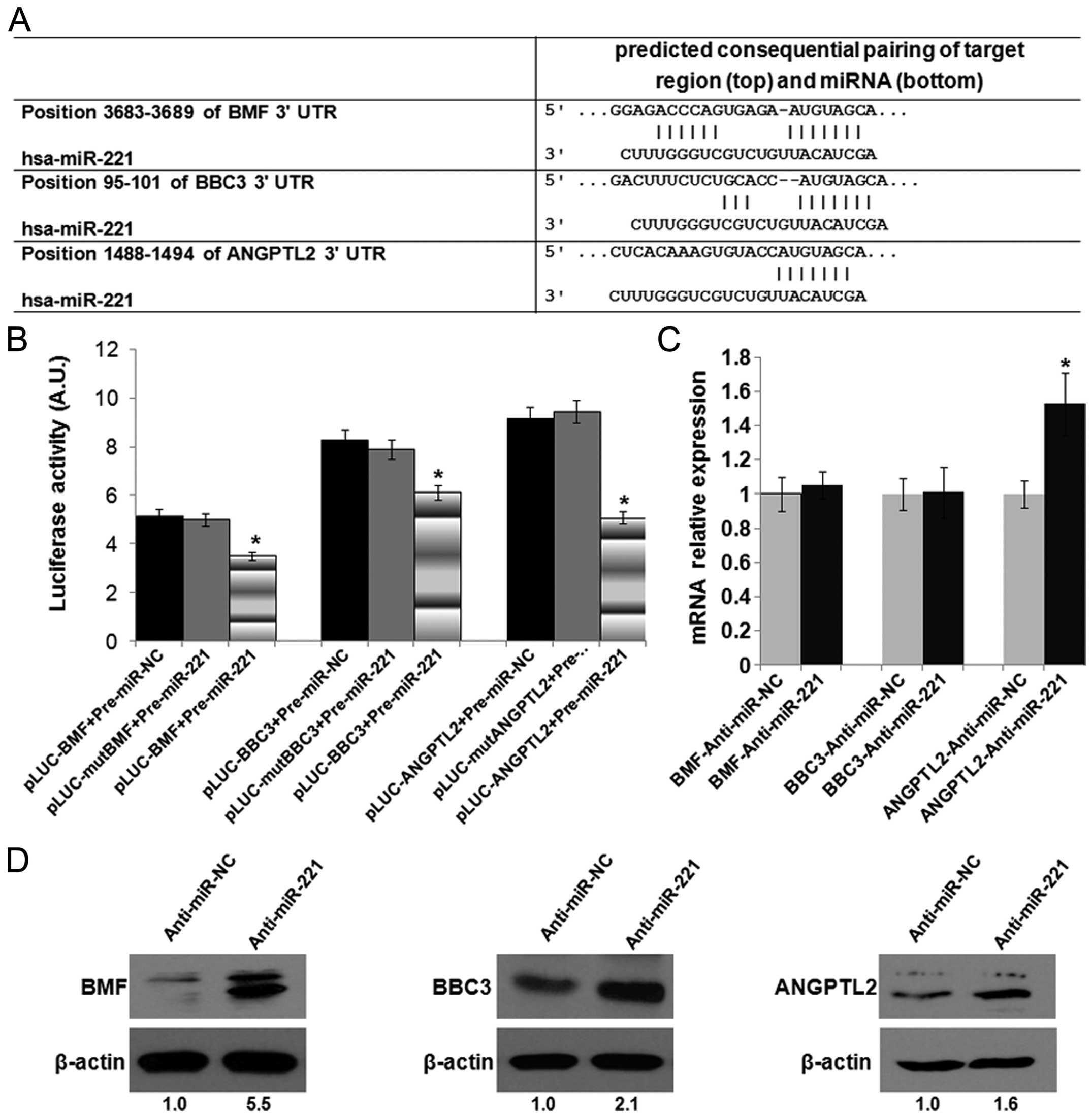
miR-221 targets BMF, BBC3 and
ANGPTL2
To elucidate the tumorigenic role of miR-221 in HCC,
we further verified several putative targets of miR-221 from
Fig. 3 (Fig. 5A). As shown in Fig. 5B, luciferase assay indicated that
miR-221 could directly aim at its predicted binding sites of BMF,
BBC3 and ANGPTL2, leading to the suppression of luciferase
expression, whereas when the binding site was mutated by
site-specific mutagenesis, the luciferase activity was reserved
comparable with the matched negative control group. At the mRNA
level, miR-221 inhibition elicited an obvious upregulation of
ANGPTL2 but not of BMF and BBC3 (Fig.
5C). While at the protein level, BMF, BBC3 and ANGPTL2 protein
were apparently increased in anti-miR-221-treated cells (Fig. 5D). These data suggest that miR-221
may suppress its targets BMF, BBC3 and ANGPTL2 mainly through a
translational inhibition at the protein level with or without mRNA
degradation.
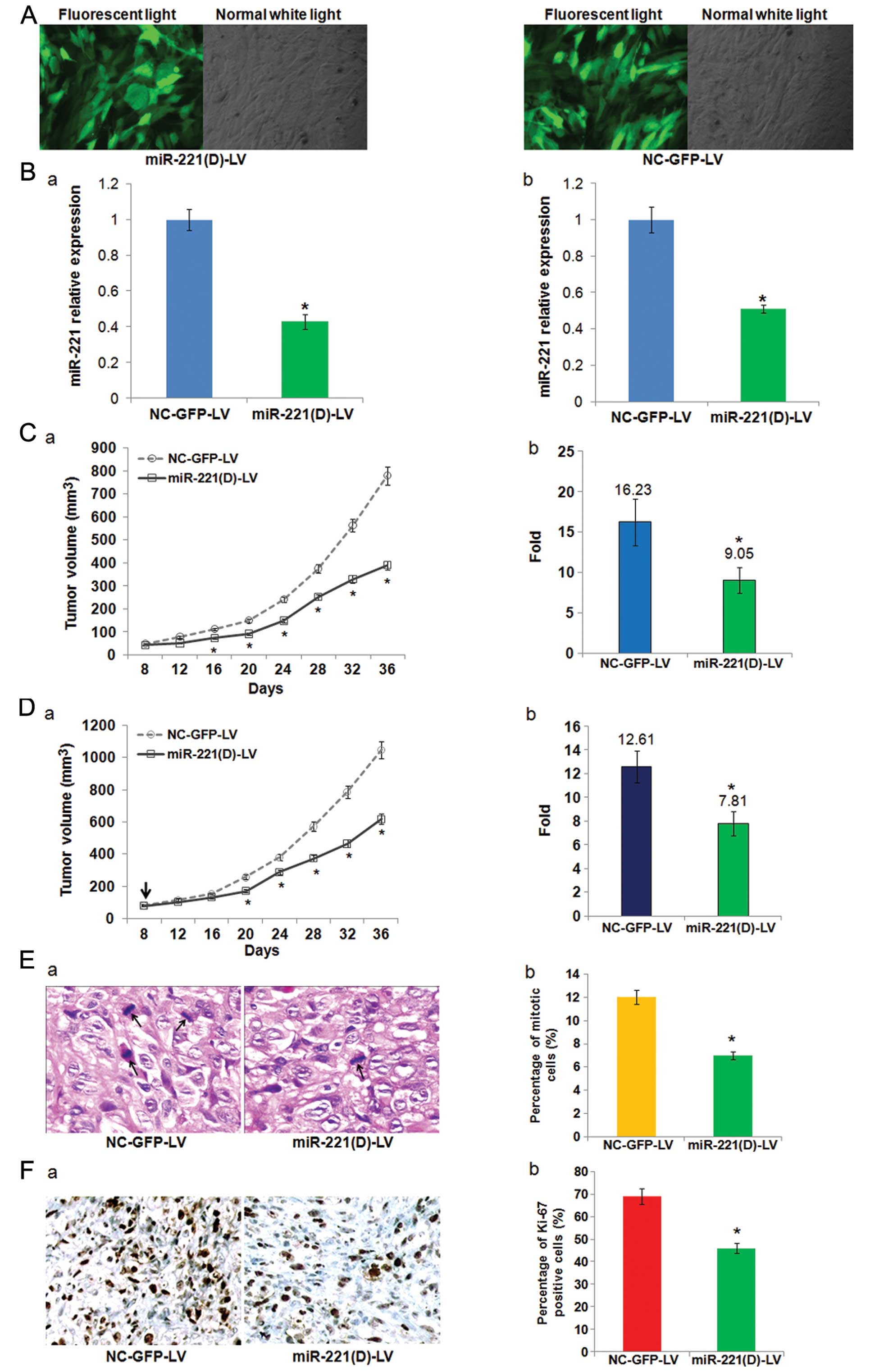
Lentivirus-mediated-anti-miR-221 inhibits
tumor formation and growth of hepatoma xenografts in vivo
Given the importance of miR-221 in HCC
carcinogenesis, it is not surprising that miR-221 is an attractive
target for developing novel therapies. Lentiviral vectors are the
most recently developed virus-derived vectors for gene therapy
applications, and have demonstrated the ability to transduce
dividing and non-dividing cells, and sustain long-term transgene
expression, which makes them uniquely attractive as gene therapy
vectors (37). Thus, we
successfully constructed a recombinant lentiviral vector termed
miR-221(D)-LV (and NC-GFP-LV as control) to transduce SK-HEP-1
cells (Fig. 6A) and inhibit the
expression of miR-221 (Fig. 6B-a).
First, we employed SK-HEP-1 cells pre-infected with miR-221(D)-LV
to establish subcutaneous tumors in nude mice, and measured the
tumor growth and significant features. As depicted in Fig. 6C, depletion of miR-221 in
vitro by miR-221(D)-LV renders SK-HEP-1 cells less efficient in
establishing tumors in vivo. The average fold increase of
tumor volumes at the sacrifice with respect to the first
measurements was much smaller in miR-221(D)-LV pre-infected tumors
vs. control tumors (9.05±1.62 vs. 16.23±2.89, P=0.034). Next, we
further assessed the effects of miR-221 inhibition on
pre-established SK-HEP-1 xenografts. Consistent with our
expectation, a single intratumor injection with 2×108 TU
miR-221(D)-LV on day 8 (when the tumor volume reached ~80
mm3), reduced the growth of tumors. As shown in Fig. 6D, the growth curves of miR-221(D)-LV
and NC-GFP-LV-treated tumors became divergent on day 16 after the
inoculation of SK-HEP-1 in mice, and this trend became more obvious
and continued to the end of the experiment. Relative to the first
measurements on day 8, the average fold increase of tumor volumes
at the sacrifice of miR-221(D)-LV-treated groups was significantly
smaller than that of the negative control (7.81±1.02 vs.
12.61±1.37, P=0.041).
To clarify the cellular mechanisms underlying
miR-221(D)-LV mediated tumor suppression, resected tissues from
those treated xenograft tumors were analyzed to verify miR-221
expression, and measured for mitotic index and expression of Ki67
as markers of proliferation. At the end of the experiment,
miR-221(D)-LV effectively reduced the expression of miR-221
(Fig. 6B-b). Consistent with the
above results, HE and Ki67 staining revealed markedly reduced
proliferation index in miR-221(D)-LV-treated tumors (Fig. 6E and F). These results indicate that
miR-221 silencing inhibits liver cancer cell growth in
vivo.
Discussion
The rationale for using miRNAs as potential
therapeutic targets for cancer is based on many facts: i) miRNAs
are natural antisense interactors; ii) miRNA expression profiles
have been shown to be related to disease state and treatment
response and the dysregulated miRNAs contribute to the initiation
and development of cancer; iii) the small size (22–24 nucleotides
in length) makes them very attractive for drug development
(38).
In the present study, we found that miR-221 plays a
central role in HCC signaling pathway by bioinformatics analysis.
It is common in biology for gene expression and important processes
to have multiple layers of regulation and control. TFs and miRNAs
are key regulators of gene expression at the transcription and
post-transcription level. They may regulate the same target gene
and regulate mutually to form complex regulatory module and network
including FFLs, thus carry out the subtle regulation of the target
gene expression (39). It has been
predicted that there are hundreds of FFLs in human genome (40) and some of them have been
experimentally verified such as TP53/miR-106b/E2F (15) and NF-κB/miR-19/CYLD (13) in cancer cell proliferation. Many TFs
and miRNAs have been reported to be involved in the progress of
HCC, which is a complex process (41). Therefore, the TF and miRNA
regulations will be the key regulation of HCC progress. In this
study, we started from the top differentially expressed miRNAs and
constructed the TF-miRNA co-regulatory FFLs in HCC using the same
strategy we used for schizophrenia and T-cell acute lymphoblastic
leukemia (13,42). Figs.
1–3 show that miR-221 was a
core miRNA with the most targets of HCC-related genes and formed
many FFL regulations with some key TFs. We also showed that miR-221
may target CDKN1B, CDKN1C, BMF, DDIT4, PTEN, TIMP3, E2F3, TP53BP2,
BBC3, ANGPTL2 and many other genes related to HCC (Fig. 3). Some of these targets have been
experimentally confirmed. Furthermore, Fig. 3 also provides insight into FFL
regulation of miR-221 involved in HCC. TFs c-Myc, c-Jun and STAT3
were predicted to target many important HCC genes and formed FFLs
with miR-221. Particularly, c-Myc and miR-221 formed 13 FFLs with
HCC genes, including FFLs c-Myc/miR-221/E2F3 and
c-Myc/miR-221/CDKN1C. c-Myc is a widely studied oncogene and it
plays a critical role in human pathogenesis (43). Thus, the feed-forward loop
regulatory modules among c-Myc, miR-221 and HCC genes warrant
further exploration.
Previous studies have demonstrated that
overexpression of miR-221 augments cell proliferation, colony
formation, invasion and increase the number of cells in S phase,
while inhibiting cell apoptosis (8,17,18,26,27).
Here, we re-confirmed the role of miR-221 as an oncogene through
miR-221 silencing functional analysis including cell proliferation,
cell cycle, apoptosis, cell migration, invasion and clone formation
and found that inhibition of miR-221 in liver cancer cells
decreased cell proliferation, clonogenicity, migration/invasion and
also induced G1 arrest and apoptosis (Fig. 4). To investigate the molecular
mechanism of miR-221-mediated phenotype in hepatoma cells described
above, we further validated that BMF, BBC3 and ANGPTL2 are direct
targets of miR-221 and showed that miR-221 suppressed the
expression of BMF, BBC3 and ANGPTL2 by binding directly to the
3′-UTR of these genes (Fig. 5),
which support the findings of our bioinformatics analysis. Among
them, BMF is a member of the Bcl-2 family belonging to
pro-apoptotic BH3-only proteins. Our result that BMF is a target of
miR-221 is in accordance with the result published by Gramantieri
et al (27). BBC3, also
known as PUMA, is an essential mediator of cell death in response
to various apoptotic signals, notable by its pivotal role in
p53-induced apoptotic pathway (44). Markedly, Pineau et al found
that BBC3 is a putative target of miR-221 and that there is an
inverse correlation between miR-221 and BBC3 expression in HCC, but
failed to validate BBC3 as a direct target of miR-221 by luciferase
assay in the supporting information of their publication (8). However, we have verified that BBC3 is
a direct target of miR-221, and this is similar to the results
observed in human epithelial cancers and glioblastoma (45,46).
ANGPTL2 belongs to the angiopoietin family for its limited sequence
homology with angiopoietins (47).
It has been reported that ANGPTL2 works as a functional tumor
suppressor gene repressing cell growth and impairing cell clone
formation in ovarian cancer (48).
To the best of our knowledge, that miR-221 directly targets ANGPTL2
has not previously been reported. These results indicate that
miR-221 silencing could inhibit liver cancer malignant properties
in vitro through regulating many HCC-related genes including
BMF, BBC3 and ANGPTL2.
The above findings strongly underscore the
possibility of miR-221 as an ideal target for therapeutic
intervention. In Fig. 6, we showed
that in vitro depletion of miR-221 by
lentivirus-mediated-anti-miR-221 renders liver cancer cells less
efficient in the establishment of in vivo xenografts, while
in vivo intratumoral knockdown of miR-221 reduces tumor
growth of liver cancer cell xenografts. In the present study,
lentiviral vectors provided an efficient gene delivery and gave us
a stable loss of function of miR-221 for study in vitro and
in vivo. Hence, lentivirus-mediated antagomir expression for
stable loss of function of a specific miRNA may be a useful
laboratory tool to study miRNA functions and may be considered for
clinical gene therapy applications (34,49).
Our animal studies indicated that lentivirus-mediated-anti-miR-221
treatment could suppress the growth of hepatoma xenografts in
vivo. Similarly, Park et al showed miR-221 silencing by
chol-anti-miR-221 blocks HCC and promotes survival in a valid
orthotopic mouse model of HCC (50). These results support that miR-221
may be an ideal target for HCC therapy and future studies to
determine the relative efficacy of targeting miR-221 compared with
other miRNA delivery methods, such as nanoparticles, may be
required to identify the optimal miRNA delivery methods for further
clinical translation.
In conclusion, we demonstrated that miR-221 is a
critical modulator in HCC and miR-221 silencing could inhibit liver
cancer malignant properties in vitro and in vivo
through regulation of its targets including BMF, BBC3, ANGPTL2,
emphasizing the promising potential of miR-221 inhibition for HCC
therapy.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81101824, 31171271,
81000874 and 81070333), the Outstanding Youth Science Foundation of
Tongji Hospital (no. YXQN005), and the Youth Sciences and
Technology Chenguang Planning of Wuhan (no. 2014070404010219).
References
|
1
|
Roberts LR: Sorafenib in liver cancer -
just the beginning. N Engl J Med. 359:420–422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murakami Y, Yasuda T, Saigo K, et al:
Comprehensive analysis of microRNA expression patterns in
hepatocellular carcinoma and non-tumorous tissues. Oncogene.
25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Varnholt H, Drebber U, Schulze F, et al:
MicroRNA gene expression profile of hepatitis C virus-associated
hepatocellular carcinoma. Hepatology. 47:1223–1232. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ladeiro Y, Couchy G, Balabaud C, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hou J, Lin L, Zhou W, et al:
Identification of miRNomes in human liver and hepatocellular
carcinoma reveals miR-199a/b-3p as therapeutic target for
hepatocellular carcinoma. Cancer Cell. 19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pineau P, Volinia S, McJunkin K, et al:
miR-221 overexpression contributes to liver tumorigenesis. Proc
Natl Acad Sci USA. 107:264–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuen MF, Wu PC, Lai VC, Lau JY and Lai CL:
Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma.
Cancer. 91:106–112. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kawate S, Fukusato T, Ohwada S, Watanuki A
and Morishita Y: Amplification of c-myc in hepatocellular
carcinoma: correlation with clinicopathologic features,
proliferative activity and p53 overexpression. Oncology.
57:157–163. 1999.
|
|
11
|
Lin CP, Liu CR, Lee CN, Chan TS and Liu
HE: Targeting c-Myc as a novel approach for hepatocellular
carcinoma. World J Hepatol. 2:16–20. 2010.PubMed/NCBI
|
|
12
|
Suzuki H, Fujita H, Mullauer L, et al:
Increased expression of c-jun gene during spontaneous
hepatocarcinogenesis in LEC rats. Cancer Lett. 53:205–212. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye H, Liu X, Lv M, et al: MicroRNA and
transcription factor co-regulatory network analysis reveals miR-19
inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids
Res. 40:5201–5214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Wang C, Wang M, et al: A cyclin
D1/microRNA 17/20 regulatory feedback loop in control of breast
cancer cell proliferation. J Cell Biol. 182:509–517. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brosh R, Shalgi R, Liran A, et al:
p53-Repressed miRNAs are involved with E2F in a feed-forward loop
promoting proliferation. Mol Syst Biol. 4:2292008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He XX, Chang Y, Meng FY, et al:
MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and
suppresses liver cancer cell growth in vitro and in vivo. Oncogene.
31:3357–3369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garofalo M, Di Leva G, Romano G, et al:
miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar
|
|
18
|
le Sage C, Nagel R, Egan DA, et al:
Regulation of the p27Kip1 tumor suppressor by miR-221
and miR-222 promotes cancer cell proliferation. EMBO J.
26:3699–3708. 2007.
|
|
19
|
Ciafre SA, Galardi S, Mangiola A, et al:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee EJ, Gusev Y, Jiang J, et al:
Expression profiling identifies microRNA signature in pancreatic
cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He H, Jazdzewski K, Li W, et al: The role
of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad
Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chun-Zhi Z, Lei H, An-Ling Z, et al:
MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun K, Wang W, Zeng JJ, Wu CT, Lei ST and
Li GX: MicroRNA-221 inhibits CDKN1C/p57 expression in human
colorectal carcinoma. Acta Pharmacol Sin. 32:375–384. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Callegari E, Elamin BK, Giannone F, et al:
Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic
model. Hepatology. 56:1025–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fornari F, Gramantieri L, Ferracin M, et
al: MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human
hepatocellular carcinoma. Oncogene. 27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gramantieri L, Fornari F, Ferracin M, et
al: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar
|
|
30
|
Sethupathy P, Corda B and Hatzigeorgiou
AG: TarBase: a comprehensive database of experimentally supported
animal microRNA targets. RNA. 12:192–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang Q, Wang Y, Hao Y, et al:
miR2Disease: a manually curated database for microRNA deregulation
in human disease. Nucleic Acids Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Landt SG, Marinov GK, Kundaje A, et al:
ChIP-seq guidelines and practices of the ENCODE and modENCODE
consortia. Genome Res. 22:1813–1831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shannon P, Markiel A, Ozier O, et al:
Cytoscape: a software environment for integrated models of
biomolecular interaction networks. Genome Res. 13:2498–2504. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scherr M, Venturini L, Battmer K, et al:
Lentivirus-mediated antagomir expression for specific inhibition of
miRNA function. Nucleic Acids Res. 35:e1492007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Scherr M, Battmer K, Ganser A and Eder M:
Modulation of gene expression by lentiviral-mediated delivery of
small interfering RNA. Cell Cycle. 2:251–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Naldini L, Blömer U, Gallay P, et al: In
vivo gene delivery and stable transduction of nondividing cells by
a lentiviral vector. Science. 272:263–267. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
D’Costa J, Mansfield SG and Humeau LM:
Lentiviral vectors in clinical trials: current status. Curr Opin
Mol Ther. 11:554–564. 2009.
|
|
38
|
Tili E, Michaille JJ, Gandhi V, Plunkett
W, Sampath D and Calin GA: miRNAs and their potential for use
against cancer and other diseases. Future Oncol. 3:521–537. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Re A, Corá D, Taverna D and Caselle M:
Genome-wide survey of microRNA-transcription factor feed-forward
regulatory circuits in human. Mol Biosyst. 5:854–867. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zeng L, Yu J, Huang T, et al: Differential
combinatorial regulatory network analysis related to venous
metastasis of hepatocellular carcinoma. BMC Genomics. 13(Suppl 8):
S142012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo AY, Sun J, Jia P and Zhao Z: A novel
microRNA and transcription factor mediated regulatory network in
schizophrenia. BMC Syst Biol. 4:102010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zimonjic DB and Popescu NC: Role of DLC1
tumor suppressor gene and MYC oncogene in pathogenesis of human
hepatocellular carcinoma: Potential prospects for combined targeted
therapeutics (Review). Int J Oncol. 41:393–406. 2012.
|
|
44
|
Jeffers JR, Parganas E, Lee Y, et al: Puma
is an essential mediator of p53-dependent and -independent
apoptotic pathways. Cancer Cell. 4:321–328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang C, Zhang J, Zhang A, et al: PUMA is
a novel target of miR-221/222 in human epithelial cancers. Int J
Oncol. 37:1621–1626. 2010.PubMed/NCBI
|
|
46
|
Zhang CZ, Zhang JX, Zhang AL, et al:
MiR-221 and miR-222 target PUMA to induce cell survival in
glioblastoma. Mol Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim I, Moon SO, Koh KN, et al: Molecular
cloning, expression, and characterization of angiopoietin-related
protein. Angiopoietin-related protein induces endothelial cell
sprouting. J Biol Chem. 274:26523–26528. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kikuchi R, Tsuda H, Kozaki K, et al:
Frequent inactivation of a putative tumor suppressor,
angiopoietin-like protein 2, in ovarian cancer. Cancer Res.
68:5067–5075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feng SY, Dong CG, Wu WK, Wang XJ, Qiao J
and Shao JF: Lentiviral expression of anti-microRNAs targeting
miR-27a inhibits proliferation and invasiveness of U87 glioma
cells. Mol Med Rep. 6:275–281. 2012.PubMed/NCBI
|
|
50
|
Park JK, Kogure T, Nuovo GJ, et al:
miR-221 silencing blocks hepatocellular carcinoma and promotes
survival. Cancer Res. 71:7608–7616. 2011. View Article : Google Scholar : PubMed/NCBI
|