Introduction
A correlation between regulation of apoptosis and a
number of human pathologies, such as cancer, autoimmune diseases
and neurodegenerative disorders has been identified (1,2).
Alterations of apoptosis are important in the development of
hepatocellular carcinoma cells and many pro- or anti-apoptotic
factors are involved in this process.
Omi/HtrA2 is a mitochondrial serine protease that is
released into the cytoplasm by apoptotic stimuli and induces cell
death through its own serine protease activity in addition to
attenuating the activity of inhibitor of apoptosis proteins (IAPs).
In the cytoplasm, the released mature Omi/HtrA2 binds to and blocks
the activity of IAPs, leading to the activation of caspases and
execution of cell death. In addition to this caspase-dependent
pathway, the cytosolic mature Omi/HtrA2 can induce cell death in a
caspase-independent manner that is exclusively dependent on its
ability to function as a protease even when caspases are
inactivated (3–11).
Previous studies have described Omi/HtrA2 mRNA
expression in cancer cells (12–18).
Results of previous studies showed that primary hepatocellular
carcinoma requires Omi/HtrA2 expression for cell apoptosis
(19). It has been reported that
the Omi/HtrA2 pro-apoptotic marker differs in some cell types
(20,21). However, the manner in which the
Omi/HtrA2 pro-apoptotic marker reacts during the process of
hepatocellular carcinoma cell apoptosis remains to be determined.
Thus, the aim of this study was to investigate the expression
status of Omi/HtrA2 in hepatocellular carcinoma cells to predict
cell apoptosis alteration and explore the possible mechanism of its
effect on hepatocellular carcinoma cell apoptosis. The experimental
outcome may provide a theoretical and experimental basis for
molecular therapy targeting Omi/HtrA2.
Materials and methods
Cell culture
Normal L02 hepatocellular and HepG2, Hep3B and PLC
hepatocellular carcinoma cells (prepared from the Experimental
Center of Hepatobiliary Surgery, Qilu Hospital, Shandong
University, Jinan, China) were maintained as monolayers in standard
medium comprising Dulbecco’s modified Eagle’s medium (DMEM: 4.5 g/l
of glucose) containing 10% heat-inactivated fetal calf serum and
supplemented with 20 mM HEPES, 100 μg/ml penicillin and 100 μg/ml
streptomycin (Merck, Darmstadt, Germany). The cells were incubated
at 37°C, 5% CO2 and saturated humidity.
Omi/HtrA2 expression vector pEGFP-Omi and
Omi/HtrA2 small-interference RNA expression vector psiRNA-Omi
construction
After RNA extraction from hypoxic HepG2 cells, cDNA
was synthesized with reverse transcriptase as described in the
introduction of cDNA Synthesis kit (Gibco-Life Technologies,
Carlsbad, CA, USA). For amplification of human Omi/HtrA2, the
primers used were: Omi/HtrA2 sense:
5′-ATATTATAGATCTATGGCTGCGCCGAGGGC-3′, antisense:
5′-AGTTAGTCGACTCTTCTGTGACCTCAGGG GTC-3′. The Omi/HtrA2 product was
digested with SalI and BglII (Fermentas, Shenzhen,
China) and ligated into the SalI and BglII linearized
target vector pEGFP-N1 (Clontech, Shanghai, China) to obtain
pEGFP-Omi containing the full-length human Omi/HtrA2. Positive
clones were confirmed by restriction analysis as well as sequencing
of the insert. Expression of Omi/HtrA2 RNA and protein was detected
to determine whether to successfully construct Omi/HtrA2 expression
vector pEGFP-Omi.
siRNAs were synthesized and purified by GenePharma,
Inc. (Shanghai, China). One specific siRNA against the positions of
Omi/HtrA2 open reading frame (795-814) was designed. The sequence
of Omi/HtrA2 siRNA was: sense: 5′-TCCCACTGCAG
AACACGATCACATTCAAGAGATGTGATCGTGTTCTGC AGTTT-3′ and antisense:
5′-CAAAAAACTGCAGAACACG ATCACATCTCTTGAATGTGATCGTGTTCTGCAGT-3′. A
scrambled version of Omi/HtrA2 siRNA was synthesized and used as a
control siRNA (sense: 5′-TCCCACTGCAGGACTT
GCCATGCTTAGCTCGTACGTGAGCTAGGCATTT-3′ and antisense:
5′-CAAAAAACTGGGAAATGTACACATCTCTC TTGGCTTAACGTTGTTCTGCAGT-3′). The
Omi/HtrA2 siRNA and control siRNA product were digested with
BbsI (Fermentas, Shenzhen, China) and ligated into
BbsI linearized target vector psiRNA-Hh1neo (Invitrogen Life
Technologies, Carlsbad, CA, USA) to obtain Omi/HtrA2
small-interference RNA expression vector psiRNA-Omi and psiRNA.
After expression vector psiRNA and psiRNA-Omi/HtrA2 were
transfected into HepG2 cells by Lipofectamine 2000 (Invitrogen,
Shanghai, China), the expression of Omi/HtrA2 mRNA and protein was
analyzed by A quantitative RT-PCR (RT-qPCR) and western blotting to
assess silencing efficiency of Omi/HtrA2 small-interference RNA
expression vector psiRNA-Omi/HtrA2.
Quantitative RT-PCR analysis
Total RNA was isolated from normal L02
hepatocellular and HepG2, Hep3B and PLC hepatocellular carcinoma
cells using TRIzol reagent (Gibco-Life Technologies). cDNA was
synthesized with reverse transcriptase as described in the
introduction of the cDNA Synthesis kit (Fermentas).
RT-qPCR analysis was carried out using the
SYBR-Green PCRMaster Mix (Applied Biosystems, Foster City, CA, USA)
using the Applied Biosystems 7500 RT-PCR system according to the
manufacturer’s instructions. Omi/HtrA2 primers used were sense:
5′-GGGCAGTGCTGTTGTTGTT-3′ and antisense: 5′-GCAGGTGCTGTCTTCTCCA-3′,
GAPDH sense: 5′-GAC CCCTTCATTGACCTCAAC-3′ and antisense: 5′-CTTCTC
CATGGTGGTGAAGA-3′. PCR was carried out at a final volume of 25 ml
with a SYBR-Green PCR Master Mix, using 1 ml cDNA and 900 nmol of
each primer for the respective genes. Cycling conditions were: 50°C
for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15
sec and 60°C for 1 min.
Comparison of Omi/HtrA2 mRNA expression was based on
the comparative CT method, where L02 cell Omi/HtrA2 mRNA expression
value was 1 and GAPDH expression was β-actin.
Western blot analysis
Adherent and floating cells were homogenized in
radioimmunoprecipitation assay buffer (10 mmol/l Tris-HCl, pH 8.0,
10 mmol/l EDTA, 0.15 mol/l NaCl, 1% NP-40, 0.5% sodium dodecyl
sulphate (SDS), 1 mg/ml Aprotinin, 1 mmol/l phenyl methyl sulphonyl
fluoride). The protein content was measured using the BCA protein
assay reagent and 10 μg was electrophoresed on a 12.5% SDS-PAGE gel
under non-reducing conditions. Prior to loading the sample on the
gel, the protein loads were equalized and the electrophoresed
proteins were transferred to nitrocellulose membranes. An
equivalent protein loading for each lane was reconfirmed by
staining the nitrocellulose membrane with Ponceau using the 36-kDa
GAPDH band present in the supplement as a reference marker. The
membranes were then probed with primary polyclonal antibody against
Omi/HtrA2, XIAP, ped/pea-15, HAX-1 (Santa Cruz Biotechnology, Santa
Cruz, CA, USA), followed by peroxidase-labeled secondary
antibodies. Western blot analyses were visualized using the
enhanced chemiluminescence detection system according to the
manufacturer’s instructions.
RNA extraction and RT-PCR analysis
Total mRNA was isolated from hepatocellular
carcinoma cells using TRIzol reagent following the manufacturer’s
instructions. cDNA was synthesized with reverse transcriptase as
described in the introduction of the cDNA Synthesis kit. For the
amplification of Omi/HtrA2 (978 bp), the primers used were:
Omi/HtrA2 sense: 5′-GCCATGATGGTGCATGACCGGTTAG-3′ and antisense:
5′-AGTCCGTAAATTGGGGCCATGC-3′. β-actin (245 bp) sense:
5′-GCATGCTCTATAGGAACGCG-3′ and antisense:
5′-CGATGGCAATCCTTACGTAAC-3′. The PCR regimen for Omi/HtrA2 and
β-actin involved an initial denaturation step of 94°C for 30 sec,
followed by 30 cycles at 94°C for 40 sec, 55°C for 40 sec and 72°C
for 60 sec on a GeneAmp PCR system 9700 (Perkin-Elmer). The
densitometric analysis of PCR products was performed using
MAGIAS-1000 software and normalized relative to the β-actin
expression for each sample. The same experiments were performed
three times.
Detecting the effect of Omi/HtrA2 gene
silencing by RNA interference on hepatocellular carcinoma cell
viability using MTT assay
Cell viability was determined by measuring cell
metabolism using the 3-(4, 5-dimethylthiazol-2-yl)-2,
5-diphenyltetrazolium bromide (MTT) assay. In brief, HepG2 cells,
HepG2 cells transfected into psiRNA and HepG2 cells transfected
into psiRNA-Omi were plated at 5×103 cells/well in
96-well tissue culture plates. After treatment with doxorubicin at
concentrations of 0, 10−4, 10−3,
10−2 and 10−1 mol/l for 36 h, MTT was added
to each well at a final concentration of 5 mg/ml and the cells were
incubated for 4 h at 37°C. The medium was then removed and the
cells were dissolved with dimethyl sulfoxide (DMSO; Sigma-Aldrich,
St. Louis, MO, USA). Absorbance was measured at 570 nm (referenced
to 650 nm) in a microplate reader (Bio-Tech Instruments Inc.,
Winooski, VT, USA).
As described earlier, the effect of Omi/HtrA2 gene
silencing by RNA interference on Hep3B and PLC cell viability was
determined using an MTT assay.
Detection of the effect of Omi/HtrA2 gene
silencing by RNA interference on hepatocellular carcinoma cell
apoptosis by flow cytometric analysis
HepG2 cells transfected into psiRNA and HepG2 cells
transfected into psiRNA-Omi were incubated with doxorubicin at a
concentration of 10−5 mol/l for 12 h, at which point the
control group without doxorubicin was established. Each group of
cells was detached by 0.25% trypsin and washed twice with cold
phosphate-buffered saline. The cells were stained with Annexin
V-FITC (Sigma) according to the manufacturer’s instructions. The
cells were analyzed on a FACS Calibur flow cytometer and data
analysis was performed with CellQuest software (Becton Dickinson,
Franklin Lakes, NJ, USA). Since necrotic cells also exhibited
phosphatidylserine due to the loss of membrane integrity, DNA
staining with propidium iodide, added to the Annexin V-FITC
solution, was used to distinguish necrotic cells from the Annexin
V-positive stained cell clusters.
The effect of Omi/HtrA2 serine protease activity on
Hep3B and PLC cell apoptosis was detected using an MTT assay.
Detection of the effect of ucf-101
(Omi/HtrA2 protease inhibitor) on hepatocellular carcinoma cell
viability using MTT assay
Cell viability was determined by measuring cell
metabolism using an MTT assay. Briefly, HepG2 cells were plated at
5×103 cells/well in 96-well tissue culture plates.
Following exposure to ucf-101 at concentrations of 0,
10−4, 10−3, 10−2 and
10−1 mol/l) (Merck) for 36 h, the cells were exposed to
doxorubicin at a concentration of 10−3 mol/l, followed
by the addition of MTT to each well at a final concentration of 5
mg/ml. The cells were subsequently incubated for 4 h at 37°C. The
medium was then removed and the cells were dissolved with DMSO.
Absorbance was measured at 570 nm (referenced to 650 nm) in a
microplate reader (Bio-Tech Instruments).
As described earlier, the effect of ucf-101 on Hep3B
and PLC cell viability was determined using an MTT assay.
Detecting the effect of ucf-101
(Omi/HtrA2 protease inhibitor) on cell apoptosis through flow
cytometric analysis
HepG2 cells were incubated with doxorubicin and
ucf-101 at concentrations of 10−3 and 10−1
mol/l, respectively, for 12 h, at which point the control group
without ucf-101 was established. Each group of cells was detached
by 0.25% trypsin and washed twice with cold phosphate-buffered
saline. The cells were stained with Annexin V-FITC (Sigma-Aldrich)
according to the manufacturer’s instructions. The cells were
analyzed on a FACS Calibur flow cytometer and data analysis was
performed with CellQuest software (Becton Dickinson). Since
necrotic cells also exhibited phosphatidylserine due to the loss of
membrane integrity, DNA staining with propidium iodide, added to
the Annexin V-FITC solution, was used to distinguish necrotic cells
from the Annexin V-positive stained cell clusters.
The effect of ucf-101 on Hep3B and PLC cell
apoptosis were detected using an MTT assay.
Detecting the effect of HepG2 and Hep3B
cell viability when cells transfected with Omi/HtrA2 expression
vector pEGFP-Omi were exposed to ucf-101
Cell viability was determined by measuring cell
metabolism MTT assay. Briefly, HepG2 cells, HepG2 cells transfected
into pEGFP and HepG2 cells transfected into pEGFP-Omi were plated
at 5×103 cells/well in 96-well tissue culture plates.
Following treatment with doxorubicin at a concentration of 0,
10−4, 10−3, 10−2 and
10−1 mol/l for 36 h, MTT was added to each well at a
final concentration of 5 mg/ml and the cells were incubated for 4 h
at 37°C. The medium was then removed and the cells were dissolved
with (DMSO; Sigma-Aldrich). Absorbance was measured at 570 nm
(referenced to 650 nm) in a microplate reader (Bio-Tech
Instruments).
After HepG2 cells, HepG2 cells transfected into
pEGFP and HepG2 cells transfected into pEGFP-Omi were exposed to
doxorubicin at concentrations of 0, 10−4,
10−3, 10−2 and 10−1 mol/l) for 36
h and the cells were exposed to ucf-101 at a concentration of
10−1 mol/l, the above experiment was repeated.
The effect of Hep3B cell viability when cells
transfected with Omi/HtrA2 expression vector pEGFP-Omi were exposed
to ucf-101 was determined using an MTT assay.
Detecting the protein expression of XIAP,
ped/pea-15 and HAX-1 in HepG2 and Hep3B
HepG2, Hep3B and PLC cells were divided into the
control group, the group treated with ucf-101, and the group
transfected into pEGFP-Omi. The control groups were incubated with
doxorubicin at a concentration of 10−3 mol/l for 24 h,
the groups treated with ucf-101 were cultured at a concentration of
10−3 mol/l and ucf-101 at 10−1 mol/l for 24
h, and the groups transfected into pEGFP-Omi were incubated with
docxorubicin at a concentration of 10−3 mol/l for 24 h.
The protein expression of XIAP, ped/pea-15 and HAX-1 was detected
by western blotting.
Statistical analysis
Values of each target gene were presented as the
mean ± SD. Silencing efficiency of psiRNA-Omi/HtrA2 in HepG2 cells
and the effect of Omi/HtrA2 RNA interference on HepG2 cell
apoptosis were evaluated using the paired-samples t-test and
subsequent statistical analysis was carried out using one-way
ANOVA. Statistical analyses were carried out using SSPS 13.0 for
Windows. P<0.05 was considered significant.
Results
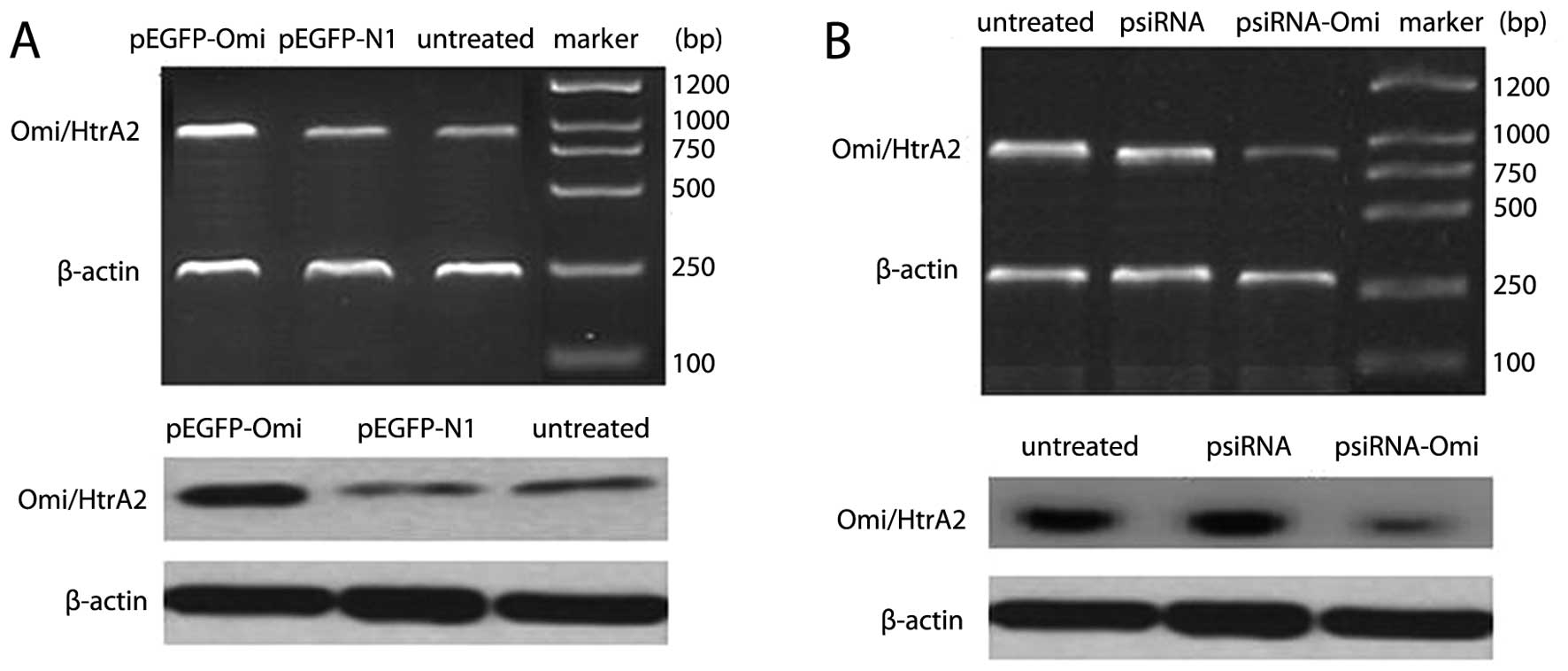
Successful construction of Omi/HtrA2
expression vector pEGFP-Omi and Omi/HtrA2 small-interference RNA
expression vector psiRNA-Omi
After HepG2 cells were transfected into pEGFP-Omi,
Omi/HtrA2 mRNA and protein expression in HepG2 cells was
effectively increased (Fig. 1A,
P<0.05), whereas no change was observed for Omi/HtrA2 mRNA and
protein expression in HepG2 cells transfected into pEGFP.
After HepG2 cells were transfected into psiRNA-Omi,
Omi/HtrA2 mRNA and protein expression in HepG2 cells was
effectively suppressed (Fig. 1B,
P<0.05), whereas no change was observed for Omi/HtrA2 mRNA and
protein expression in HepG2 cells transfected into psiRNA.
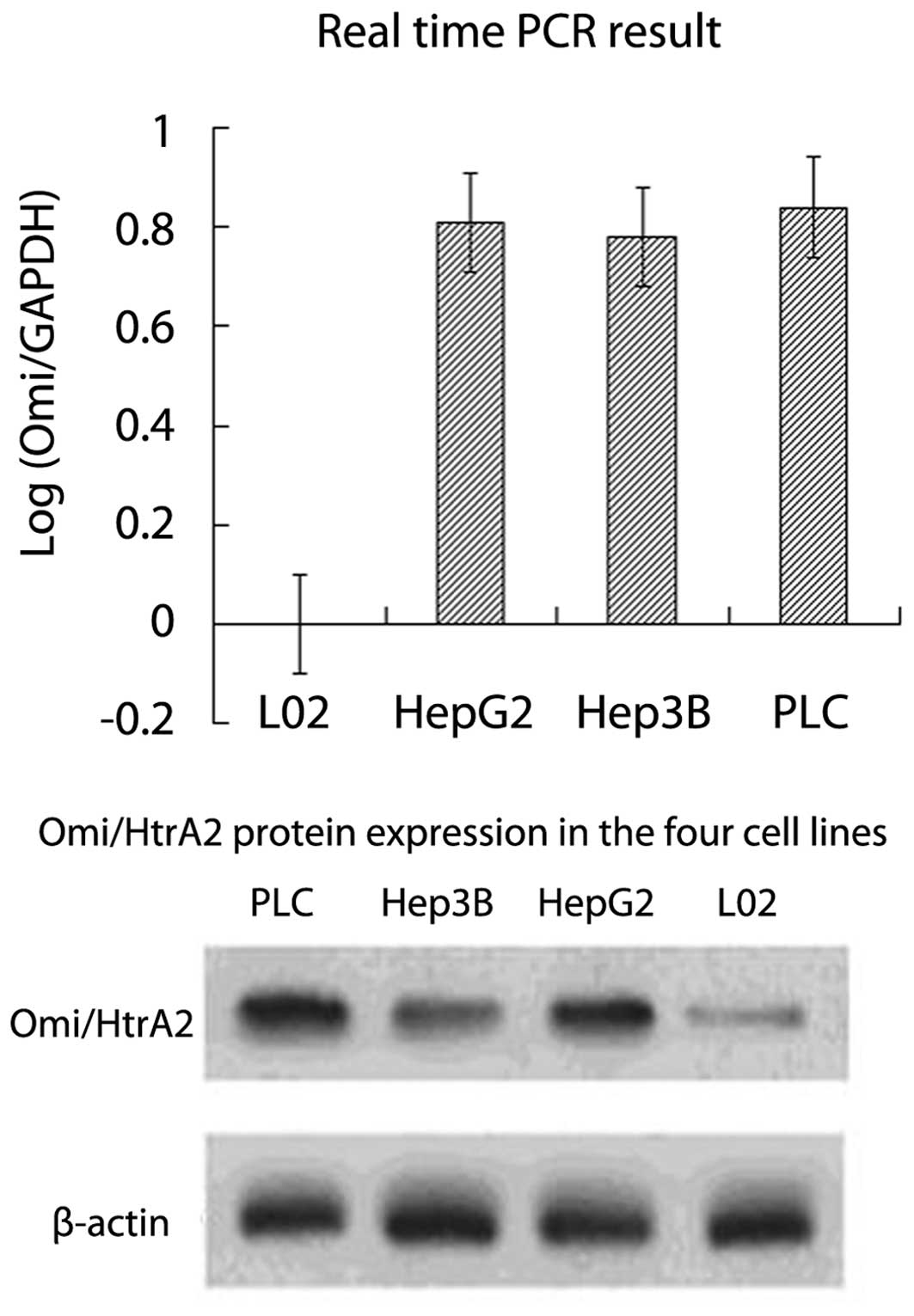
Omi/HtrA2 mRNA and protein overexpress in
hepatocellular carcinoma cells
The RT-qPCR assay showed that Omi/HtrA2 mRNA
expression level in the HepG2, Hep3B and PLC hepatocellular
carcinoma cells was ~8 times that of the normal L02 hepatocellular
cells (Fig. 2, P<0.01). However,
no significant difference was identified for the Omi/HtrA2 mRNA
expression level for the HepG2, Hep3B and PLC hepatocellular
carcinoma cells (P>0.05). Western blot analysis revealed that
the Omi/HtrA2 protein expression was significantly higher in
hepatocellular carcinoma cells than that in normal L02
hepatocellular cells (Fig. 2,
P<0.05), which was consistent with the result obtained from
RT-qPCR.
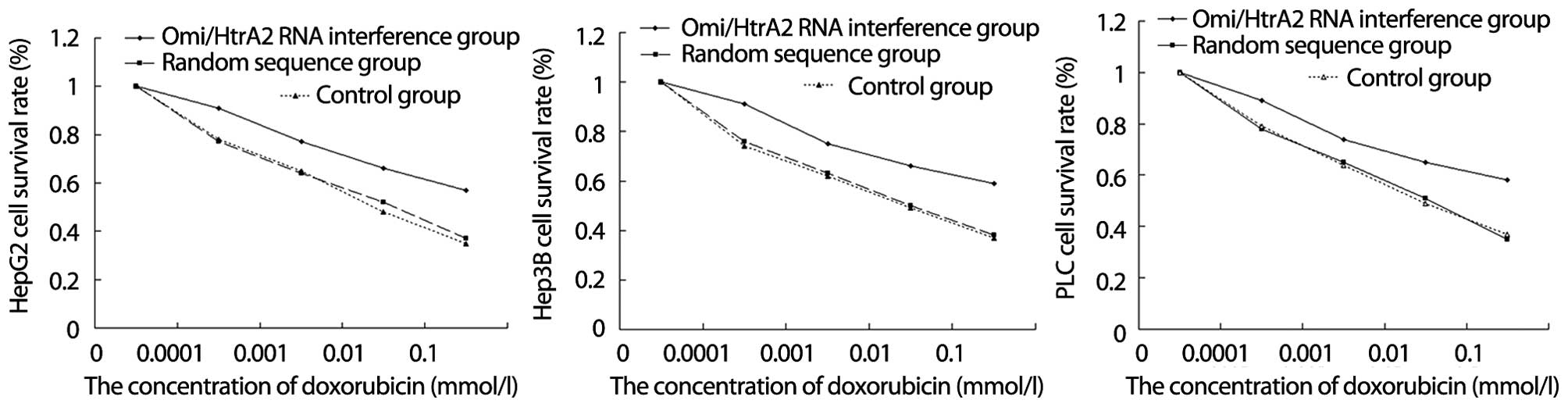
Effect of silencing of Omi/HtrA2
expression on hepatocellular carcinoma cell viability
MTT assay showed that, after HepG2 cells, HepG2
cells transfected into psiRNA and HepG2 cells transfected into
psiRNA-Omi/HtrA2 were incubated with doxorubicin at varying
concentrations for 36 h, the cell survival rate of three
experimental groups was decreased as the doxorubicin concentration
increased. However, the reduced amplitude of HepG2 cells
transfected into psiRNA-Omi (RNA interference) was significantly
less than that of the other experimental groups (Fig. 3A, P<0.05). Similarly, the reduced
amplitude of Hep3B (Fig. 3B,
P<0.05) and PLC (Fig. 3C,
P<0.05) cell viability after Omi/HtrA2 gene silencing by RNA
interference was significantly less than that of the relative
experimental groups.
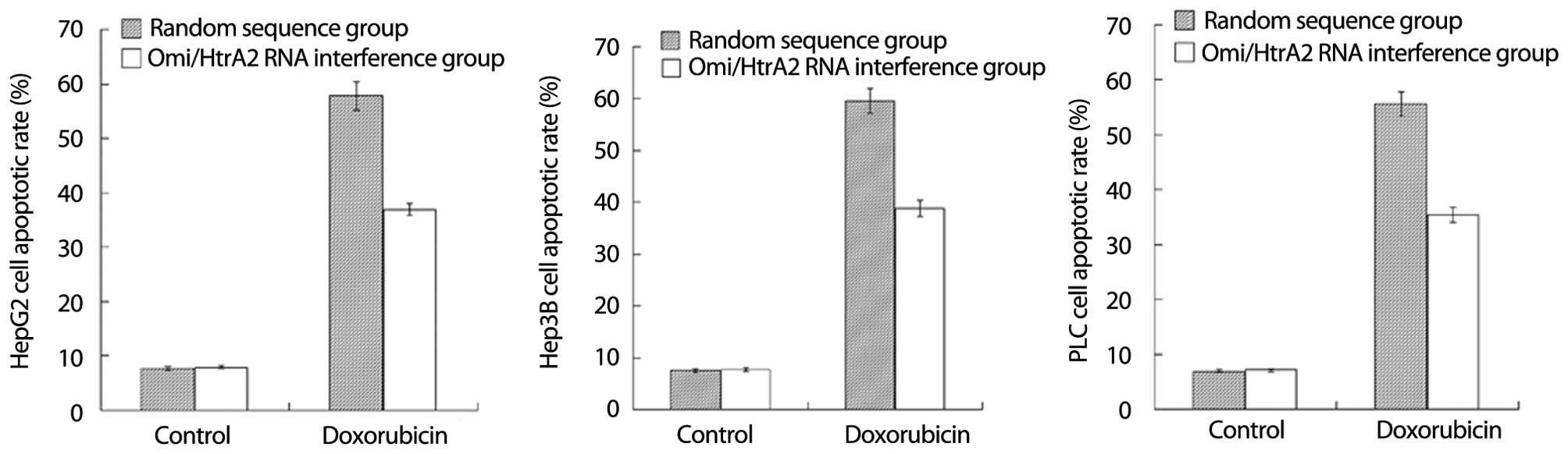
Effect of silencing of Omi/HtrA2
expression on hepatocellular carcinoma cell apoptosis
After HepG2 cells transfected into psiRNA and HepG2
cells transfected into psiRNA-Omi were incubated with doxorubicin
at a concentration of 10−5 mol/l for 12 h, the apoptotic
rate of HepG2 cells transfected into psiRNA and HepG2 cells
transfected into psiRNA-Omi was 57.8 and 36.9%, respectively
(Fig. 4A, P<0.05). The result
showed that HepG2 cell apoptosis was downregulated after Omi/HtrA2
gene silencing by RNA interference. Similarly, we observed the same
effect on Hep3B (Fig. 4B,
P<0.05) and PLC (Fig. 4C,
P<0.05) cell apoptosis after Omi/HtrA2 gene silencing by RNA
interference.
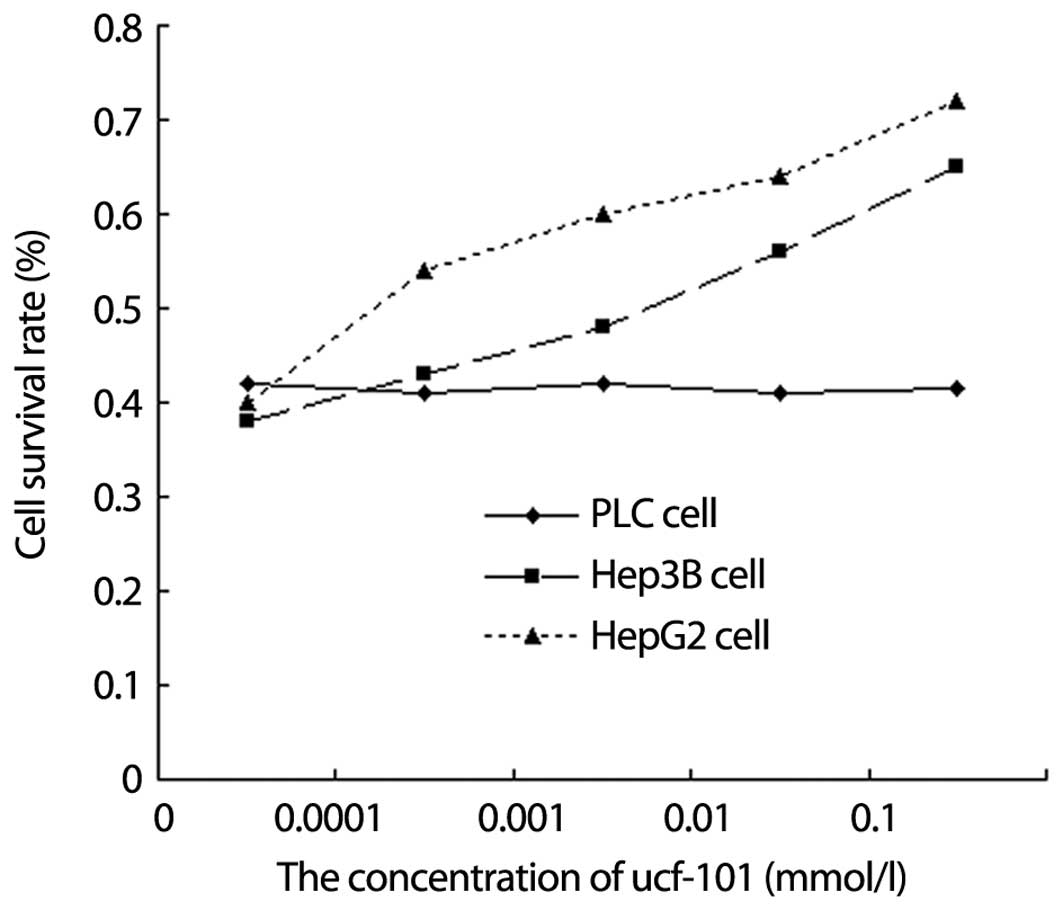
Different effect of ucf-101 (Omi/HtrA2
protease inhibitor)on hepatocellular carcinoma cell viability
MTT assay showed that, after HepG2, Hep3B and PLC
cells were incubated with ucf-101 at different concentrations for
36 h, the cell survival rate of HepG2 and Hep3B was increased with
the increase in ucf-101 concentration (Fig. 5, P<0.05). The increased amplitude
of HepG2 cells was significantly more than that of Hep3B cells,
although no change was evident in the cell survival rate of PLC
cells.
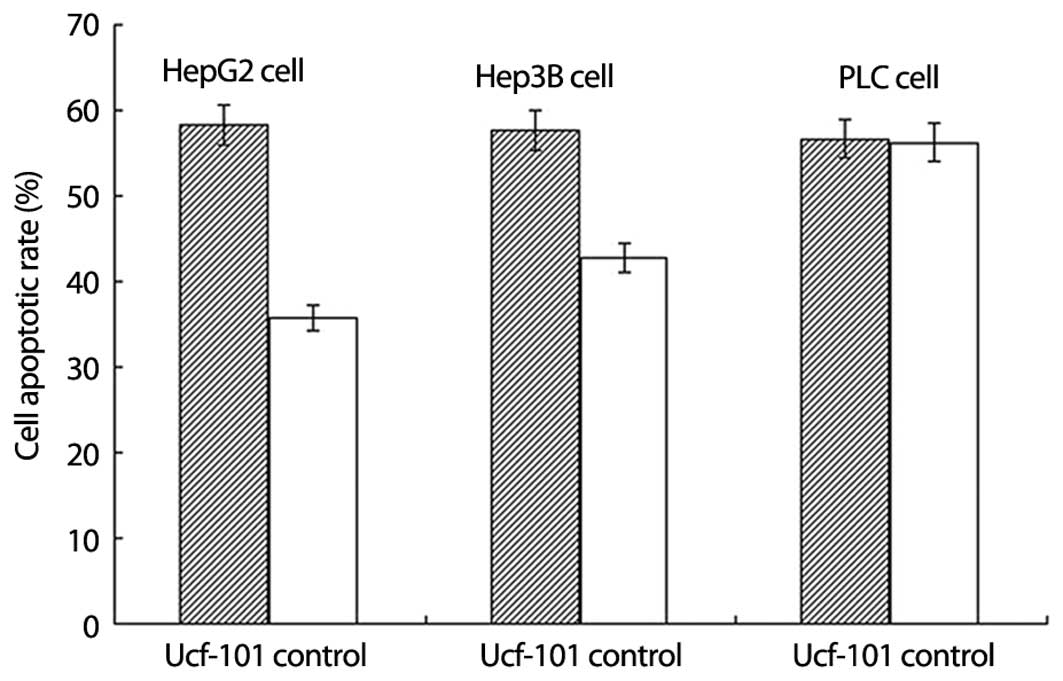
Different effect of ucf-101 (Omi/HtrA2
protease inhibitor) on hepatocellular carcinoma cell apoptosis
After HepG2 cells were incubated with doxorubicin at
a concentration of 10−3 mol/l and ucf-101 at
10−1 mol/l for 12 h, the apoptotic rate of HepG2 cells
exposed to doxorubicin and ucf-101 and HepG2 cells only exposed to
doxorubicin was 35.7 and 58.3%, respectively (Fig. 6, P<0.05) and that of Hep3B cells
exposed to doxorubicin and ucf-101 and Hep3B cells only exposed to
doxorubicin was 42.8 and 57.7%, respectively (Fig. 6, P<0.05). The result showed that
HepG2 and Hep3B cell apoptosis was downregulated following exposure
to ucf-101. We also found that PLC cell apoptosis did not change
following the exposure of cells to ucf-101.
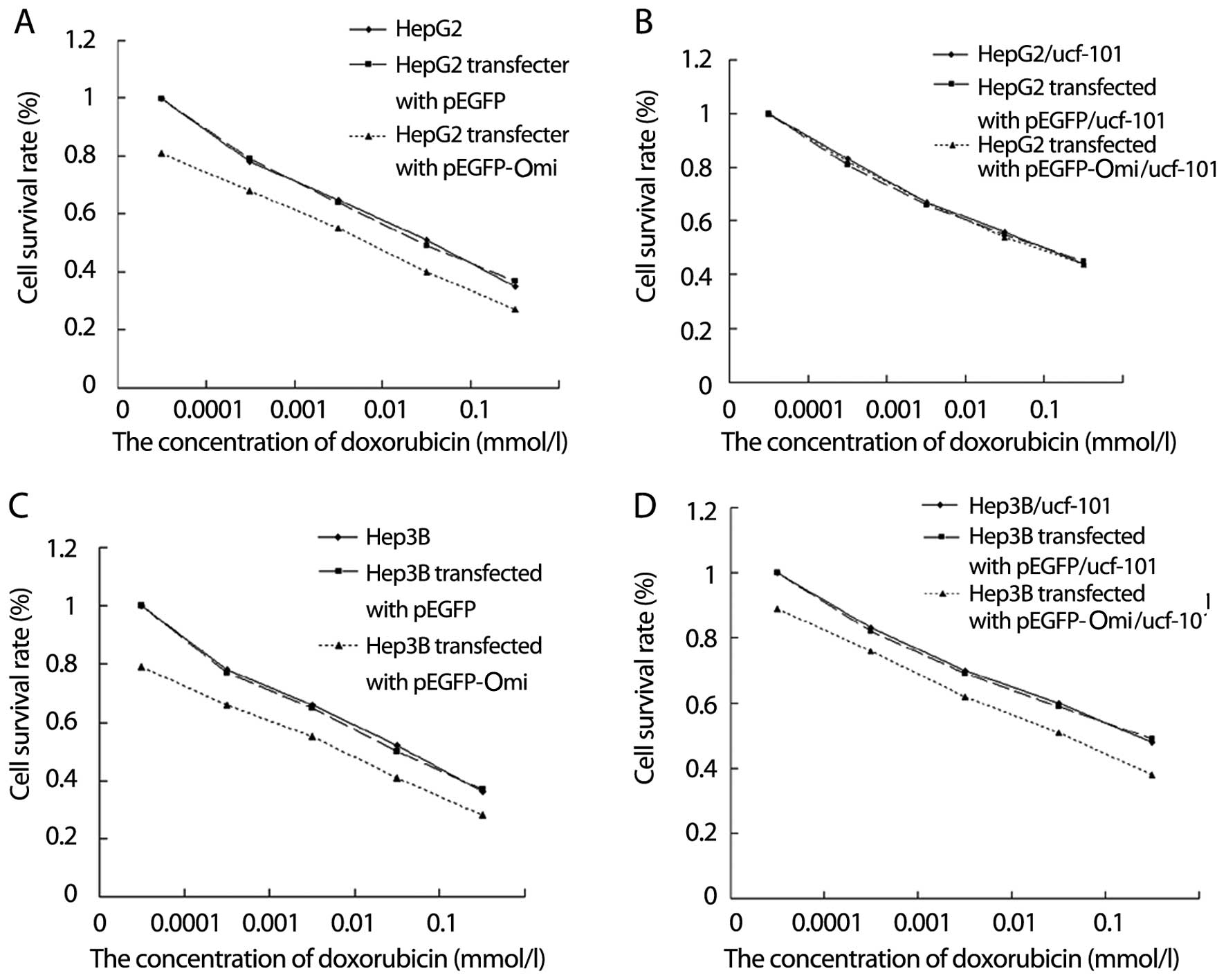
Effect of HepG2 and Hep3B cell viability
when cells transfected with Omi/HtrA2 expression vector pEGFP-Omi
were exposed to ucf-101
MTT assay showed that after HepG2, HepG2 transfected
into pEGFP-N1 and HepG2 transfected into pEGFP-Omi were incubated
under different concentrations of doxorubicin for 36 h, the cell
survival rate of the three experimental groups was decreased with
an increase in the concentration of doxorubicin. However, the
reduced amplitude of HepG2 cells transfected into pEGFP-Omi was
significantly more than that of other experimental groups (Fig. 7A, P<0.05). When simultaneous
treatment with ucf-101 at 10−1 mol/l was administered to
the three cell groups for 36 h, almost no difference was identified
in the survival rate of the three groups (Fig. 7B). Compared with the above
observation, when Hep3B, Hep3B transfected into pEGFP-N1 and Hep3B
transfected into pEGFP-Omi were exposed to doxorubicin under
different concentrations, similar results for the change of the
survival rate of the three groups were obtained (Fig. 7C, P<0.05). However, when
simultaneous treatment with ucf-101 at 10−1 mol/l was
administered to the three cell groups for 36 h, the survival rates
of the groups increased to a certain degree, but maintained the
original difference when cells were not treated with ucf-101
(Fig. 7D). These results suggested
that ucf-101 almost offset the effect of Omi/HtrA2 expression
vector pEGFP-Omi on HepG2 cell viability. By contrast, for Hep3B
cells, the experiment outcome showed that ucf-101 partly
counteracted the effect of Omi/HtrA2 expression vector pEGFP-Omi on
cell viability.
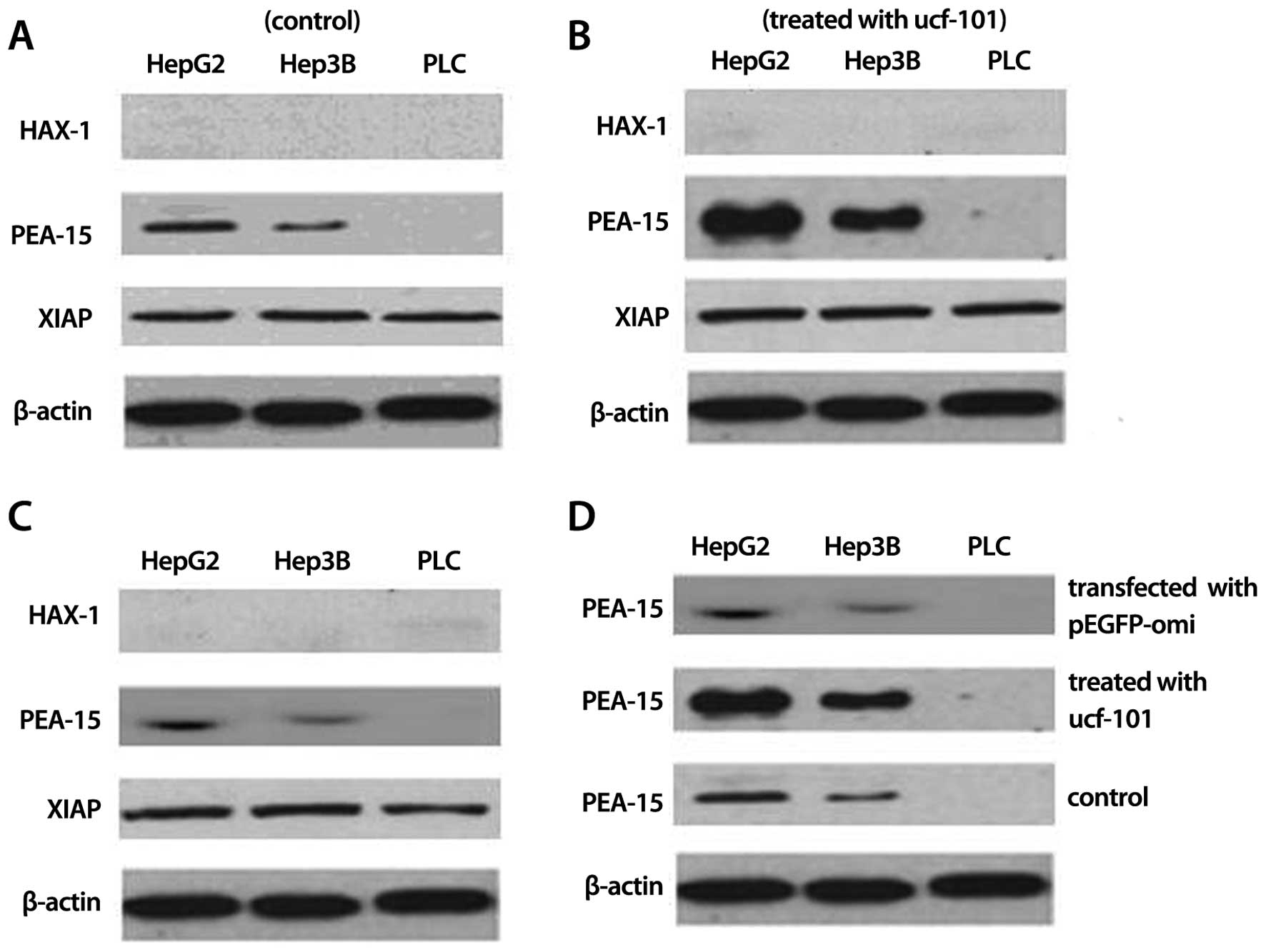
Protein expression of XIAP, ped/pea-15
and HAX-1 in HepG2, Hep3B and PLC cells
To clarify the possible mechanism of the difference
of Omi/HtrA2 pro-apoptotic marker in various hepatocellular
carcinoma cell lines, we detected the protein expression of XIAP,
ped/pea-15 and HAX-1 in HepG2, Hep3B and PLC cells. Western blot
analysis found that XIAP was overexpressed in HepG2, Hep3B and PLC
cells but its expression had no difference among the cell lines,
while HAX-1 was not expressed in HepG2, Hep3B and PLC cells. PLC
cells were devoid of ped/pea-15 expression while ped/pea-15 was
overexpressed in HepG2 and Hep3B cells and ped/pea-15 expression
was higher in HepG2 cells than that in Hep3B cells (P<0.05)
(Fig. 8A). To identify the above
detection, we examined the protein expression of XIAP, ped/pea-15
and HAX-1 in HepG2, Hep3B and PLC cells treated with ucf-101 or
transfected into pEGFP-Omi for 24 h. The results showed no change
in the protein expression of XIAP and HAX-1 in HepG2, Hep3B and PLC
cells. However, the ped/pea-15 protein expression increased when
HepG2 and Hep3B cells were exposed to ucf-101 (P<0.05) (Fig. 8B) but decreased when HepG2 and Hep3B
cells were transfected into pEGFP-Omi (P<0.05) (Fig. 8C). At the same time, the change of
ped/pea-15 protein expression in HepG2 cells was more obvious than
that in Hep3B cells (Fig. 8D).
Discussion
In the present study, we analyzed Omi/HtrA2
expression in normal L02 hepatocellular and HepG2, Hep3B and PCL
hepatocellular carcinoma cells through RT-qPCR and western blot
analysis. The results showed that Omi/HtrA2 was overexpressed in
the hepatocellular carcinoma cells. Our study also demonstrated
that hepatocellular carcinoma cell apoptosis was downregulated
after Omi/HtrA2 gene silencing by RNA interference and induced when
cells were transfected with Omi/HtrA2 expression vector pEGFP-Omi.
Thus, we suggest that Omi/HtrA2 overexpression promotes
hepatocellular carcinoma cell apoptosis. We previously reported
that Omi/HtrA2 was overexpressed in hepatocellular carcinoma
tissues and Omi/HtrA2 expression was closely correlated with tumor
size, tumor differentiation, clinical stage and lymph-node
metastasis (19). The present
finding that Omi/HtrA2 induces hepatocellular carcinoma cell
apoptosis confirmed and extended the observations made in our
previous study.
The serine protease Omi/HtrA2 is released from
mitochondria into the cytosol in response to apoptotic stimuli,
inducing cell death in a caspase-dependent manner by interacting
with the IAP as well as in a caspase-independent manner that relies
on its protease activity. Omi/HtrA2 proapoptotic function presents
a ‘dual’ nature of Omi/HtrA2 proapoptotic activity, which may be
dependent on IAP binding or on the serine protease properties.
Previous studies have demonstrated that the manner
in which (for IAP-binding or serine protease activity) Omi/HtrA2
participates in cell death may differ in some cell types. Blink
et al (20) found that, for
the usual apoptotic process in neutrophil inactivation of IAPs was
sufficient for Omi/HtrA2 to realize its proapoptotic potential and
the serine protease component was not essential, although under
certain conditions Omi/HtrA2 may mediate cell death through its
serine protease properties from within the mitochondria.
Srinivasula et al (20,21)
found that in 293 cells transfected with the active site mutant
Omi/HtrA2, which had no protease activity, caspase activation and
cell death were markedly reduced, whereas transfection of the
wild-type protein induced both events and resulted in a significant
reduction in the amount of XIAP in the transfected cells, which
appeared to be a substrate of Omi/HtrA2.
The abovementioned studies provided important
findings. However, whether the Omi/HtrA2 dual pro-apoptotic marker
(the way of IAP-binding or serine protease activity) demonstrates a
difference in the various hepatocellular carcinoma cell lines
remained to be elucidated. In our experiments, the effect of
Omi/HtrA2 on hepatocellular carcinoma cell viability and apoptosis
through using ucf-101 (Omi/HtrA2 protease inhibitor) was examined.
We found that HepG2 and Hep3B cell viability and apoptosis were
suppressed by Omi/HtrA2 serine protease inhibitor ucf-101 while PLC
cell viability and apoptosis was not affected by ucf-101. Omi/HtrA2
serine protease activity was involved in the process of HepG2 and
Hep3B cell apoptosis, whereas Omi/HtrA2 serine protease activity
did not participate in PLC cell apoptosis. Therefore, we suggest
that in PLC cells the only manner in which Omi/HtrA2 induces cell
death may be dependent on IAP-binding under some conditions.
In addition, our study showed that the increased
amplitude of HepG2 cell viability was higher than that of Hep3B
cells following cell exposure to ucf-101. Thus, Omi/HtrA2 induction
of cell apoptosis in HepG2 and Hep3B cells is also different and
Omi/HtrA2 induction of HepG2 cell apoptosis may mainly be dependent
on its serine protease activity while IAP-binding and its serine
protease activity participate in Hep3B cell apoptosis. To identify
how Omi/HtrA2 induces HepG2 and Hep3B cell apoptosis, we detected
the change of HepG2 and Hep3B cell viability when cells transfected
with Omi/HtrA2 expression vector pEGFP-Omi were exposed to ucf-101
(Omi/HtrA2 protease inhibitor). Our study results demonstrate that
ucf-101 almost offset the effect of Omi/HtrA2 expression vector
pEGFP-Omi on HepG2 cell viability. By contrast, for Hep3B cells,
the experiment outcome showed that ucf-101 partly counteracted the
effect of Omi/HtrA2 expression vector pEGFP-Omi on cell viability.
This result confirms the above supposition that the way that
Omi/HtrA2 induces HepG2 cell apoptosis may be mainly dependent on
Omi/HtrA2 serine protease activity while IAP-binding and its serine
protease activity participate in Hep3B cell apoptosis. Based on the
above data, we consider that the Omi/HtrA2 marker induces cell
apoptosis differently in various hepatocellular carcinoma cell
lines under varying conditions.
Of note, which factors cause Omi/HtrA2 pro-apoptotic
marker to differ in various hepatocellular carcinoma cell lines
remains to be elucidated. Yang et al (22) reported that the overexpression of
Omi/HtrA2 causes caspase-independent cell death, depending on the
protease activity of Omi/HtrA2 alone, independent of IAP
inhibition, although no functional substrates were identified.
Subsequently, ped/pea-15 and HAX-1 (the mitochondrial
anti-apoptotic protein HS1-associated protein X-1) were identified
as Omi/HtrA2 substrates by Trencia et al (23) and Cilenti et al (24), respectively, and the two substrates
were involved in the apoptotic process. Therefore, we postulate
that ped/pea-15 and HAX-1 as the substrates of Omi/HtrA2 serine
protease may be important in the Omi/HtrA2 pro-apoptotic marker
difference. Thus, we detected ped/pea-15, HAX-1 and XIAP protein
expression in HepG2, Hep3B and PLC cells. The results show that
ped/pea-15 protein expression in HepG2, Hep3B and PLC cells has a
significant difference. Moreover, PLC cells have no ped/pea-15
protein expression while ped/pea-15 was overexpressed in HepG2 and
Hep3B cells and ped/pea-15 protein expression in HepG2 cells was
higher than that in Hep3B cells. This outcome provides a reasonable
explanation for the above experiment that cell viability and
apoptosis of HepG2, Hep3B and PLC cells treated with ucf-101 had an
obvious difference. Thus, ped/pea-15 expression level causes the
difference of Omi/HtrA2 pro-apoptotic marker in various
hepatocellular carcinoma cell lines. Nevertheless, additional
experiments are required to validate the findings.
Taken together, our study results show that
Omi/HtrA2 overexpression facilitates hepatocellular carcinoma cell
apoptosis and that, for IAP-binding (caspase-dependent pathway) or
serine protease activity (caspase-independent pathway) of
Omi/HtrA2, the manner in which Omi/HtrA2 induces cell apoptosis
differs in the various hepatocellular carcinoma cell lines and
ped/pea-15 expression level results in this difference of Omi/HtrA2
pro-apoptotic marker. However, the role and possible mechanisms of
Omi/HtrA2 expression on hepatocellular carcinoma cell apoptosis may
provide a novel option for deploying Omi/HtrA2 to carry out
targeting therapy in hepatocellular carcinoma.
Acknowledgements
This study was supported by the Chinese Foundation
for Hepatitis Prevention and Control, TianQing Liver Disease
Research Fund (no. TQGB2011019), the Jiangxi Provincial Natural
Sciences Foundation Research Grant (no. 20132 BAB205048), the
Jiangxi Province Science and Technology Support Program (no.
20122BBG70119), and the National Natural Sciences Foundation
Research Grant of China (no. 81460442).
Reference
|
1
|
Kroemer G and Reed JC: Mitochondrial
control of cell death. Nat Med. 6:513–519. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thompson CB: Apoptosis in the pathogenesis
and treatment of disease. Science. 267:1456–1462. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suzuki Y, Imai Y, Nakayama H, Takahashi K,
Takio K and Takahashi R: A serine protease, HtrA2, is released from
the mitochondria and interacts with XIAP, inducing cell death. Mol
Cell. 8:613–621. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hegde R, Srinivasula SM, Zhang Z, et al:
Identification of Omi/HtrA2 as a mitochondrial apoptotic serine
protease that disrupts inhibitor of apoptosis protein-caspase
interaction. J Biol Chem. 277:432–438. 2002. View Article : Google Scholar
|
|
5
|
Martins LM, Iaccarino I, Tenev T, et al:
The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP
through a reaper-like motif. J Biol Chem. 277:439–444. 2002.
View Article : Google Scholar
|
|
6
|
Verhagen AM, Silke J, Ekert PG, et al:
HtrA2 promotes cell death through its serine protease activity and
its ability to antagonize inhibitor of apoptosis proteins. J Biol
Chem. 277:445–454. 2002. View Article : Google Scholar
|
|
7
|
Liu Z, Sun C, Olejniczak ET, et al:
Structural basis for binding of Smac/DIABLO to the XIAP BIR3
domain. Nature. 408:1004–1008. 2000. View
Article : Google Scholar
|
|
8
|
Wu G, Chai J, Suber TL, Wu JW, Du C, Wang
X and Shi Y: Structural basis of IAP recognition by Smac/DIABLO.
Nature. 408:1008–1012. 2000. View
Article : Google Scholar
|
|
9
|
Faccio L, Fusco C, Viel A and Zervos AS:
Tissue specific splicing of Omi stress-regulated endoprotease leads
to an inactive protease with a modified PDZ motif. Genomics.
68:343–347. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li W, Srinivasula SM, Chai J, et al:
Structural insights into the pro-apoptotic function of
mitochondrial serine protease HtrA2/Omi. Nat Struct Biol.
9:436–441. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki Y, Takahashi-Niki K, Akagi T,
Hashikawa T and Takahashi R: Mitochondrial protease Omi/HtrA2
enhances caspase activation through multiple pathways. Cell Death
Differ. 11:208–216. 2004. View Article : Google Scholar
|
|
12
|
Faccio L, Fusco C, Chen A, Martinotti S,
Bonventre JV and Zervos AS: Characterization of a novel human
serine protease that has extensive homology to bacterial heat shock
endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem.
275:2581–2588. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee SH, Lee JW, Kim HS, et al:
Immunohistochemical analysis of Omi/HtrA2 expression in stomach
cancer. APMIS. 111:586–590. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu XY, Xu YM, Chen XC, Ping H, Chen ZH and
Zeng FQ: Immunohistochemical analysis of Omi/HtrA2 expression in
prostate cancer and benign prostatic hyperplasia. APMIS.
114:893–898. 2006. View Article : Google Scholar
|
|
15
|
Wu SJ, Ng LT, Lin DL, Huang SN, Wang SS
and Lin CC: Physalis peruviana extract induces apoptosis in human
HepG2 cells through CD95/CD95L system and the mitochondrial
signaling transduction pathway. Cancer Lett. 215:199–208. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamaguchi H, Bhalla K and Wang HG: Bax
plays a pivotal role in the apsigargin-induced apoptosis of human
colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2
release from mitochondria. Cancer Res. 63:1483–1489.
2003.PubMed/NCBI
|
|
17
|
Kempkensteffen C, Hinz S, Christoph F, et
al: Expression levels of the mitochondrial IAP antagonists
Smac/DIABLO and Omi/HtrA2 in clear-cell renal cell carcinomas and
their prognostic value. J Cancer Res Clin Oncol. 134:543–550. 2008.
View Article : Google Scholar
|
|
18
|
Zhou H, Chen J, Lu X, Shen C, Zeng J, Chen
L and Pei Z: Melatonin protects against rotenone-induced cell
injury via inhibition of Omi and Bax-mediated autophagy in Hela
cells. J Pineal Res. 52:120–127. 2012. View Article : Google Scholar
|
|
19
|
Xu Z, Chen X, Peng C, Liu E, Li Y, Li C
and Niu J: The expression and clinical significance of Omi/Htra2 in
hepatocellular carcinoma. Hepatogastroenterology. 60:6–13.
2012.
|
|
20
|
Blink E, Maianski NA, Alnemri ES, Zervos
AS, Roos D and Kuijpers TW: Intramitochondrial serine protease
activity of Omi/HtrA2 is required for caspase-independent cell
death of human neutrophils. Cell Death and Differ. 11:937–939.
2004. View Article : Google Scholar
|
|
21
|
Srinivasula SM, Gupta S, Datta P, et al:
Inhibitor of apoptosis proteins are substrates for the
mitochondrial serine protease Omi/HtrA2. J Biol Chem.
278:31469–31472. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang QH, Church-Hajduk R, Ren JY, Newton
ML and Du CY: Omi/HtrA2 catalytic cleavage of inhibitor of
apoptosis (IAP) irreversibly inactivates IAPs and facilitates
caspase activity in apoptosis. Genes Dev. 17:1487–1496. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trencia A, Fiory F, Maitan MA, et al:
Omi/HtrA2 promotes cell death by binding and degrading the
anti-apoptotic protein ped/pea-15. J Biol Chem. 279:46566–46572.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cilenti L, Soundarapandian MM, Kyriazis
GA, et al: Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2
protease during cell death. J Biol Chem. 279:50295–50301. 2004.
View Article : Google Scholar : PubMed/NCBI
|