Introduction
Hepatocellular carcinoma (HCC) is a malignancy with
dysregulated differentiation (1).
Differentiation-related genes in HCC have been enumerated; for
example, the well-known biomarker α-fetoprotein (AFP), only
expressed during early stages of liver development, is always
highly expressed in HCC. Differentiation therapy has been quite
successful for various malignancies, particularly acute
promyelocytic leukemia (2).
However, effective differentiation therapy is lacking for HCC
(3,4), partly due to the limited knowledge of
dysregulated differentiation in HCC (5).
During normal liver development, key transcription
factors, including hepatocyte nuclear factor 4α (HNF4α), HNF1α,
HNF1β, CCAAT/enhancer binding protein α (C/EBPα) and C/EBPβ,
control differentiation (6). HNF4α
plays a critical role in hepatocyte differentiation (7) and controls the expression of more than
40% of hepatocyte genes. Recently, the critical roles of HNF4α in
the dysregulated differentiation and carcinogenesis in HCC have
been identified (8,9). A nude mouse model of HCC showed that a
recombinant adenovirus carrying HNF4α potently promoted the
differentiation of HCC into normal hepatocytes and suppressed
tumorigenesis (4).
CXCR7 was formerly known as RDC1 or orphan receptor
since its ligands were unknown. Knowledge concerning CXCR7
increased after the functional ligands SDF-1α (or CXCL12) and ITAC
(or CXCL11) were found (10,11).
CXCR7 is a G-protein-coupled seven-transmembrane receptor. However,
it also mediates β-arrestin-biased signaling (12). Functionally, CXCR7 has been shown to
be involved in cardiac development (13). In addition, evidence suggests links
between CXCR7 and tumor proliferation or invasion (14). Moreover, our previous research
demonstrated the critical role of CXCR7 in HCC (15,16),
which is related to differentiation (unpublished data).
Additionally, during the differentiation of embryonic stem cells,
the epigenetic suppression of CXCR7 by SUZ12 is lost, and the
expression of CXCR7 can be increased 20-fold (17), suggesting a critical role for CXCR7
in stem cell differentiation. Therefore, we directed our attention
to the potential role of CXCR7 in the dysregulation of the
differentiation in HCC.
Here, CXCR7 was identified to be closely associated
with the differentiation of HCC, and a close relationship between
CXCR7 and HNF4α was found, which was confirmed by the
immunohistochemical (IHC) staining of tissue microarrays (TMAs).
High CXCR7 levels and low HNF4α levels were correlated with poor
survival. Furthermore, ligand activation, inhibition assays, and
RNA interference (RNAi) demonstrated that the regulation of HNF4α
by CXCR7 was mitogen-activated protein kinase (MAPK)-dependent.
Materials and methods
Reagents and antibodies
Oncostatin M (OSM), SDF-1α and CXCL11 were purchased
from Peprotech (Rocky Hill, NJ, USA). U0126 and dimethyl sulfoxide
(DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Monoclonal antibodies used in the flow cytometric
analysis were mouse anti-human monoclonal antibodies (mAbs)
CD44-PE, CD133-APC, CD90-PE, CD24-FITC, the IgG-PE isotype, the
IgG-APC isotype and the IgG-FITC isotype (all purchased from
Miltenyi Biotec, Bergisch Gladbach, Germany). Antibodies used for
immunofluorescence, immunoblotting and immunohistochemistry were as
follows: mouse anti-human monoclonal CD90 (Abcam, Cambridge, MA,
USA), rabbit anti-human polyclonal CD133 (Abnova, Walnut, CA, USA),
mouse anti-human monoclonal CD44, rabbit anti-human albumin (ALB),
phospho-p44/42 MAPK (Erk1/2), and rabbit anti-human HNF4α mAb (Cell
Signaling Technologies, Danvers, MA, USA), rabbit anti-human
polyclonal CD24, rabbit anti-human β-actin mAb (Epitomics,
Burlingame, CA, USA), rabbit anti-human polyclonal transferrin (TF)
(Proteintech Group Inc., Chicago, IL, USA), rabbit anti-human AFP
mAb, rabbit anti-human C/EBPα and C/EBPβ mAb, Erk2/p42 MAPK
antibody (Epitomics), rabbit anti-human CXCR7 IgG (Novus
Biologicals, Littleton, CO, USA), and horseradish
peroxidase-conjugated goat anti-rabbit IgG F(ab’)2 antibodies
(Jackson ImmunoResearch, West Grove, PA, USA).
Cell lines and culture
Human HCC cell lines with elevated lung metastatic
potential (MHCC97L, MHCC97H, and HCCLM3) were established at the
Liver Cancer Institute of Fudan University. The human HCC cell
lines with low metastatic potential were SMMC-7721 (established at
the Second Military Medical University), Huh7 and HepG2 (obtained
from the American Type Culture Collection). These cell lines were
cultured in high-glucose DMEM (Gibco-BRL, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (Hyclone, Logan, UT,
USA).
To induce differentiation, 10 to 40 ng/ml OSM and 1
or 5 μM dexamethasone (Dex) were used. Chemical differentiation
inducer DMSO was also used to help establish a model of
differentiation induction. For the CXCR7 stimulation assay, 100
ng/ml recombinant human SDF-1α and 200 ng/ml CXCL11 were used. For
the inhibition assay, cells were serum-starved for at least 8 h
before the MEK1/2 inhibitor U0126 (10 μM) was added.
Flow cytometry
The expression levels of cancer stem cell
(CSC)-related markers were determined by flow cytometry. Briefly,
tumor cells were grown to 80% confluency. After trypsin digestion,
the cells were re-suspended in medium at a concentration of
1×106 cells/ml and incubated with antibodies against
CD44, CD133, CD90 and CD24 (diluted 1:11) at 4°C for 15 min. After
washing 3 times with PBS, the cells were analyzed using a FACS flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Immunofluorescence
Cell surface expression of CSC-related markers and
CXCR7 was also determined by immunofluorescence analysis. Cells
were grown on glass coverslips to 40–50% confluency and fixed,
permeabilized, blocked, and incubated with primary monoclonal
antibodies overnight at 4°C. Slides were washed and incubated with
an anti-mouse or an anti-rabbit Cy3-conjugated secondary antibody
(Jackson ImmunoResearch). Cells were counterstained with
4–6-diamidino-2-phenylindole to visualize cell nuclei and for
inspection by fluorescence microscopy (Olympus).
Western blot analysis
Western blotting was performed according to the wet
transfer protocol using the Bio-Rad Transfer Cell System (Bio-Rad,
Ontario, Canada). Analyses of protein expression were performed
according to the manufacturer’s instructions. Images were examined
using Image Lab Software® (Bio-Rad).
RNA interference
Small interfering RNAs (siRNAs; GenePharma Corp.,
Shanghai, China) demonstrated to be effective for the knockdown of
CXCR7 expression (15) were used.
siRNA transfection of the cells was performed using the
Lipofectamine 2000 protocol (18).
A negative control siRNA, 5′-UUC UCC GAA CGU GUC ACG UTT-3′, was
also used. A FAM-labeled negative control siRNA was used to monitor
transfection efficiency.
Gene microarray
The Oligo GEArray Human Chemokine and Chemokine
Receptors Chip (SuperArray Bioscience, Frederick, MD, USA) was used
to compare profiles of the HCCLM3, MHCC97-L, and SMMC-7721 cells
according to the manufacturer’s protocol. Total-RNA was extracted,
isolated, and purified according to the TRIzol reagent (Invitrogen
Corp., Carlsbad, CA, USA) protocol. GEArray® Analyzer
Software (SuperArray Bioscience) was used for data analysis. The
microarray analysis was performed twice with similar results.
Patients and follow-up
Ethical approval was obtained from the Zhongshan
Hospital Research Ethics Committee, and informed consent was
obtained from each patient. Data from 112 patients were retrieved
from the prospectively designed database. These patients underwent
hepatectomy by the same surgical team from January 2000 to May
2004. The hepatectomy for HCC was carried out as described
previously (19). All patients were
classified as Child-Pugh A, and all tumors were identified as HCC
by histological analysis.
Regular follow-up procedures in our clinic include
the following: AFP assay and liver ultrasonography every 3 months
during the first year and every 6 months thereafter; and magnetic
resonance imaging or computed tomography scanning after 1 month and
every 6 months thereafter. Chest computed tomography scanning is
regularly used to identify lung metastasis. Lung metastasis was
confirmed by biopsy through endoscopy or histology after partial
pulmonary resection. Until May 2009, lung metastasis was found in
56 patients. Ten patients with resectable lung metastases received
partial resection of the lung.
TMA and IHC
Hematoxylin and eosin-stained slides were screened
for optimal tumor content and tissue adjacent to the tumor (TAT)
with a distance of 2 cm. The TMA was constructed in accordance with
standard procedures (20) based on
112 tumor tissues and 46 TATs. Two cores were taken from each
formalin-fixed, paraffin-embedded HCC sample by using punch cores
that measured 1.0 mm in diameter from the center of the tumor foci
and TAT. A two-step method of IHC including heat-induced
antigen-retrieval procedure was performed as previously described
(20). Detection without the
primary antibody was considered as the negative control.
Scoring and categorization of CXCR7
expression
CXCR7 expression was defined by staining intensity
and the percentage of positive tumor cells as described previously
(21). Two pathologists observed
the results independently. We simplified the assay results of CXCR7
into CXCR7Low (weak staining) and CXCR7High
(strong staining). Similarly, HNF4α expression was categorized as
HNF4αLow or HNF4αHigh, except that
HNF4αLow included the negatively stained population due
to the generally weak staining.
Statistical analysis
When two groups of cells or tissues were compared,
analysis was performed with the Student’s t-test. The Pearson
χ2 test was used to compare qualitative variables in the
clinical pathology analysis. When expected sample values were
<5, Fisher’s exact test was used. Spearman’s rank test was used
to detect the correlation between variables. Overall survival (OS),
time to progression (TTP), and time to extrahepatic metastasis were
observed in the survival analysis. OS was calculated from the date
of hepatectomy to the date of death regardless of cause. TTP was
calculated from the date of hepatic resection to the date of
recurrence. Time to lung metastasis was calculated from the date of
hepatectomy to the date of lung metastasis with definite clinical
diagnosis. The patients lost in the follow-up and the patients who
had not achieved the desired results at the end of this study were
recognized as censored cases. The Kaplan-Meier method was used to
describe the survival curves, and the log-rank test was used to
compare survival distributions between groups. The Breslow test was
also used when survival curves indicated greater differences during
the early follow-up period. All P-values were obtained using
two-tailed tests and the statistical significance was set at 0.05.
Statistical analyses were carried out by SPSS 18.0 Software (SPSS
Inc., Chicago, IL, USA).
Results
Screening of the HCC cell lines for
differentiation induction
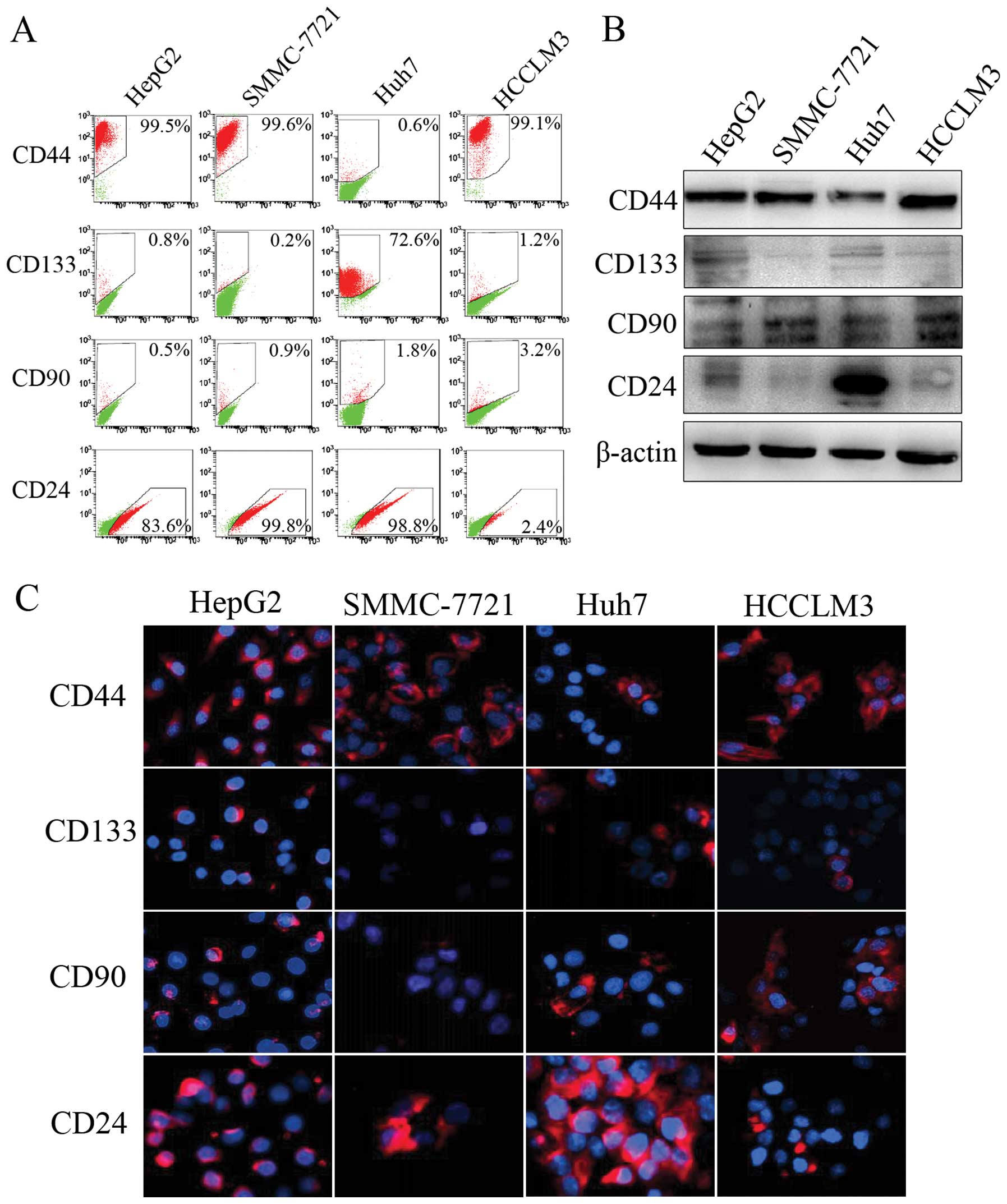
Due to the highly heterogenetic character of HCC, we
first detected HCC-related stem cell biomarkers in four HCC cell
lines (HepG2, SMMC-7721, Huh7 and HCCLM3) with different
characteristics and backgrounds. Flow cytometric analysis indicated
that Huh7 cells had the highest ratios of CD133 (72.6%) and CD24
(98.8%), which was confirmed by western blot and immunofluorescence
analyses (Fig. 1). HCCLM3 cells had
the highest CD90-positive ratio (3.2%) and a CD44-positive ratio of
>90%. Since these CSC-related markers have been linked to HCC
progression and dysregulated differentiation potential (22–24),
Huh7 and HCCLM3 cells were selected for further analysis.
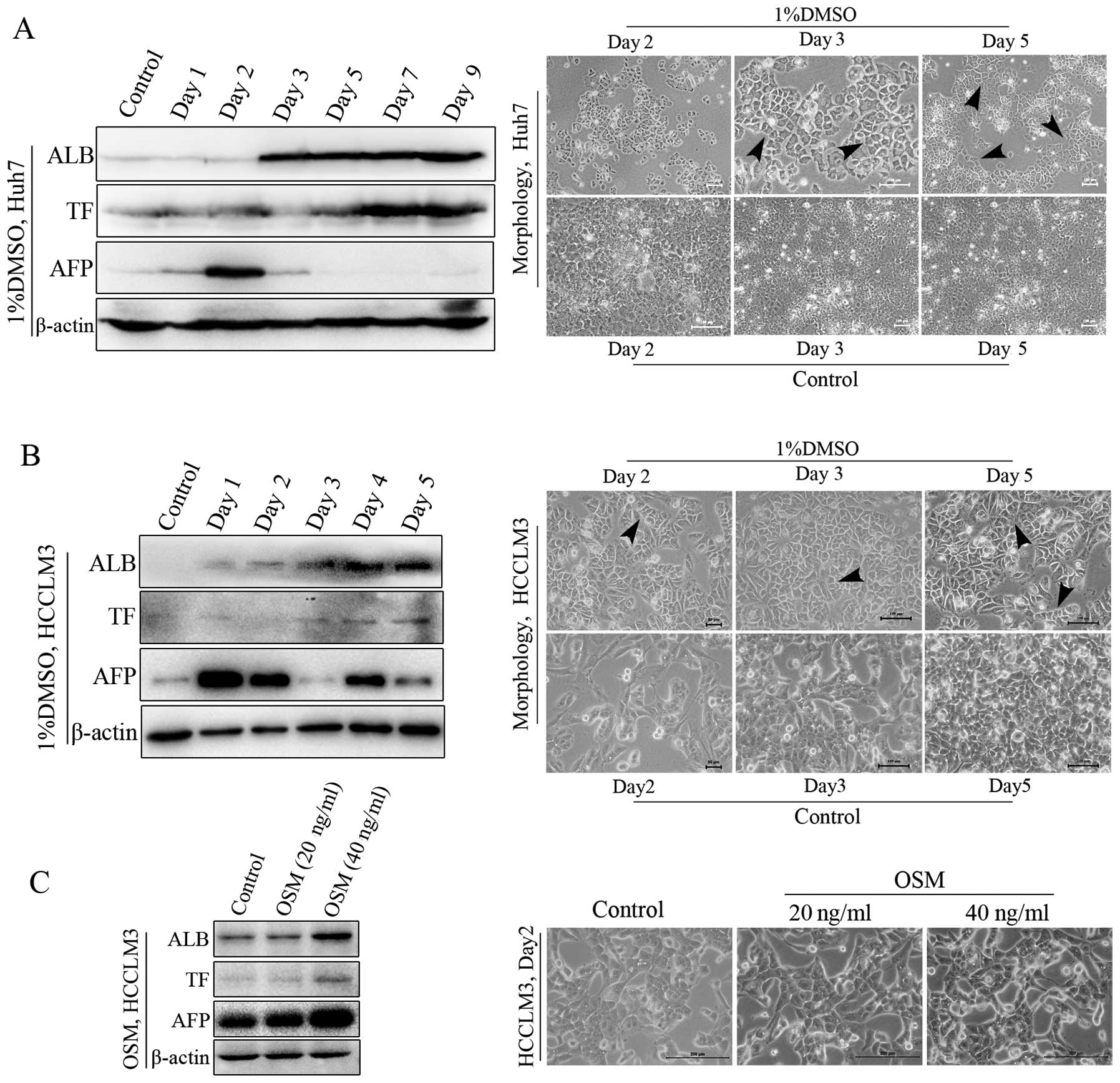
HCC cells differentiate into
hepatocyte-like cells upon induced differentiation
The differentiation induction model for HCC was
established using potential differentiation inducers. DMSO was used
as a chemical inducer of differentiation. OSM, which has the
ability to maintain the maturation and differentiation capacity of
hepatocytes (25), was also used.
Both in the Huh7 and in the HCCLM3 cells, DMSO showed a strong
ability to induce HCC differentiation into normal hepatocytes,
based on hepatocyte differentiation markers or elevated expression
of plasma proteins ALB and TF, and the typical morphological
features of differentiated hepatocytes (Fig. 2A and B). The typical morphology of
differentiation was maintained from day 2 until late stages of the
induction process. Meanwhile, results from HCCLM3 cells revealed
that OSM had the ability to induce elevated expression of ALB and
TF (Fig. 2C). However, no typical
morphological features were noted, which indicated the main role of
OSM in maintaining the maturation of cells. Dex was unable to
induce differentiation when used alone (data not shown). In
addition, AFP was also increased early during induction, which was
consistent with previously reported results (26). Next, AFP levels decreased gradually
accompanied by the elevated ALB and TF, and the occurrence of
hepatocyte-like morphology.
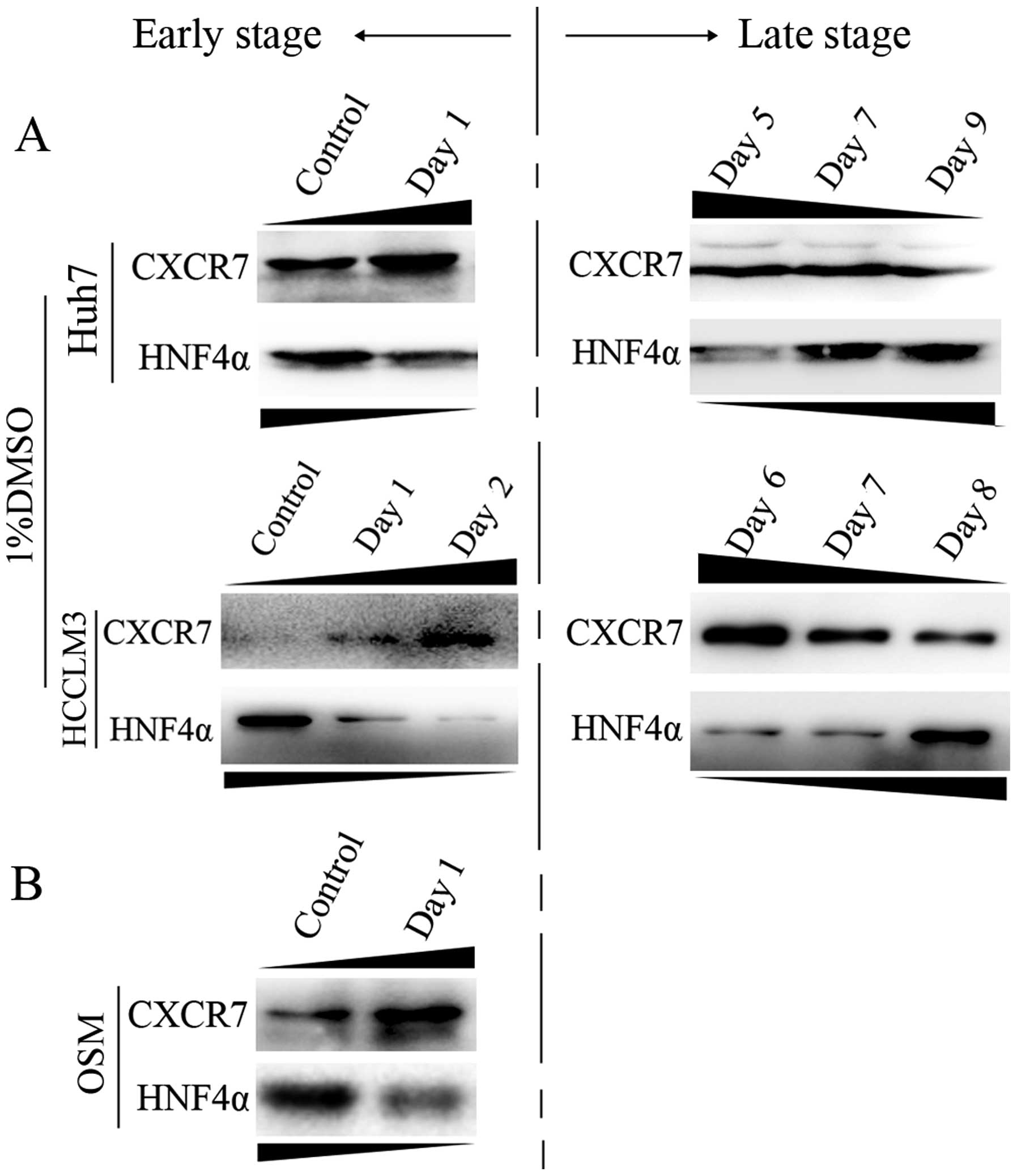
CXCR7 is inversely correlated with HNF4α
upon induced differentiation
Based on the model of HCC differentiation in
vitro, we further observed CXCR7 levels following induced
differentiation by 1% DMSO and 40 ng/ml OSM. After DMSO treatment,
CXCR7 expression was increased early and was decreased during the
late stages of differentiation, which showed an inverse expression
trend to HNF4α, which has a critical role in liver-specific gene
expression. As shown in Fig. 3A,
during early stages of induced differentiation, CXCR7 was elevated
and the HNF4α level was decreased in both Huh7 and HCCLM3 cells.
Whereas during the late stage, CXCR7 levels were decreased and
HNF4α levels were elevated. In addition, CXCR7 levels were elevated
while the opposite trend was observed for HNF4α during the early
stage of differentiation in response to 40 ng/ml OSM treatment
(Fig. 3B).
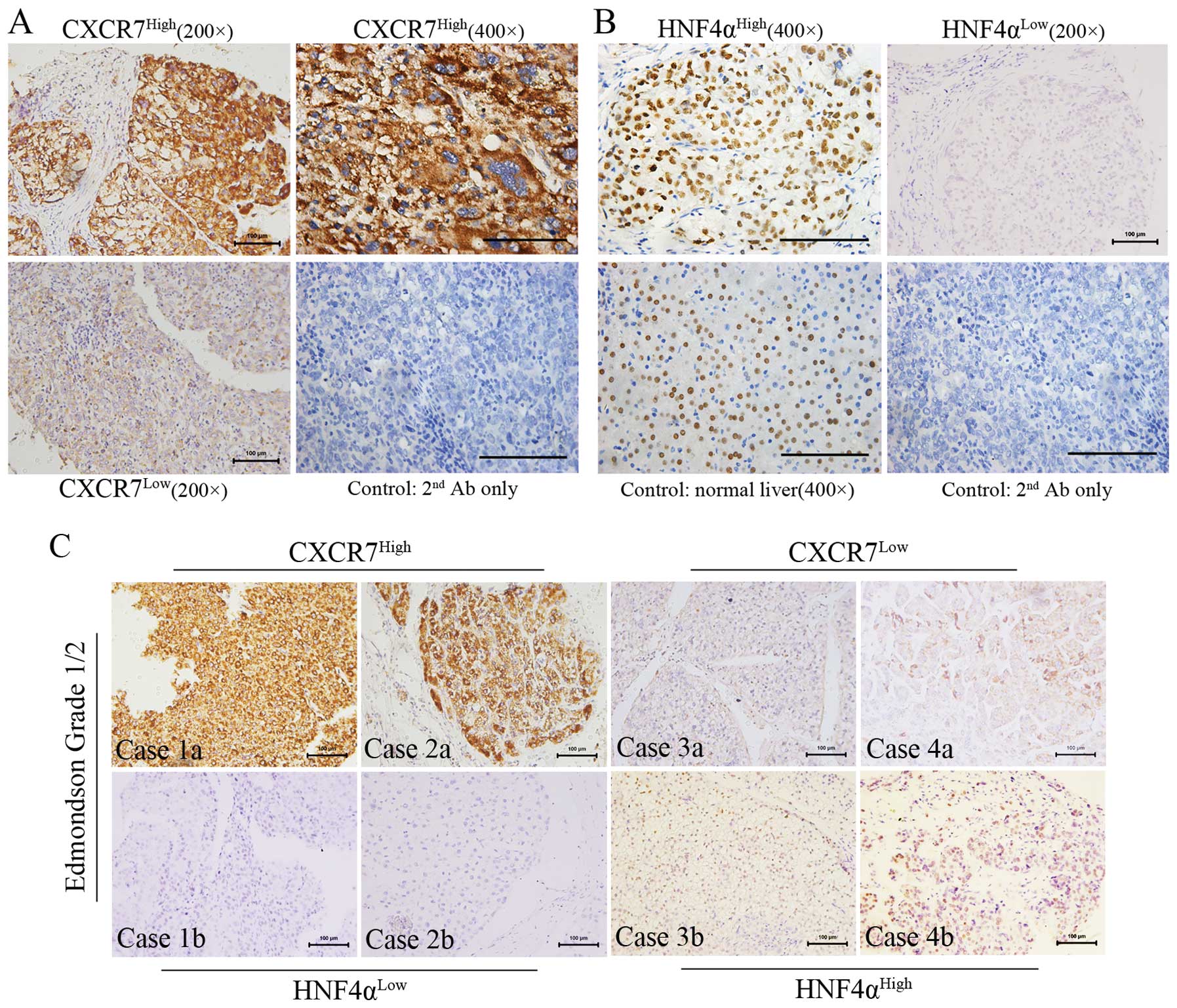
High expression of CXCR7 is correlated
with decreased HNF4α expression
Furthermore, IHC analysis of the TMA was performed
to confirm the relationship between CXCR7 and HNF4α.
Immunopositivity for CXCR7 was mainly observed at the membrane or
in the cytoplasm of the HCC cells, whereas for HNF4α it was
expressed mainly in the nucleus. The expression intensity and
localization of CXCR7 and HNF4α are shown in Fig. 4A and B, respectively. Immunostaining
of normal hepatocytes indicated the specificity and selectivity of
anti-HNF4α. Stratum analysis based on cell differentiation
(Edmondson grade 1/2; n=57) indicated that CXCR7 was negatively
correlated with HNF4α expression (Spearman’s rho=−0.296; P=0.025).
Among the tumors with low HNF4α expression (n=48), 62.5% were
CXCR7High, whereas only 22.2% of the tumors with high
HNF4α expression (n=9) were CXCR7High. Furthermore,
immunostaining analyses indicated that the distribution and
expression levels of CXCR7 were inversely associated with those of
HNF4α on a relatively well-differentiated background (Fig. 4C). In addition,
CXCR7HighHNF4αLow tumors tended to be larger
(Fisher’s exact test, P=0.002), suggesting a relationship between
differentiation and proliferation of HCC (Table I).
 | Table IClinicopathological factors and
combined CXCR7 and HNF4α expression. |
Table I
Clinicopathological factors and
combined CXCR7 and HNF4α expression.
| No. of
patients | |
|---|
|
| |
|---|
| Clinicopathological
characteristics |
CXCR7High/HNF4αLow
(n=30) |
CXCR7Low/HNF4αHigh
(n=7) | P-value |
|---|
| Age, (years) | | | 1.000 |
| ≤60 | 24 | 6 | |
| >60 | 6 | 1 | |
| Gender | | | 0.570 |
| Male | 26 | 7 | |
| Female | 4 | 0 | |
| HBsAg | | | 0.571 |
| Negative | 6 | 0 | |
| Positive | 24 | 7 | |
| Cirrhosis | | | 0.327 |
| Absent | 24 | 4 | |
| Present | 6 | 3 | |
| AFP, μg/l | | | 0.360 |
| ≤20 | 7 | 3 | |
| >20 | 23 | 4 | |
| Tumor size
(cm) | | | 0.002 |
| ≤5 | 6 | 6 | |
| >5 | 24 | 1 | |
| No. of tumor
nodules | | | 0.213 |
| Single | 21 | 3 | |
| Multiple | 9 | 4 | |
| Tumor
encapsulation | | | 0.217 |
| Well
encapsulated | 17 | 6 | |
| Poorly
encapsulated | 13 | 1 | |
| Microvascular
invasion | | | 0.674 |
| Negative | 16 | 5 | |
| Positive | 14 | 2 | |
| Portal lymphatic
status | | | 1.000 |
| No | 28 | 7 | |
| Yes | 2 | 0 | |
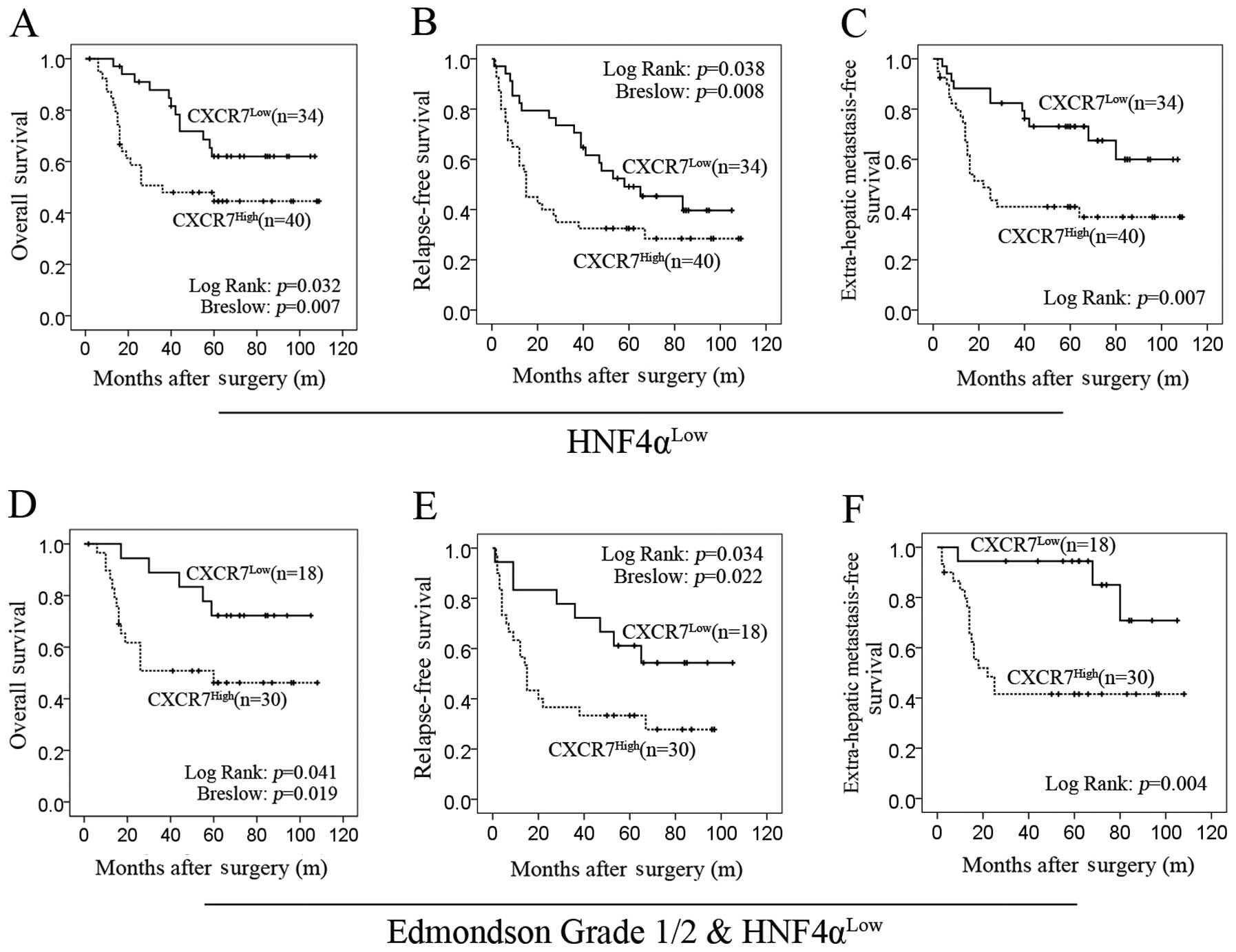
High expression of CXCR7 predicts poor
prognosis when combined with HNF4α
There was a statistically significant difference in
OS between the CXCR7High and CXCR7Low groups
on the HNF4αLow background (log-rank test; P=0.032). In
particular, there was a strong difference during the early stages
of follow-up (Breslow test; P=0.007; Fig. 5A). The median OS of
CXCR7High patients was much shorter than that of
CXCR7Low patients (36 vs. 81.2 months). Similarly, on
the HNF4αLow background, the TTP of the
CXCR7High patients was significantly shorter than that
for the CXCR7Low patients (log-rank test; P=0.038),
particularly during the early stages of follow-up (Breslow test;
P=0.008; Fig. 5B). In addition, the
time that relapsed before observing lung metastasis was shorter for
CXCR7High patients than for CXCR7Low patients
(log-rank test; P=0.007) (Fig. 5C).
In addition, in patients with high HNF4α expression in tumors,
there was no significant difference between CXCR7High
patients and CXCR7Low patients (data not shown).
Since there was a greater correlation between CXCR7
and HNF4α in relatively well-differentiated HCC, the prognostic
value of CXCR7 was further evaluated in patients with Edmondson
grade 1/2 HCC (n=48) (Fig. 5D and
E). We found that CXCR7 expression had a strong prognostic
value in the time-to-metastasis analysis (log-rank test; P=0.004;
Fig. 5F).
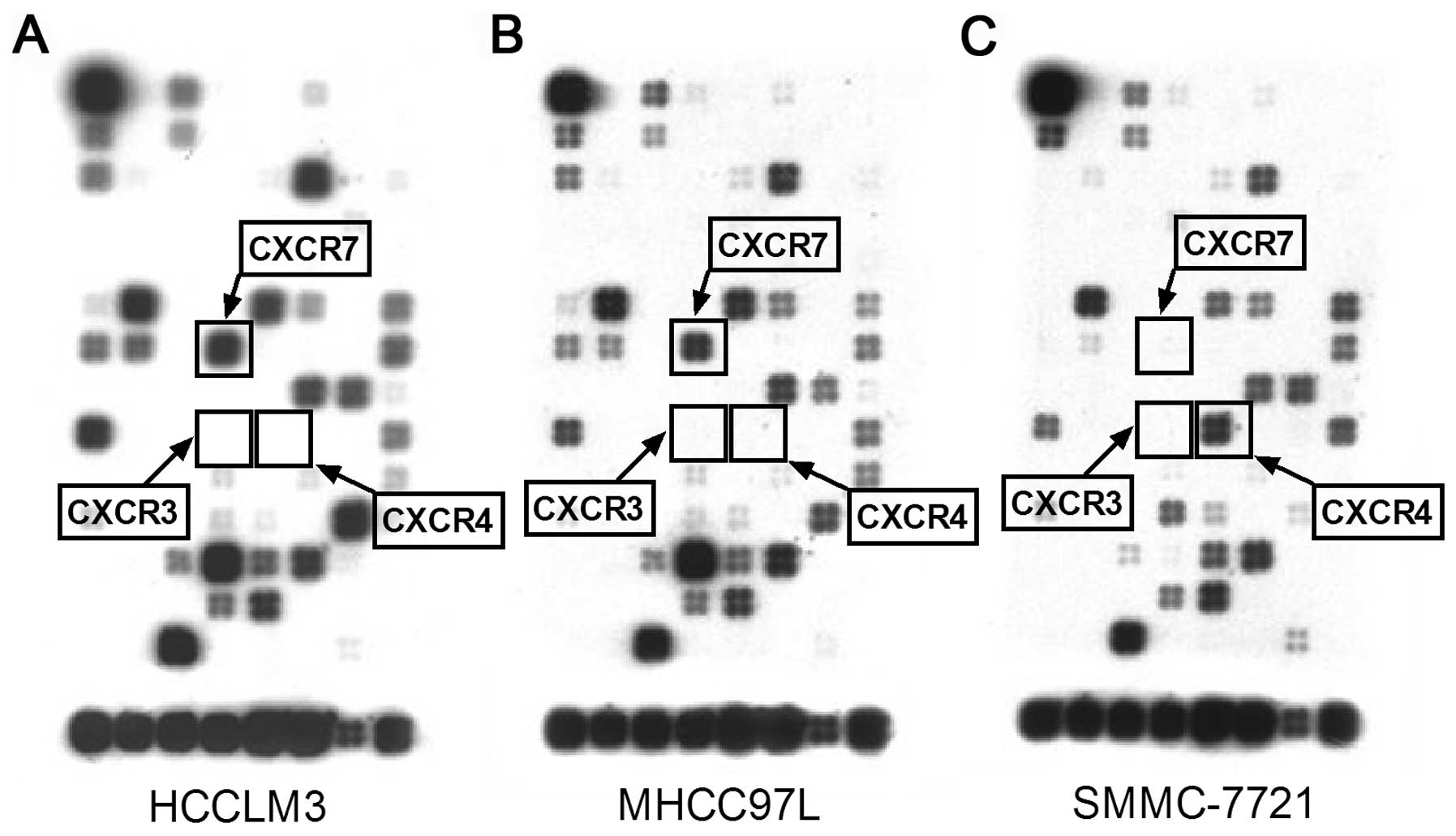
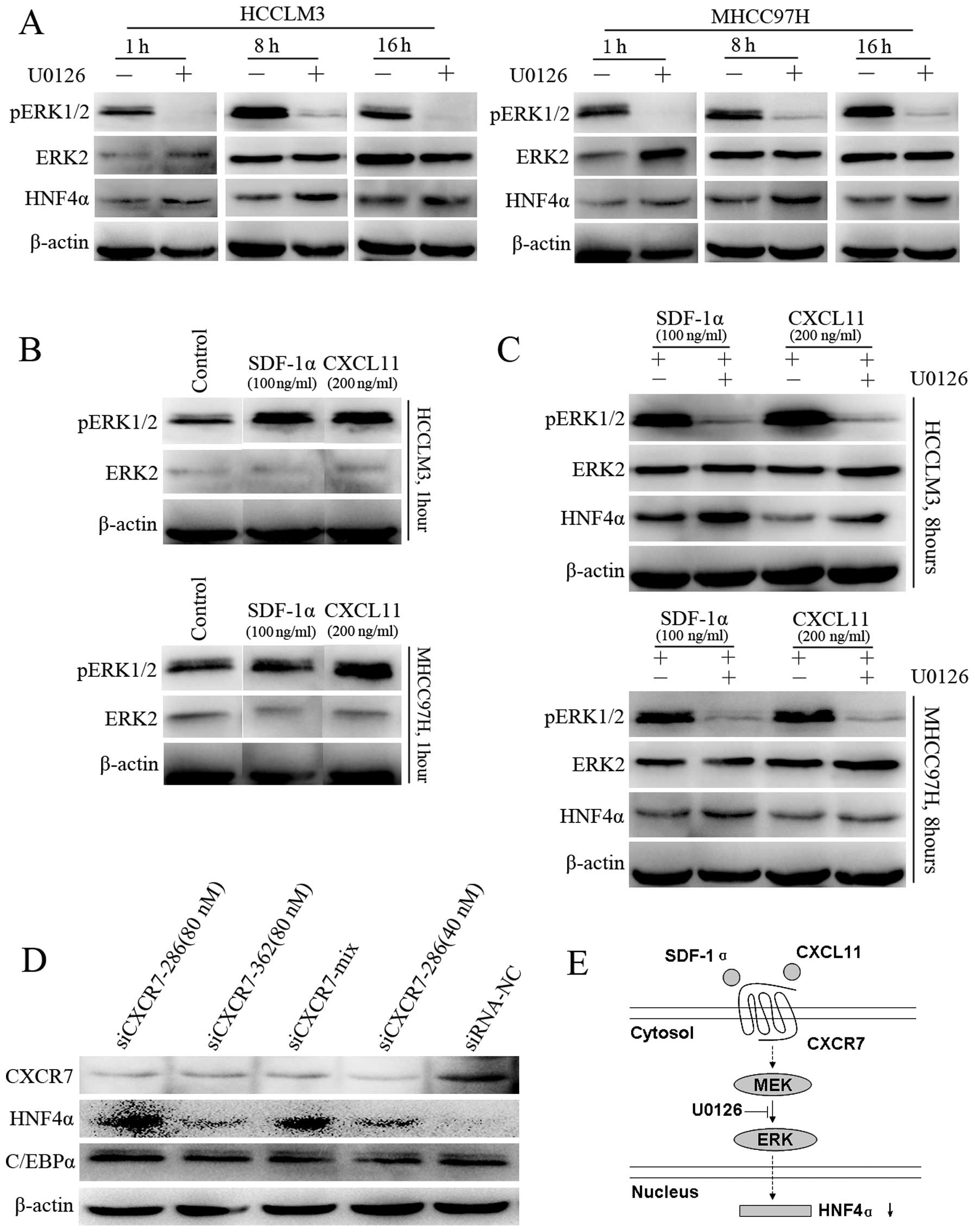
Activated CXCR7 suppresses HNF4α through
ERK-dependent signaling
Since CXCR7 is an atypical G-protein-coupled
receptor that has been reported to mediate extracellular regulated
protein kinase (ERK) signaling, we explored the possibility of a
CXCR7-ERK-HNF4α pathway. Gene chip analysis indicated that HCCLM3
cells highly expressed CXCR7. CXCR4 and CXCR3, however, were nearly
undetectable (Fig. 6). Therefore,
we utilized HCCLM3 cells as the
CXCR7+CXCR4−CXCR3− model to
observe CXCR7 signaling stimulated by SDF-1α and CXCL11.
Phosphorylation of MAPKs ERK1/2 was decreased at 1, 8 or 16 h, and
was accompanied by increased HNF4α expression (Fig. 7A), which indicated suppression of
the ERK pathway by HNF4α. Then, following stimulation with SDF-1α
or CXCL11, elevated phosphorylation of ERK1/2 was observed
(Fig. 7B). Meanwhile, western
blotting indicated elevated HNF4α levels after exposure to U0126
(Fig. 7C), which indicated that the
use of U0126 abrogated the inhibitory effect of CXCR7 ligands on
HNF4α expression. Furthermore, after downregulating CXCR7 in the
HCCLM3 cells, HNF4α was increased in all positive siRNA groups
(Fig. 7D). However, another
hepatic-enriched nuclear factor C/EBPα was not obviously affected.
Another cell line, MHCC97H, with the same genetic background as
HCCLM3 and also expressing high levels of CXCR7 and extremely low
levels of CXCR4 and CXCR3 was used to confirm these results. The
results with this cell line were similar to those using the HCCLM3
cell line.
Discussion
As a chemokine receptor, CXCR7 plays roles in crest
cell movement in development. Similarly, the role of CXCR7 in
invasion and metastasis has been demonstrated in tumors. However,
as an atypical G-protein-coupled receptor, other functions of CXCR7
have just begun to be unveiled. Evidence suggests the potential
role of CXCR7 during the differentiation of embryonic stem cells
(17). In the present study, the
close correlation between CXCR7 and differentiation of HCC was
found, including its opposite expression pattern with HNF4α upon
induced differentiation. Moreover, the relationship between CXCR7
and HNF4α was confirmed by histological analysis of HCC samples
after hepatic resection. Furthermore, high CXCR7 expression levels
were closely correlated with poor survival and extrahepatic
metastasis in tumors with low HNF4α expression. These findings
strongly suggest a critical role for CXCR7 in the differentiation
of HCC.
Differentiation therapy is a useful therapeutic
strategy for malignancy. However, effective differentiation therapy
is lacking for HCC. In this study, a new pathway mediated by CXCR7
in de-differentiated HCC was demonstrated, as summarized in
Fig. 7E. Although the screened HCC
cell lines have distinct genetic backgrounds and different CSC-like
biomarkers, the typical differentiation phenotype can be observed
in each, indicating that the CXCR7-MAPK-HNF4α cascade is the
general pathway in the differentiation of HCC. In addition,
adenoviral targeting of HNF4α has achieved success in mouse models
of HCC (4). Therefore, the
CXCR7-MAPK-HNF4α pathway discovered in this study represents a
promising target for differentiation therapy of HCC.
In this study, the suppression of HNF4α by CXCR7
signaling was ERK-dependent, which was inhibited by a specific MEK
inhibitor. Similar to our finding, the MAPK pathway has been found
to control HNF4α directly in hepatoma cells (27). Moreover, mucroporin-M1 was found to
selectively activate MAPK signaling and lead to the down-regulation
of HNF4α expression (28). On the
other hand, as an atypical G-protein-coupled receptor, CXCR7 has
been shown to mediate β-arrestin-biased signaling. As a
polyfunctional adaptor molecule, β-arrestin can mediate multiple
downstream pathways (29).
Functionally, CXCR7 has been shown to activate MAPK through
β-arrestin. Meanwhile, it is well known that the MAPK pathway is
one of the main pathways to control differentiation (30). Therefore, β-arrestin may participate
in the CXCR7-ERK-HNF4α signaling cascade, which warrants further
study.
In addition to our finding that CXCR7 controlled
HNF4α expression, HNF4α can also be modified at the
post-translational level (31).
Additionally, cyclin D1 inhibits the function of HNF4α (32), whereas GSK3β promotes the function
of HNF4α (33). In addition, TGFβ
has been found to impair the efficiency of HNF4α through GSK3β
inactivation (34). Recently, the
Wnt/β-catenin pathway was reported to converge with HNF4α-driven
transcription in liver zone specification (35). However, unlike these previous
reports, our findings clearly suggest a signaling pathway involved
in the differentiation of HCC.
The best proof of principle for differentiation
therapy has been the treatment of acute promyelocytic leukemia with
all-trans-retinoic acid. However, retinoic acid has a very
limited role in HCC differentiation, which suggests that HCC has
its own differentiation features. The HCC cell lines used in the
present study have distinct genetic backgrounds and different
CSC-like biomarkers. However, the selected HCC cell lines can be
induced successfully to differentiate into hepatocyte-like cells,
which suggests the existence of CSC-like cells or progenitor-like
cells in HCC tumor cells and the possibility for differentiation
therapy. In addition, upregulation of AFP and downregulation of
HNF4α were observed during the early stages of induced
differentiation, similar to a report concerning the differentiation
of hepatocytes from embryonic stem cells (26). These findings suggest that HCC cells
reside in different de-differentiated stages. Additionally, BMP4
administration has been found to induce the differentiation of
CD133+ hepatic CSCs and block their contributions to
HCC. Therefore, differentiation therapy based on hepatic CSCs may
be a promising strategy.
In conclusion, our data indicate that the
transmembrane receptor CXCR7 is closely associated with the
differentiation of HCC. Activated CXCR7 signaling can suppress the
key transcriptional factor HNF4α through ERK activation,
implicating the CXCR7-ERK-HNF4α cascade as a potential target for
HCC differentiation therapy.
Acknowledgements
This study was supported by the State Key Project on
Infectious Diseases of China (no. 2012ZX10002-016) and the Youth
Backbone Fund from Fudan University (B-233).
References
|
1
|
Ker CG, Chen HY, Chen KS, et al: Clinical
significance of cell differentiation in hepatocellular carcinoma.
Hepatogastroenterology. 50:475–479. 2003.PubMed/NCBI
|
|
2
|
Nowak D, Stewart D and Koeffler HP:
Differentiation therapy of leukemia: 3 decades of development.
Blood. 113:3655–3665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Sun H, Zhao F, et al: BMP4
administration induces differentiation of CD133+ hepatic
cancer stem cells, blocking their contributions to hepatocellular
carcinoma. Cancer Res. 72:4276–4285. 2012.PubMed/NCBI
|
|
4
|
Yin C, Lin Y, Zhang X, et al:
Differentiation therapy of hepatocellular carcinoma in mice with
recombinant adenovirus carrying hepatocyte nuclear factor-4alpha
gene. Hepatology. 48:1528–1539. 2008. View Article : Google Scholar
|
|
5
|
Ishiyama T, Kano J, Minami Y, Iijima T,
Morishita Y and Noguchi M: Expression of HNFs and C/EBP alpha is
correlated with immunocytochemical differentiation of cell lines
derived from human hepatocellular carcinomas, hepatoblastomas and
immortalized hepatocytes. Cancer Sci. 94:757–763. 2003. View Article : Google Scholar
|
|
6
|
Costa RH, Kalinichenko VV, Holterman AX
and Wang X: Transcription factors in liver development,
differentiation, and regeneration. Hepatology. 38:1331–1347. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
DeLaForest A, Nagaoka M, Si-Tayeb K, et
al: HNF4A is essential for specification of hepatic progenitors
from human pluripotent stem cells. Development. 138:4143–4153.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Walesky C, Edwards G, Borude P, et al:
Hepatocyte nuclear factor 4 alpha deletion promotes
diethylnitrosamine-induced hepatocellular carcinoma in mice.
Hepatology. 57:2480–2490. 2013. View Article : Google Scholar
|
|
9
|
Ning BF, Ding J, Yin C, et al: Hepatocyte
nuclear factor 4 alpha suppresses the development of hepatocellular
carcinoma. Cancer Res. 70:7640–7651. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Balabanian K, Lagane B, Infantino S, et
al: The chemokine SDF-1/CXCL12 binds to and signals through the
orphan receptor RDC1 in T lymphocytes. J Biol Chem.
280:35760–35766. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burns JM, Summers BC, Wang Y, et al: A
novel chemokine receptor for SDF-1 and I-TAC involved in cell
survival, cell adhesion, and tumor development. J Exp Med.
203:2201–2213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rajagopal S, Kim J, Ahn S, et al:
Beta-arrestin - but not G protein-mediated signaling by the ‘decoy’
receptor CXCR7. Proc Natl Acad Sci USA. 107:628–632. 2010.
|
|
13
|
Yu S, Crawford D, Tsuchihashi T, Behrens
TW and Srivastava D: The chemokine receptor CXCR7 functions to
regulate cardiac valve remodeling. Dev Dyn. 240:384–393. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maksym RB, Tarnowski M, Grymula K, et al:
The role of stromal-derived factor-1-CXCR7 axis in development and
cancer. Eur J Pharmacol. 625:31–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xue TC, Chen RX, Han D, et al:
Down-regulation of CXCR7 inhibits the growth and lung metastasis of
human hepatocellular carcinoma cells with highly metastatic
potential. Exp Ther Med. 3:117–123. 2012.PubMed/NCBI
|
|
16
|
Xue TC, Chen RX, Ren ZG, Zou JH, Tang ZY
and Ye SL: Transmembrane receptor CXCR7 increases the risk of
extrahepatic metastasis of relatively well-differentiated
hepatocellular carcinoma through upregulation of osteopontin. Oncol
Rep. 30:105–110. 2013.
|
|
17
|
Pasini D, Bracken AP, Hansen JB, Capillo M
and Helin K: The polycomb group protein Suz12 is required for
embryonic stem cell differentiation. Mol Cell Biol. 27:3769–3779.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dalby B, Cates S, Harris A, et al:
Advanced transfection with Lipofectamine 2000 reagent: primary
neurons, siRNA, and high-throughput applications. Methods.
33:95–103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun HC, Zhuang PY, Qin LX, et al:
Incidence and prognostic values of lymph node metastasis in
operable hepatocellular carcinoma and evaluation of routine
complete lymphadenectomy. J Surg Oncol. 96:37–45. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Simon R, Mirlacher M and Sauter G:
Immunohistochemical analysis of tissue microarrays. Methods Mol
Biol. 664:113–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lugli A, Spichtin H, Maurer R, et al:
EphB2 expression across 138 human tumor types in a tissue
microarray: high levels of expression in gastrointestinal cancers.
Clin Cancer Res. 11:6450–6458. 2005. View Article : Google Scholar
|
|
22
|
Liu LL, Fu D, Ma Y and Shen XZ: The power
and the promise of liver cancer stem cell markers. Stem Cells Dev.
20:2023–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee TK, Castilho A, Cheung VC, Tang KH, Ma
S and Ng IO: CD24(+) liver tumor-initiating cells drive
self-renewal and tumor initiation through STAT3-mediated NANOG
regulation. Cell Stem Cell. 9:50–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang ZF, Ho DW, Ng MN, et al: Significance
of CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008.
|
|
25
|
Yamashita T, Honda M, Nio K, et al:
Oncostatin m renders epithelial cell adhesion molecule-positive
liver cancer stem cells sensitive to 5- Fluorouracil by inducing
hepatocytic differentiation. Cancer Res. 70:4687–4697. 2010.
View Article : Google Scholar
|
|
26
|
Hay DC, Zhao D, Fletcher J, et al:
Efficient differentiation of hepatocytes from human embryonic stem
cells exhibiting markers recapitulating liver development in vivo.
Stem Cells. 26:894–902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hatzis P, Kyrmizi I and Talianidis I:
Mitogen-activated protein kinase-mediated disruption of
enhancer-promoter communication inhibits hepatocyte nuclear factor
4alpha expression. Mol Cell Biol. 26:7017–7029. 2006. View Article : Google Scholar
|
|
28
|
Zhao Z, Hong W, Zeng Z, et al:
Mucroporin-M1 inhibits hepatitis B virus replication by activating
the mitogen-activated protein kinase (MAPK) pathway and
down-regulating HNF4alpha in vitro and in vivo. J Biol Chem.
287:30181–30190. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shenoy SK and Lefkowitz RJ:
β-Arrestin-mediated receptor trafficking and signal transduction.
Trends Pharmacol Sci. 32:521–533. 2011.
|
|
30
|
Basson MA: Signaling in cell
differentiation and morphogenesis. Cold Spring Harb Perspect Biol.
4:2012.pii: a008151. View Article : Google Scholar
|
|
31
|
Yokoyama A, Katsura S, Ito R, et al:
Multiple post-translational modifications in hepatocyte nuclear
factor 4α. Biochem Biophys Res Commun. 410:749–753. 2011.
|
|
32
|
Hanse EA, Mashek DG, Becker JR, et al:
Cyclin D1 inhibits hepatic lipogenesis via repression of
carbohydrate response element binding protein and hepatocyte
nuclear factor 4α. Cell Cycle. 11:2681–2690. 2012.PubMed/NCBI
|
|
33
|
Sakamaki J, Daitoku H, Kaneko Y, Hagiwara
A, Ueno K and Fukamizu A: GSK3β regulates gluconeogenic gene
expression through HNF4α and FOXO1. J Recept Signal Transduct Res.
32:96–101. 2012.
|
|
34
|
Cozzolino AM, Alonzi T, Santangelo L, et
al: TGFβ overrides HNF4α tumor suppressing activity through GSK3β
inactivation: implication for hepatocellular carcinoma gene
therapy. J Hepatol. 58:65–72. 2013.
|
|
35
|
Colletti M, Cicchini C, Conigliaro A, et
al: Convergence of Wnt signaling on the HNF4alpha-driven
transcription in controlling liver zonation. Gastroenterology.
137:660–672. 2009. View Article : Google Scholar : PubMed/NCBI
|