Introduction
Lung cancer is the leading cause of cancer-related
mortality-worldwide (1), and the
5-year survival rate is low. Non-small cell lung cancer (NSCLC) is
subdivided into three subtypes: squamous cell carcinoma (SCC,
~28%), large-cell carcinoma (LCC, ~24%) and adenocarcinoma (~48%)
(2). Most of the mortality
associated with lung adenocarcinoma arises from uncontrolled
metastases. Exploring the molecular mechanisms underlying lung
adenocarcinoma metastasis is necessary to overcome lung
adenocarcinoma metastasis and recurrence.
Scaffold protein neural precursor cell expressed,
developmentally downregulated 9 (NEDD9), also known as HEF1 and
Cas-L (3,4), belongs to the family of Crk-associated
substrate (CAS) proteins that regulate protein complexes
controlling cell attachment, migration, invasion, cell cycle,
apoptosis and oncogenic signal transduction (5–7).
Overexpression of the NEDD9 protein is strongly linked to poor
prognosis in cancer, as well as resistance to first-line
chemotherapeutics in multiple tumor types including breast cancer
(8), glioblastoma (9), melanoma (10) and gastrointestinal carcinoma
(11,12). High levels of NEDD9 mRNA and protein
have been shown to be present in human lung carcinoma tissues, and
are highly related to overall survival (OS) and to progression-free
survival (PFS) (13–16).
Gene therapy for tumors has focused on gene
replacement, antisense nucleic acid techniques, cytokine gene
therapy and RNA interference (RNAi). RNAi is a post-transcriptional
regulation method that provides a rapid means of depleting mRNA by
introducing double-stranded RNA homologous to a particular message,
causing its sequence-specific degradation. Using small interfering
RNA (siRNA) to silence target genes is simple, specific and
effective (17).
In the present study, NEDD9-specific lentiviral
particles were chemically synthesized and transfected into the
human lung adenocarcinoma A549 cell line. The inhibitory effect of
siRNA on the expression of NEDD9 mRNA and protein was detected by
RT-PCR and western blotting, and biological characteristics of A549
cells in vitro and in vivo including proliferation,
cell cycle, migration and invasion were investigated.
Materials and methods
NEDD9 siRNA lentivirus
siRNA targeting NEDD9 (GenBank Accession No.
NM_182966) and a non-targeting RNA were chemically synthesized by
GeneChem Technology Co., Ltd. (Shanghai, China). The target
sequence was GTGTCCTAT TTCTTAGTGA, as determined by our previous
research (18). The vehicle was
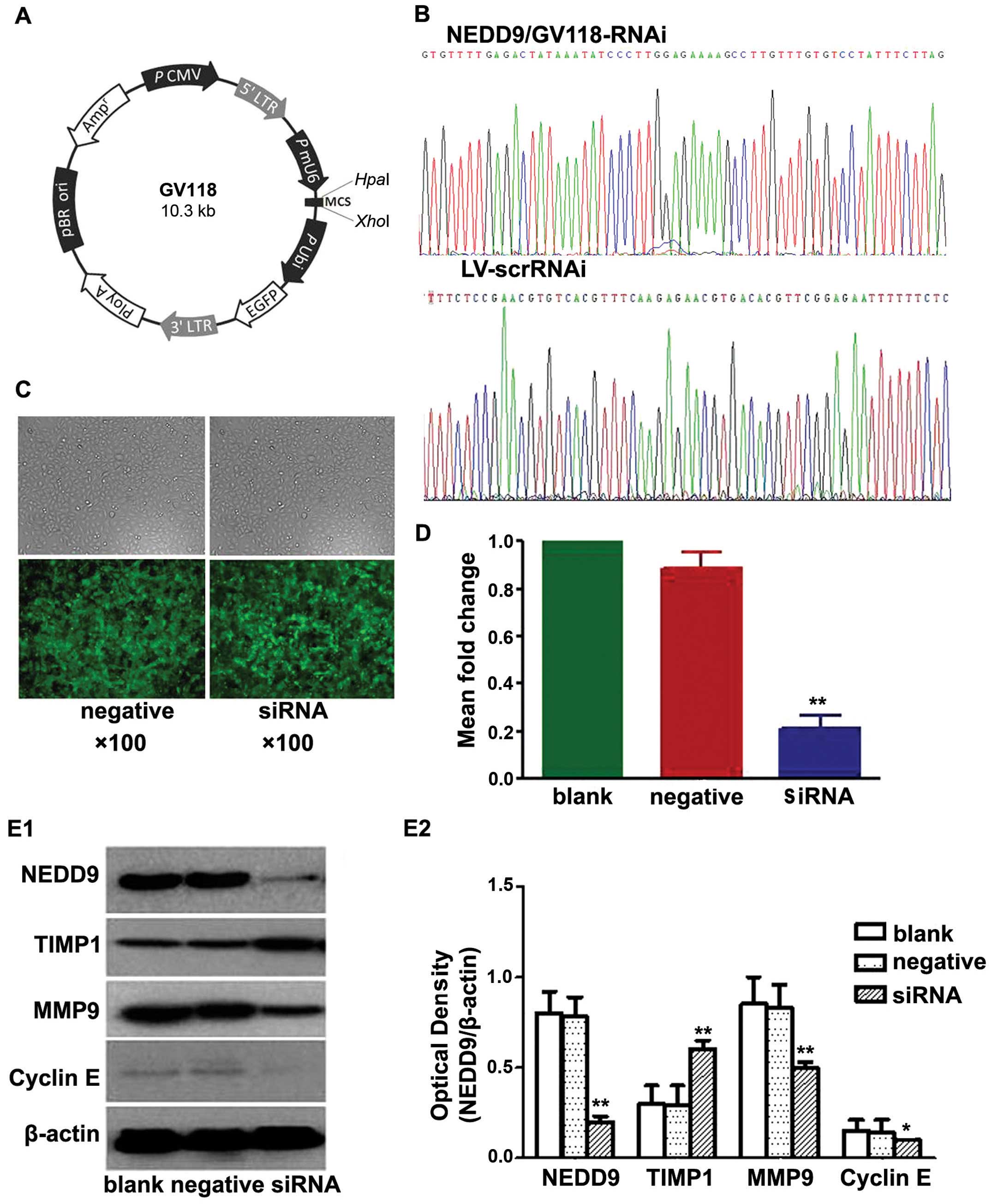
GV118 (Fig. 1A). A negative control
siRNA sequence (TTCTCCGAACGTGTCACGT) was used as a control for
NEDD9 siRNA.
Lentiviral vector transduction
A549 cells were transduced with siRNA-expressing
lentiviruses at a multiplicity of infection of 20 particles/cell in
Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT, USA)
containing 5 μg/ml polybrene and incubated at 37°C in 5%
CO2 in 6-well plates. GFP expression was observed by
fluorescence microscopy three days after transduction, and cells
were harvested at 4 days after transduction for RT-PCR or western
blot analysis.
RNA isolation and establishment of RT-PCR
detection of NEDD9 mRNA
Total RNA from three groups (blank control group,
negative control group and siRNA group) was purified from cells
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instructions. Firststrand cDNA was synthesized
using 2.5 μg total RNA and AMV retroviridase (Promega). Specific
primers were designed for the mRNA sequence of the NEDD9 gene, and
NEDD9 and GAPDH segments were amplified. Primers were from Shanghai
Bioengineering Co. (Shanghai, China). NEDD9 gene primers were
upstream 5′-CGTGGGTAAAAAGGTGTT CC-3′ and downstream
5′-CAAGCCTCCAAACTCAGGAC-3′ (amplified segment 124 bp); GAPDH
primers were upstream 5′-TCGTGGAAGGACTCATGACC-3′ and downstream
5′-AG GGATGATGTTCTGGAGAG-3′ (amplified segment 97 bp).
NEDD9 mRNA was quantified by FQ-PCR using an ABI
PRISM 7500 Sequence Detection System (Applied Biosystems, Foster
City, CA, USA). Reactions consisted of 20 μl 2x real-time PCR
buffer, 0.5 μl of upstream and downstream primers for NEDD9, 2 μl
reverse transcription product, 0.2 μl Taq DNA polymerase,
and ddH2O to 40 μl. FQ-PCR reaction conditions consisted
of 95°C for 3 min; 95°C for 15 sec, 65°C for 45 sec, for 40 cycles
and 72°C for 2 min. GAPDH was simultaneously amplified as an
internal reference. All reactions were performed in triplicate.
Results were analyzed by calculating the Ct values for NEDD9 and
GAPDH in samples, and the relative expression of NEDD9 mRNA in each
group (the relative fold, RF) using the 2−ΔΔCt value was
calculated (19).
Protein isolation and western blot
analysis
NEDD9 protein expression was examined in A549 cells
by western blot analysis. Cells were harvested in lysis buffer (2%
SDS, 50 mM Tris, pH 7.4, 1 mM EDTA, protease inhibitor mixture).
The supernatant was collected after centrifugation and protein
concentrations were determined using a BCA protein assay kit
(Pierce, Rockford, IL, USA). Equal amounts of protein (40 μg) were
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis on 8% gels and transferred onto nitrocellulose
membranes (Hyclone). The membranes were blocked with 5% (v/v)
skimmed milk and probed with phospho-rabbit anti-human polyclonal
primary antibodies for NEDD9 (93 kDa; Abcam, San Francisco, CA,
USA). TIMP1 was detected using the rabbit monoclonal anti-human
TIMP1 antibody (28.5 kDa; Santa Cruz Biotechnology, Santa Cruz, CA,
USA). MMP9 was detected using a rabbit polyclonal antihuman MMP9
antibody (92 kDa; Santa Cruz Biotechnology) and cyclin E was
detected using rabbit polyclonal anti-human cyclin E antibody (34
kDa; Santa Cruz Biotechnology). Incubations were carried out at 4°C
overnight. Membranes were washed and incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China),
at room temperature for 1 h. Antibodies against β-actin (43 kDa;
Santa Cruz Biotechnology) were used to measure protein loading.
Bound antibodies were visualized using an electrochemiluminescence
system (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Cell proliferation assay
Soft agar colony formation assay
Growth medium (2x DMEM) was mixed 1:1 (v/v) with
1.2% low melting agar, and 300 μl was plated in a layer in 6-well
plates. Uninfected and infected cells were trypsinized,
centrifuged, resuspended in 0.35% agar medium (equal volumes of
0.7% noble agar and culture medium), and plated onto the top agar
at an initial concentration of 1,000 cells/well. Cells were fed
every 3 days with 1.5 ml growth medium. After culturing for 14
days, adherent cells were washed twice with PBS and fixed with 4%
paraformaldehyde for 30 min at room temperature. Colonies were
visualized using cell staining with gentian violet (Beyotime
Institute of Biotechnology Co., Ltd., Shanghai, China) and counted
under a phase contrast microscopy with low magnification. All
studies were performed at least three times.
Flow cytometric analysis
To determine the impact of overexpression of NEDD9
on cell cycle kinetics, A549 cells were collected at 72 h following
transfection. Ribonuclease A (final concentration of 0.1 mg/ml) was
added to 1×106 cells, and the cells were incubated at
37°C for 30 min, then on ice for 15 min. A549 cells were
resuspended in 50 μg/ml propidium iodide and incubated for 30 min
in the dark at 4°C. Cell cycle distribution and the percentage of
cells with degraded DNA were determined by flow cytometric analysis
(FCA) at an excitation wavelength of 488 nm using a FACSCalibur
flow cytometer (Beckman Coulter, Brea, CA, USA) at the Medical and
Science Research Institute of Zhengzhou University. Cell cycle
histograms were obtained from three determinations, each with a
total of 100,000 cells/treatment.
In vitro cell migration and invasion
assays
To measure cell migration and invasion,
wound-healing assays and Transwell assays were performed. For the
wound-healing assays, cells (5×105 cells/well) were
plated in 6 -well plates. After 12 h, the confluent monolayer was
scratched manually with a plastic 200-μl pipette tip and after
washing with PBS, the wounded monolayers of the cells were allowed
to heal for 48 h. Each migration assay was conducted at least three
times independently.
Transwell cell migration assays were performed as
previously described using Transwells (8 μmol/l pore size
polycarbonate membranes) (Corning, ChemiconA, USA). Cells from the
various treatment groups (1×105) in 0.5 ml serum-free
medium were placed in the upper chamber. Lower chambers were loaded
with 0.8 ml medium containing 10% PBS. Cells that migrated into the
lower chamber were counted after 24 h of incubation at 37°C in 5%
CO2. Non-migratory cells were removed. Invasive cells
were stained with 0.2% crystal violet in 10% ethanol. To quantify
the invasive cells, three independent fields per well were
photographed under phase contrast microscopy. The number of cells
per field were counted and averaged.
In vivo study of lung adenocarcinoma
xenograft tumor models in nude mice
Thirty 4-week-old female BALB/c nude mice (weight
15–20 g) were purchased from the Beijing Lihua Experimental Animal
Center (Beijing, China). Mice were housed in a
temperature-controlled, pathogen-free animal facility with 12-h
light and dark cycles. All animal experiments were performed under
protocols approved by the Animal Center Animal Care and Use
Committee of Zhengzhou University. After two weeks, the mice were
divided into 3 groups (n=6 per group): A549 control (blank),
lentivirus negative-siRNA (negative) and lentivirus NEDD9-siRNA
(siRNA) group. Cells (5×106 in 100 μl PBS) with
lentivirus expression vectors or untransfected cells were injected
subcutaneously into the right flank of mice and cancer growth was
observed. Tumor mass (xenograft) volume (20), calculated as volume (mm3)
= width2 (mm2) × length (mm)/2 was detected
every 2–3 days using a caliper. After 5 weeks, the mice were
sacrificed and the tumors were harvested and weighed.
Statistical analysis
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) for Windows
statistical software package was used for analysis. One-way
analysis of variance (ANOVA) was used to investigate differences
between the three groups. Values are presented as mean ± standard
deviation (SD). P<0.05 indicates a significant difference, and
P<0.01 indicates a highly significant difference.
Results
NEDD9 siRNA synthesis
Recombinant plasmids were purified, confirmed by
sequencing (Fig. 1B), packed into
lentiviruses and named NEDD9/GV118-RNAi and LV-scrRNAi. Viruses
were titered for use at dilutions of ~1×108 titres U/ml
(data not shown).
Results of the transfection
A549 cells were transfected with lentiviruses
containing siRNA directed against NEDD9 or a non-targeting negative
control shRNA (Scr-siRNA). Cells showing green fluorescence were
considered to be successfully transfected. As shown in Fig. 1C, the lentiviral infection rate was
high.
Effects of lentivirus-mediated NEDD9 RNAi
on expression of NEDD9 mRNA
Results of the RT-PCR (Fig. 1D) showed no significant difference
in the levels of NEDD9 mRNA between the blank control and the
negative control group (0.89±6.05-fold). The mRNA levels in the
three siRNA groups (0.21±5.42-fold) were significantly lower than
the mRNA levels in the blank control (P<0.05) and negative
control groups (P<0.05). These data indicated that NEDD9 mRNA
level in the A549 cells decreased significantly after transfection
with NEDD9-siRNA. Transfection with NEDD9-specific siRNA resulted
in degradation of NEDD9 mRNA to silence the NEDD9 gene.
Effects of lentivirus-mediated NEDD9 RNAi
on NEDD9, TIMP1, MMP9 and cyclin E expression
Western blot analysis showed that levels of NEDD9,
MMP9 and cyclin E protein in the siRNA group were significantly
lower than levels in the blank and negative control groups, while
the level of TIMP1 protein in the siRNA group was significantly
higher than levels that in the blank and negative control groups
(P<0.05; Fig. 1E). These data
indicated that NEDD9-specific siRNA-silencing significantly reduced
the levels of NEDD9, MMP9 and cyclin E protein and increased TIMP1
protein in the A549 cells.
Silencing of the levels of NEDD9 inhibits
proliferation and alters A549 cell cycle
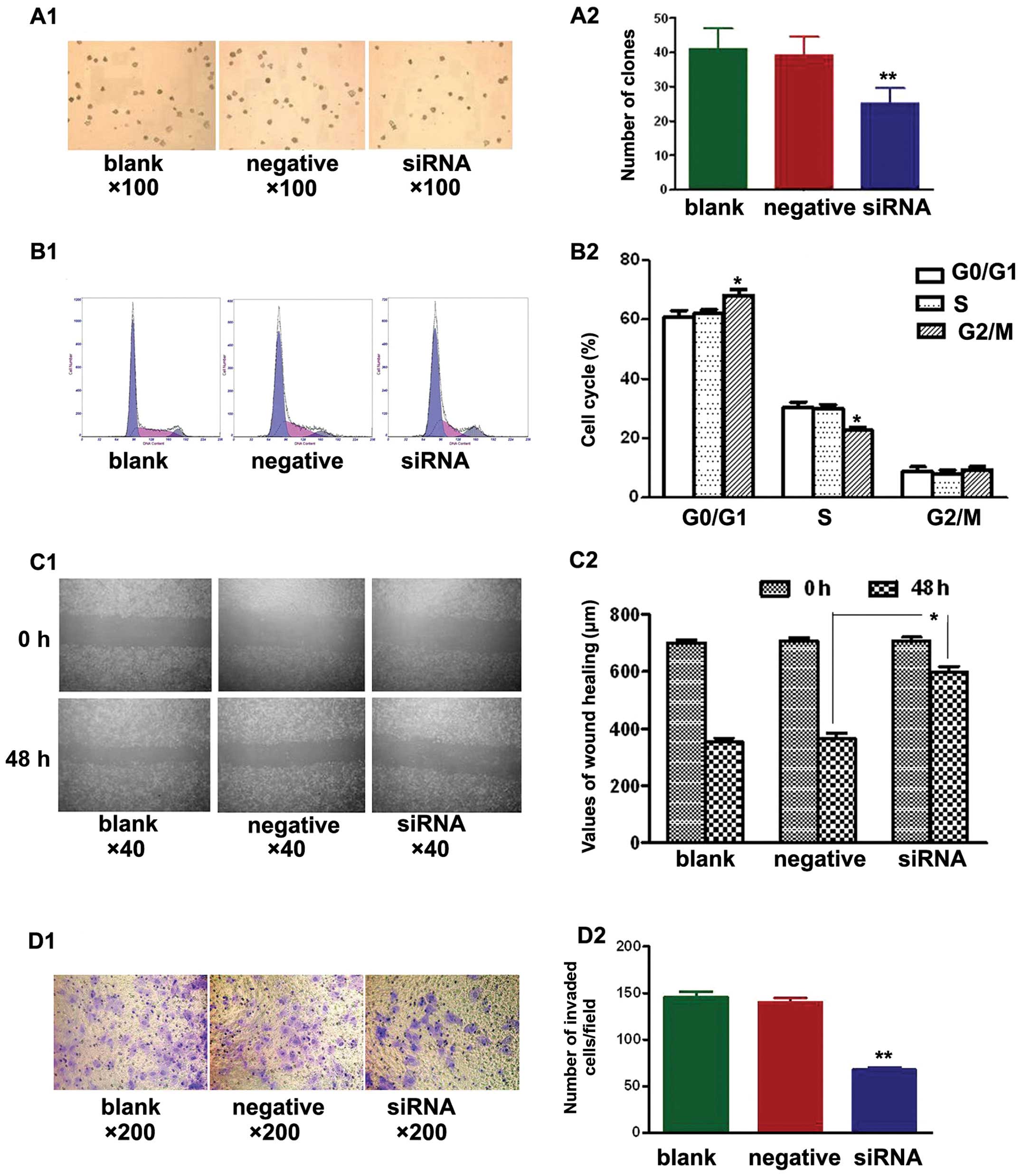
Soft agar colony formation assays were used to test
the effects of NEDD9-siRNA on the proliferation of A549 cells in
the three treatment groups. After culturing for 14 days, the
NEDD9-siRNA group showed significantly fewer clones compared with
the blank and negative control groups (P<0.01). No significant
difference was found between the blank and negative control group
(P>0.05) (Fig. 2A).
Additionally, cell cycle analysis by FCM revealed
that NEDD9-siRNA altered the cell cycle of A549 cells. The mean
values are shown in Fig. 2B. No
significant differences (P>0.05) were observed in the percentage
of cells at each cell cycle phase between the blank and negative
control group. The percentage of cells in the G0/G1 phase in the
siRNA group (68.0±2.1%) was significantly different (P<0.05)
than that of the blank (60.6±2.4%) and negative control groups
(62.0±1.4%). Similarly, a significant difference (P<0.05) was
noted in the percentage of cells in the S phase in the siRNA group
(22.6±1.1%) vs. the blank (30.5±1.7%) and negative control groups
(30.0±1.4%). However, no significant difference (P>0.05) in the
percentage of cells in the G2/M phase was observed in the siRNA
group (9.3±1.1%), relative to the blank (8.8±1.6%) and negative
control groups (8.1±1.3%). Silencing of NEDD9 increased the
percentage of cells at the G0/G1 phase and decreased the percentage
of cells at the S phase. These results suggest that siRNA treatment
arrested cells at the G1/S checkpoint and delayed S phase.
Silencing of the NEDD9 gene suppresses
A549 cell migration and invasion in vitro
A549 cells were seeded in 6-well plates and wounded
the next day. Images were captured at 0 h and 48 h after wounding.
Fig. 2C shows that cell migration
was significantly decreased in the siRNA group when compared to
that in the blank and negative control groups (P<0.05).
Transwell cell migration assays showed significantly fewer invading
cells in the siRNA group when compared to the number of invading
cells in the blank and negative control groups (Fig. 2D; P<0.01). The siRNA group had
68±10 invading cells compared with 148±23 in the blank control
group and 136±20 in the negative control group. These results
demonstrated that transfection with NEDD9 siRNA reduced the
migration and invasion of A549 cells.
NEDD9/GV118-RNAi suppresses tumor growth
in vivo
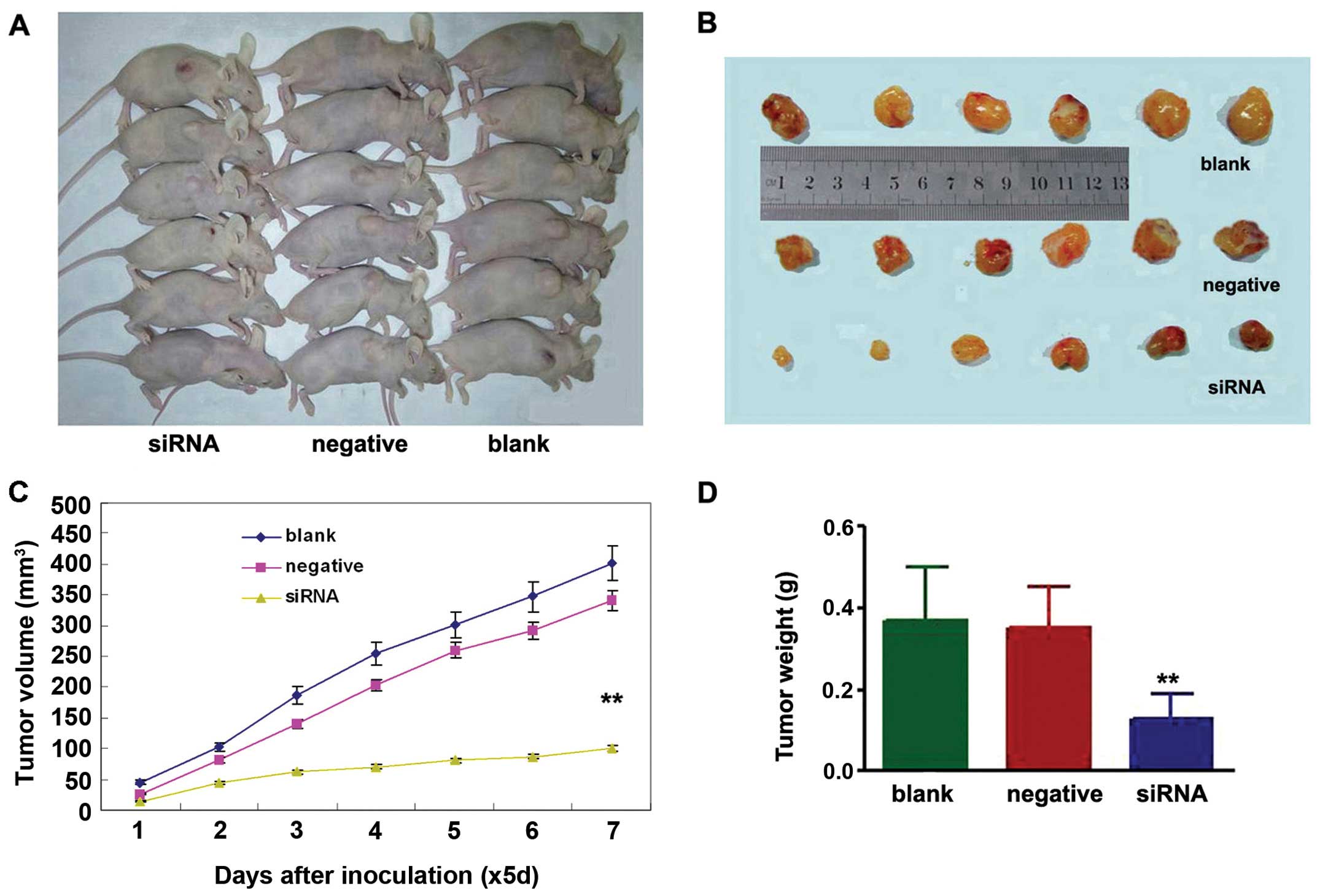
To determine whether knockdown of NEDD9 inhibits
lung tumor growth in vivo, the growth of lung adenocarcinoma
tumor xenografts in the three treatment groups was compared.
Measurement of tumor volumes began once subcutaneous tumors became
palpable and continued until tumors were excised on day 35. Then
tumor tissues were harvested and weighed. The results showed that
xenografts of the NEDD9- siRNA group had a lower tumor volume
(100.71±42.73 mm3) on day 35 than xenografts of the
negative (402.43±97.21 mm3) (P<0.01) or blank groups
(340.92±66.06 mm3) (P<0.01) (Fig. 3A–C). The average weight of tumors
derived from cell of the NEDD9-siRNA group (0.13±0.06 g) was
significantly reduced compared to the weights of tumors derived
from cells from the negative (0.35±0.10 g) (P<0.01) and blank
control groups (0.37±0.13 g) (P<0.01) (Fig. 3D).
Discussion
Human lung adenocarcinoma, a leading cause of
cancer-related mortality worldwide, exhibits features of
invasiveness and undergoes early metastasis. Human lung
adenocarcinoma is increasing in incidence and is a threat to human
health. Understanding the pathological mechanisms of lung
adenocarcinoma and identifying treatment targets are crucial.
CAS proteins mediate cell spreading (21,22)
and are important in driving cell migration (23,24).
NEDD9 is a member of the CAS protein family and an invasion-related
and metastasis-related gene found in many tumor types (8–12). Our
previous research showed that NEDD9 is overexpressed in lung
adenocarcinoma tissues (13). Kondo
et al (14) demonstrated
that tyrosine phosphorylation of NEDD9 is reduced by inhibition of
epidermal growth factor receptor protein (EGFR) in NSCLC cell
lines. They suggested that NEDD9 is a promising biomarker for NSCLC
prognosis, and its expression promotes NSCLC metastasis. Miao et
al (16) showed that NEDD9 was
overexpressed in 56.2% (59/105) of NSCLC samples compared to normal
lung tissues.
Although NEDD9 lacks known enzymatic function, it
contains many functional modules for protein interaction, leading
to its classification as a scaffolding protein. Since NEDD9 appears
to lack catalytic activity, it is not immediately promising as a
target for directed drug development, unless through agents
intended to disrupt protein-protein interactions, or through an
siRNA-based approach to deplete NEDD9 levels globally (7). siRNA-mediated knockdown of NEDD9 was
found to reduce the number of cells undergoing mitosis, and lead to
cleavage furrow regression and multinucleation (24,25).
In support of the development of NEDD9-directed drugs or siRNAs,
genetic NEDD9 knockout animals have relatively limited defects,
implying that loss of NEDD9 is well tolerated (26). RNA interference technology is an
important tool for studying gene function (27), but siRNA-mediated gene silencing is
maintained for only a short time (5–7 days) and usually
transfection efficiency is low (28). Therefore, vector-mediated RNA
interference is used. Recently, lentiviral technology has been
developed, with advantages such as low immunogenicity, wide range
of infection capabilities, and efficient integration into a host
cell genome to express stable siRNA. Lentiviral-mediated RNA
interference has become popular and can overcome the shortcomings
of chemically synthesized siRNA (29).
In the present study, we hypothesized that
downregulation of NEDD9 expression in the A549 cell line would
affect lung adenocarcinoma tumorigenesis and tumor biological
characteristics. To test this hypothesis, we used
lentivirus-mediated siRNA silencing to suppress NEDD9 expression.
RT-PCR and western blot analysis showed that NEDD9 mRNA and protein
levels were both reduced in the cultured A549 lung adenocarcinoma
cells after NEDD9 was silenced (Fig. 1C
and 1D).
MMP9, cyclin E and TIMP1 are important for cell
proliferation. We found that MMP9 and cyclin E were downregulated,
but TIMP1 was increased in the A549 cells when NEDD9 was knocked
down (Fig. 1E). This indicated that
lentivirus-mediated RNAi knockdown of NEDD9 inhibited cell
proliferation. Soft agar colony formation assays and FCM results
showed that the colony formation number was decreased and the cell
cycle was altered in the A549 cell line transfected with the
lentivirus-NEDD9-siRNA (Fig. 2A and
B). migration and in vitro invasion were also suppressed
(Fig. 2C and D). Our findings
showed that lentivirus-NEDD9-siRNA transfection of lung
adenocarcinoma cells markedly suppressed proliferation and
migration and invasion ability. Tumorigenicity was also inhibited
by lentivirus-mediated RNAi knockdown of NEDD9 expression in the
A549 cell lines in vivo. Tumor growth was greatly reduced in
the lentivirus-mediated NEDD9-siRNA transfected cell tumors while
negative siRNA-transfected xenografts and control grafts grew
aggressively in the mice (Fig.
3).
Taken together, our findings revealed that knockdown
of NEDD9 resulted in inhibition of proliferation and cell cycle
changes, and led to suppressed tumor cell migration and invasion.
Lentivirus-NEDD9-siRNA effectively inhibited human lung
adenocarcinoma cell growth in vivo and could have
therapeutic utility for human lung adenocarcinoma. However, our
results were based on a single cell line, and further research is
needed to determine the differential expression of NEDD9 in
different types of cells in vitro and in vivo.
Acknowledgements
The authors thank Dr P.J. Wang and his colleagues at
the Research Centre for Molecular Oncology of Zhengzhou University.
The present study was supported by a grant from the Science and
Technology Agency of Henan (grant no. 122300410363).
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Tuveson DA and Jacks T: Modeling human
lung cancer in mice: similarities and shortcomings. Oncogene.
18:5318–5324. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Law SF, Estojak J, Wang B, et al: Human
enhancer of filamentation 1, a novel p130Cas-Like docking protein,
associates with FAK, and induces pseudohyphal growth in
Saccharomyces cerevisiae. Mol Cell Biol. 16:3327–3337.
1996.PubMed/NCBI
|
|
4
|
Minegishi M, Tachibana K, Sato T, et al:
Structure and function of Cas-L, a 105-kD Crk-associated
substrate-related protein that is involved in β-1 integrin-mediated
signaling in lymphocytes. J Exp Med. 184:1365–1375. 1996.PubMed/NCBI
|
|
5
|
Singh M, Cowell L, Seo S, et al: Molecular
basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator
of invasion, apoptosis and cell cycle. Cell Biochem Biophys.
48:54–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O’Neill GM, Seo S, Serebriiskii IG, et al:
A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9.
Cancer Res. 67:8975–8979. 2007.PubMed/NCBI
|
|
7
|
Tikhmyanova N, Little JL and Golemis EA:
CAS proteins in normal and pathological cell growth control. Cell
Mol Life Sci. 67:1025–1048. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Minn AJ, Gupta GP, Siegel PM, et al: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Natarajan M, Stewart JE, Golemis EA, et
al: HEF1 is a necessary and specific downstream effector of FAK
that promotes the migration of glioblastoma cells. Oncogene.
25:1721–1732. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim M, Gans JD, Nogueira C, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SH, Xia D, Dubois RN, et al: Human
enhancer of filamentation 1 is a mediator of hypoxia-inducible
factor-1α-mediated migration in colorectal carcinoma cells. Cancer
Res. 70:4054–4063. 2010.PubMed/NCBI
|
|
12
|
Thaole B, Vu HA, Yasuda K, et al: Cas-L
was overexpressed in imatinib-resistant gastrointestinal stromal
tumor cells. Cancer Biol Ther. 8:683–688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang JX, Zhao GQ, Zhang GJ, et al:
Expression and clinical significance of NEDD9 in lung tissues. Med
Oncol. 29:2654–2660. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kondo S, Iwata S, Yamada T, et al: Impact
of the integrin signaling adaptor protein NEDD9 on prognosis and
metastatic behavior of human lung cancer. Clin Cancer Res.
18:6326–6338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng Y, Wang Y, Wang Z, et al: The
CRTC1-NEDD9 signaling axis mediates lung cancer progression caused
by LKB1 loss. Cancer Res. 72:6502–6511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miao Y, Li AL, Wang L, et al:
Overexpression of NEDD9 is associated with altered expression of
E-cadherin, β-catenin and N-cadherin and predictive of poor
prognosis in non-small cell lung cancer. Pathol Oncol Res.
19:281–286. 2013.PubMed/NCBI
|
|
17
|
Sen GL and Blau HM: A brief history of
RNAi: the silence of the genes. FASEB J. 20:1293–1299. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang JX, Zhao GQ, Zhang GJ, et al:
Construction and characterization of a eukaryotic expression vector
for small interfering RNA targeting the NEDD9 gene. Int J Mol Med.
30:1343–1348. 2012.PubMed/NCBI
|
|
19
|
Tanabe H, Yagihashi A, Tsuji N, et al:
Expression of survivin mRNA and livin mRNA in non-small-cell lung
cancer. Lung Cancer. 46:299–304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun BS, Dong QZ, Ye QH, et al:
Lentiviral-mediated miRNA against osteopontin suppresses tumor
growth and metastasis of human hepatocellular carcinoma.
Hepatology. 48:1834–1842. 2008. View Article : Google Scholar
|
|
21
|
Singh MK, Dadke D, Nicolas E, et al: A
novel Cas family member, HEPL, regulates FAK and cell spreading.
Mol Biol Cell. 19:1627–1636. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fashena SJ, Einarson MB, Golemis EA, et
al: Dissection of HEF1-dependent functions in motility and
transcriptional regulation. J Cell Sci. 115:99–111. 2002.PubMed/NCBI
|
|
23
|
Klemke RL, Leng J, Cheresh DA, et al:
CAS/Crk coupling serves as a ‘molecular switch’ for induction of
cell migration. J Cell Biol. 140:961–972. 1998.
|
|
24
|
Pugacheva EN and Golemis EA: The focal
adhesion scaffolding protein HEF1 regulates activation of the
Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol.
7:937–946. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dadke D, Jarnik M, Pugacheva EN, et al:
Deregulation of HEF1 impairs M-phase progression by disrupting the
RhoA activation cycle. Mol Biol Cell. 17:1204–1217. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seo S, Asai T, Saito T, et al:
Crk-associated substrate lymphocyte type is required for lymphocyte
trafficking and marginal zone B cell maintenance. J Immunol.
175:3492–3501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Elbashir SM, Harborth J, Lendeckel W, et
al: Duplexes of 21-nucleotide RNAs mediate RNA interference in
cultured mammalian cells. Nature. 411:494–498. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Echeverri CJ and Perrimon N:
High-throughput RNAi screening in cultured cells: a user’s guide.
Nat Rev Genet. 7:373–384. 2006.
|
|
29
|
Mofat J, Grueneberg DA, Yang X, et al: A
lentiviral RNAi library for human and mouse genes applied to an
arrayed viral high-content screen. Cell. 124:1283–1298. 2006.
View Article : Google Scholar : PubMed/NCBI
|