Introduction
Lung cancer is one of the most deadly diseases and
ranks first as the cause for cancer-related mortality (1). Small cell lung cancer (SCLC),
accounting for ~15% of all lung cancer cases, is a highly
aggressive subtype with neuroendocrine properties (2). Currently, for SCLC, there is no
effective treatment strategy either through surgery or chemotherapy
(3), attributed to its early
dissemination and fast development of drug resistance after
initiation of chemotherapy (4). The
5-year survival of SCLC patients is as low as 15% or less even
after aggressive treatment (5). In
light of targeted therapeutic approaches which have shown
advantages over traditional strategies, new therapeutic targets
must be explored based on the ongoing understanding of molecular
mechanisms in lung cancer development and progression.
Cancer development and progression are characterized
by deregulated gene expression networks which drive the
proliferation and metastasis of cancer cells (6). Among the deregulated cancer-related
genes, metastasis-associated gene 1 (MTA1) is one of the key
players involved in certain steps of cancer development (7). MTA1 is a component of the nuclear
remodeling and deacetylation complex (NuRD complex) that affects
gene expression ubiquitously (8).
MTA1 has been reported to be overexpressed in a series of malignant
diseases, including breast cancer (9), esophageal squamous cell carcinoma
(10), oral squamous cell carcinoma
(11), colon cancer (12) and non-small cell lung cancer (NSCLC)
(13). Moreover, it has been shown
to exert cancer progression in a number of cancer types, such as
melanoma, esophageal squamous cell carcinoma and colon cancer,
showing promise as a molecular target in cancer therapy strategy
development (14). However, whether
MTA1 also plays a key role in SCLC malignant behavior or whether it
also bears promise in SCLC treatment needs to be ascertained before
further potential MTA1-targeted therapeutics can be evaluated in
this subtype of cancer.
Thus, in the present study, we focused on the
expression status of MTA1 in SCLC and the in vitro and in
vivo biological effects of MTA1 silencing to evaluate its
potential as a therapeutic target in SCLC.
Materials and methods
Cell lines and cell culture
NCI-H446 (human SCLC), A549 and H1650 (human NSCLC)
cell lines were maintained in our laboratory. The cells were
cultured in RPMI-1640 supplemented with 10% newborn calf serum,
penicillin (100 U/ml), and streptomycin (100 mg/ml). The cells were
incubated at 37°C in a humidified atmosphere with 5%
CO2.
Small interfering RNA transfection
The MTA1 small interfering RNA (siRNA) and negative
control siRNA were purchased from Shanghai GenePharma Co., Ltd.
(Shanghai, China). The sequence of the MTA1 siRNA was
5′-GACCCTGCTGGCAGATAAA-3′. The siRNAs were dissolved in sterilized
and RNase-free water to the final concentration of 20 μM.
Lipofectamine 2000 (11668-019; Life Technologies, Grand Island, NY,
USA) was used to transfect the siRNAs into cells according to the
manufacturer’s instructions. Briefly, NCI-H446 cells were seeded in
6-well plates at a density of 3×105 cells/well in
RPMI-1640 medium in 37°C incubator until reaching 70% confluency.
After washing the cells with phosphate-buffered solution (PBS) and
serum-free RPMI-1640 medium, 5 μl siRNA was mixed with 5 μl
Lipofectamine 2000 in 1.5 ml serum-free RPMI-1640 medium at room
temperature for 20 min. Then the siRNA-Lipofectamine 2000 complex
mixture was added into the wells and was replaced with RPMI-1640
medium containing 10% fetal bovine serum after 6 h. The cells were
continuously incubated for 48 h before they were harvested for
silencing efficiency and biological assays.
Polymerase chain reaction (PCR)
The polymerase chain reaction (PCR) was performed as
previously described (13).
Briefly, total RNA was extracted from the variously treated cells
using TRIzol reagent (15596-026; Life Technologies). First Strand
cDNA was synthesized from mRNA using vigo-Script First Strand cDNA
Synthesis kit (11904-018; Life Technologies). The following primers
were used to amplify the MTA1 fragment: forward primer,
5′-CAGCTACGAGCAGCACAACG-3′ and reverse primer,
5′-TGTCCGTGGTTTGCCAGA-3′. Glyceraldehyde phosphate dehydrogenase
(GAPDH) was amplified as an internal control using forward primer,
5′-GGTGGTCTCCTCTGACTTCAACA-3′ and reverse primer,
5′-GTTGCTGTAGCCAAATTCGTTGT-3′. PCR was performed using an ABI
Veriti® 96-Well Thermal Cycler (4375786; Life
Technologies). The cycling conditions were as follows: initial
denaturation at 94°C for 60 sec, followed by 28 cycles at 94°C for
60 sec, 60°C for 60 sec and 72°C for 30 sec. The experiment was
repeated three times for each test.
Western blot analysis
Western blot analysis was performed as described in
a previous publication (15). The
protein lysis was subject to 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then
electrotransferred to polyvinylidene fluoride (PVDF) membranes.
After blocking with 5% fat-free milk in Tris-buffered saline
Tween-20, the PVDF membranes were incubated with the anti-MTA1
polyclonal antibody (ab71153; Abcam, Cambridge, MA, USA),
anti-β-actin monoclonal antibody (A5316; Sigma-Aldrich, St. Louis,
MO, USA), anti-p53 polyclonal antibody (sc-126; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-E-cadherin polyclonal
antibody (ab11512; Abcam), anti-PARP polyclonal antibody (#9542;
Cell Signaling Technology, Inc., Danvers, MA, USA), anti-PUMA
polyclonal antibody (#4976; Cell Signaling Technology), anti-cyclin
D1 or anti-cyclin E1 polyclonal antibody (bs501741, bs501085;
Bioworld Technology, Nanjing, China), overnight at 4°C, followed by
horseradish peroxidase-conjugated secondary antibody (1:5,000)
incubation for 1 h at room temperature. Signals were detected using
LAS 4000 Imaging system (Fijifilm, Tokyo, Japan). The experiment
was repeated twice.
Cell proliferation assay
3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) assay was used to detect cell proliferation. Briefly, cells
were seeded in complete growth medium in 96-well plates at a
density of 3×103 cells/well. After incubating for 1, 2,
3, 4 and 5 days, the medium in each well was replaced with 100 μl
fresh medium and 10 μl of aqueous solution of cell proliferation
MTS reagent and incubated at 37°C for 2 h. The OD values of each
well were measured with a microplate reader at λ = 490 nm.
Colony formation assay
The cells after being transfected with siRNA were
incubated for 48 h and then harvested and resuspended in complete
medium at a concentration of 1×103 cells/ml. Then, 0.5
ml of the cell suspensions was added into the 6-well plates, and
2.5 ml complete medium was added. After 10 days, the plates were
washed with PBS twice and stained with 0.2% crystal violet dye for
10 min. Finally, the cells were photographed and analyzed under an
imaging system (G:BOX, Syngene, Cambridge, UK). The detection was
repeated twice with duplicate measurements in each experiment.
Adhesion assay
A 96-well plate was precoated with 50 μl Matrigel
(cat. no 356234; BD Biosciences, Bedford, MA, USA, 10 mg/ml) in
advance. Then the three cell lines were harvested and reseeded into
96-well plates in complete growth medium at a density of
3×104 cells/well. Unattached cells were washed three
times with PBS gently at different time-points, and the cells
remaining on the wells were quantified using the MTS assay. The
percentages of adhered cells were calculated relative to the cells
in the unwashed control wells. Serial dilutions were conducted to
construct a standard curve to convert absorbance at 490 nm to cell
number.
Transwell invasion assay
The invasion assay was performed using Transwell
chambers (#3422; Corning Life Sciences, Tewksbury, MA, USA) with 50
μl Matrigel-precoated (cat. 356234; BD Biosciences, San Jose, CA,
USA, 10 mg/ml) polycarbonate membrane as described in a previous
report (15). Briefly, the three
groups of cells were collected and resuspended in serum-free
RPMI-1640 medium at a concentration of 1.5×105 cells/ml,
then the cell suspensions were added into the top chambers (200
μl/well) and the bottom chambers were filled with complete medium
(500 μl/well), followed by a 24-h incubation at 37°C. The cells
that did not penetrate the polycarbonate membrane were swabbed
using a cotton bud. The cells that transmembraned through and
adhered to the bottom of the polycarbonate membrane were stained
with 0.2% crystal violet dye for 10 min, and the number of invaded
cells was counted. The experiment was repeated twice with duplicate
measurements in each experiment. For each well, five fields were
chosen (the ones in the middle, at 3, 6, 9 and 12 time clock
positions) to count the total cell numbers. Data from three
parallel wells were averaged and compared between the groups.
Wound healing assay
Wound healing assay was performed as previously
described (16). Briefly, the same
amount of cells was seeded in a 6-well plate and incubated until
the cells grew to 80% confluencey. A sterile pipette tip was used
to scratch a line on the monolayer cells with the same width. The
detached cells were washed off with PBS and the remaining cells
were cultured in complete medium. Specific points on the scratched
lines were photographed at 0, 12 and 24 h after scratching. The
healing of the wounds was presented by the distance between the
frontline of cells moving towards the scratched area. The
experiment was repeated in triplicate.
Annexin V FITC assay and cell cycle
analysis by flow cytometry
The variously transfected cells were incubated for
48 h and then trypsinized. For theAnnexin V FITC assay, the cells
were washed with PBS twice and stained with Annexin V-FITC and PI
(cat. 556547; BD Biosciences) for 15 min at room temperature and
analyzed by flow cytometry for cells undergoing apoptosis. For cell
cycle analysis, cells were washed with PBS twice and then fixed
with cold 70% ethanol at −20°C overnight. The cells were treated
with RNase A for 30 min at 37°C, stained with propidium iodide (PI)
solution for 30 min at 4°C and then measured by flow cytometry
(cat. 643183; BD Biosciences). ModFit LT software (VMFLTWIN4,
Verity Software House Inc., Topsham, ME, USA) was used for data
acquisition and analysis.
Immunohistochemical staining of the
tissue microarray (TMA)
Paraffin-embedded small cell lung cancer TMAs (cat.
no. LC10010a) were purchased from Alenabio Biotech Ltd. (Xi’an,
China). The tissues on the TMA contained duplicate samples from 40
tumors and 10 normal lung tissues. Tumors on the TMA were
categorized based on the TNM classification of tumors, as shown in
Table I, and were verified by
adjacent H&E staining sections by two independent
pathologists.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Patient
characteristics | No. of cases | % |
|---|
| Gender (F/M) | 9/31 | 22.5/77.5 |
| Stage |
| I | 10 | 25.0 |
| II | 21 | 52.5 |
| III | 9 | 22.5 |
| Tumor |
| T1/T2 | 36 | 90 |
| T3/T4 | 4 | 10 |
| N0 | 12 | 30 |
| N1/N2 | 28 | 70 |
| Normal lung
tissue | 10 | |
The tissue microarrays were deparaffined and
antigen-retrieved routinely. Immunohistochemical staining was
performed as previously described (17). Briefly, the slides of the TMAs were
washed with PBS, quenched for 15 min in 3%
H2O2, and blocked with 2% normal goat serum
for 30 min at room temperature before the anti-MTA1 polyclonal
antibody was applied overnight at 4°C. Then the slides were
incubated with the biotinylated secondary antibody followed by
incubation with HRP-labeled streptavidin. 3,3′-Diaminobenzidine
hydrochloride (DAB) was used for staining. Then slides were
counterstained with hematoxylin before being mounted and examined
by light microscopy.
MTA1 protein levels were assessed in the TMAs. The
staining intensity scores were as follows (18): 0 (negative), 1 (weak), 2 (moderate),
and 3 (strong). The percentage of MTA1-positive cells was also
score based on four categories: 0 was scored for 0–10%, 1 for
>10–25%, 2 for >25–50%, 3 for >50–75%, and 4 for
>75–100%. Staining was scored as the product of the staining
intensity and percentage scores.
Statistical analysis
Statistically significant differences between groups
were determined using an unpaired Student’s t-test. Statistical
significance was defined at P<0.05.
Results
MTA1 is highly expressed in SCLC
specimens compared with normal lung tissues
In order to ascertain whether MTA1 plays a role in
SCLC development and progression, we assessed the expression of
MTA1 in a set of tissue microarrays containing a series of SCLC
lung cancer tissues. We analyzed the levels of MTA1 by
immunohistochemistry in the tissue microarrays (TMAs), including 40
patients with small cell lung cancer and 10 cases of normal lung
tissue (Table I). The median age of
the patients when they received surgical treatment was 52.675 years
(range, 32–76). MTA1 was scored according to the staining intensity
and percentage of positive cells in an entire cancer cell
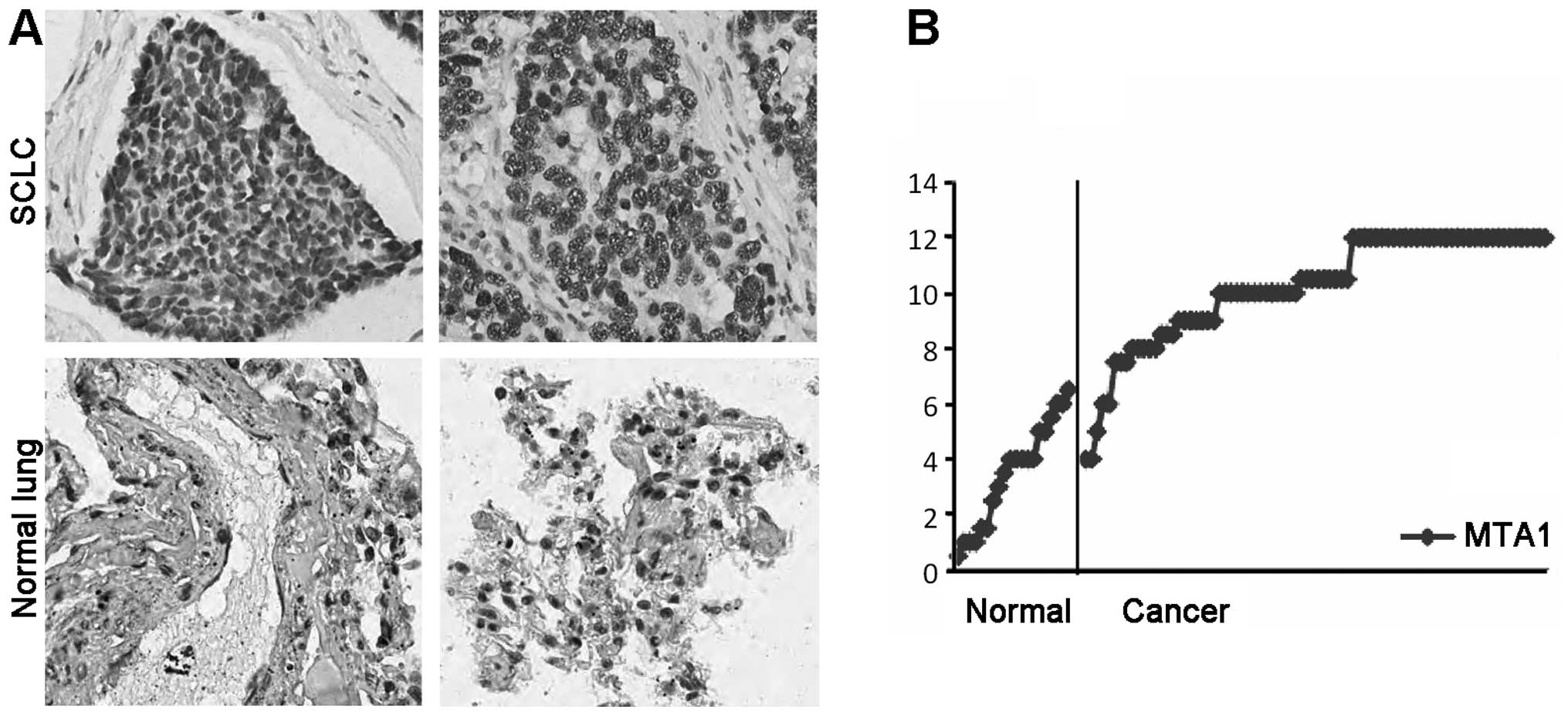
population. As shown in Fig. 1A and
B, the SCLC tissue exhibited a marked increase in MTA1
expression compared with the normal lung tissues.
MTA1 downregulation suppresses
proliferation of the lung cancer NCI-H446 cell line
To ascertain whether MTA1 plays a positive role in
SCLC proliferation, we used RNAi technique to knock down MTA1
expression in the SCLC cell lines and performed a proliferation
assay. First, we screened a series of lung cancer cell lines for
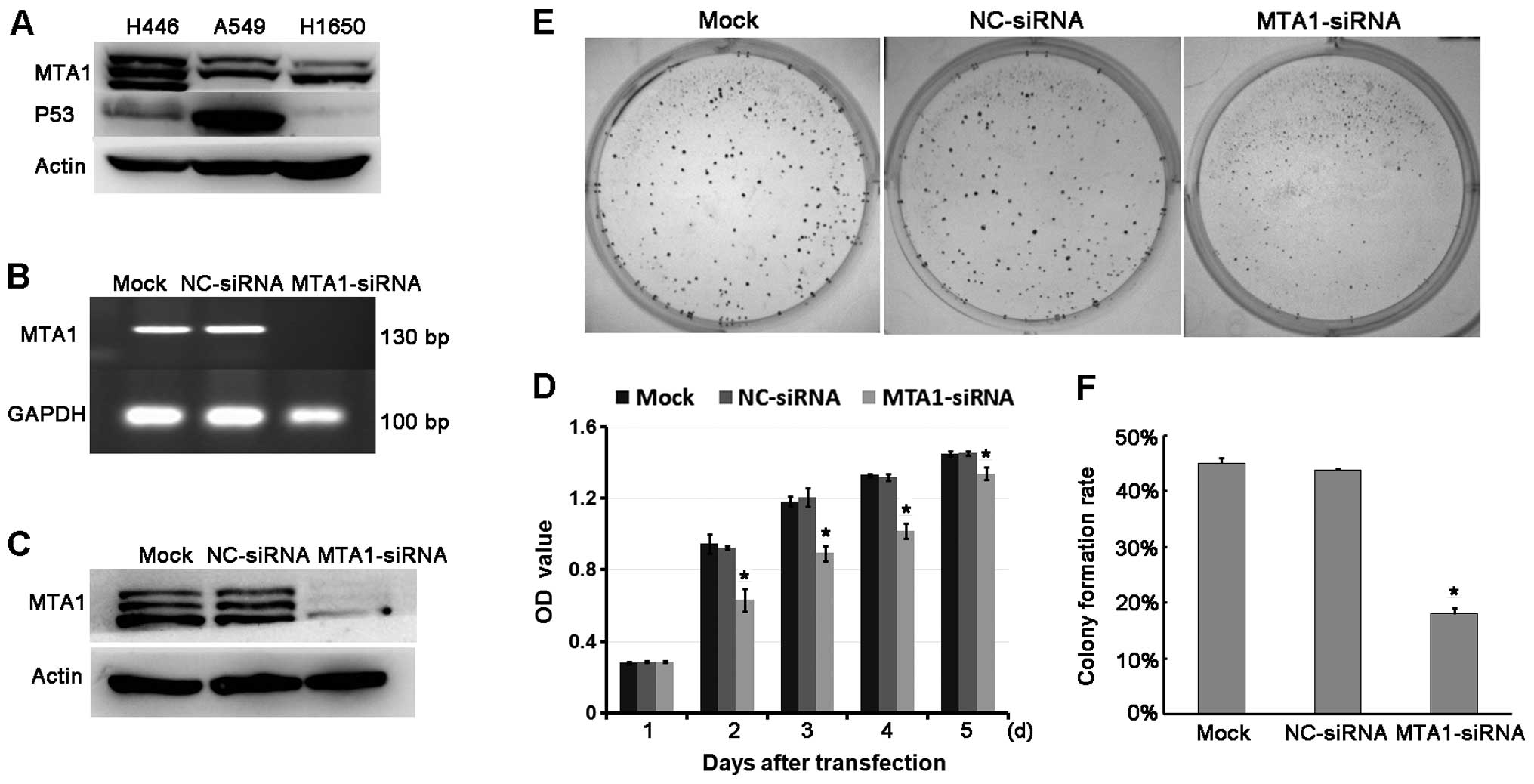
their MTA1 protein levels (Fig.
2A). We used NCI-H446 cells which express a high level of MTA1
as the model with which to evaluate the MTA1 function. After
confirming the siRNA silencing efficiency (Fig. 2B and C), we compared the
proliferation of the NCI-H446 cells between the MTA1-siRNA- and
NC-siRNA-treated groups. We found that MTA1 siRNA exerted
significant inhibition on cell growth (Fig. 2D). The colony formation ability was
also suppressed by MTA1 siRNA (Fig. 2E
and F). Therefore, downregulation of MTA1 contributes to the
tumor growth of SCLC.
Downregulation of MTA1 expression impairs
the adhesion and invasive abilities of the SCLC cell line
MTA1 has been suggested to be involved in the
process of metastasis. To ascertain whether MTA1 has an impact on
SCLC cancer metastasis, we evaluated the adhesion and invasion
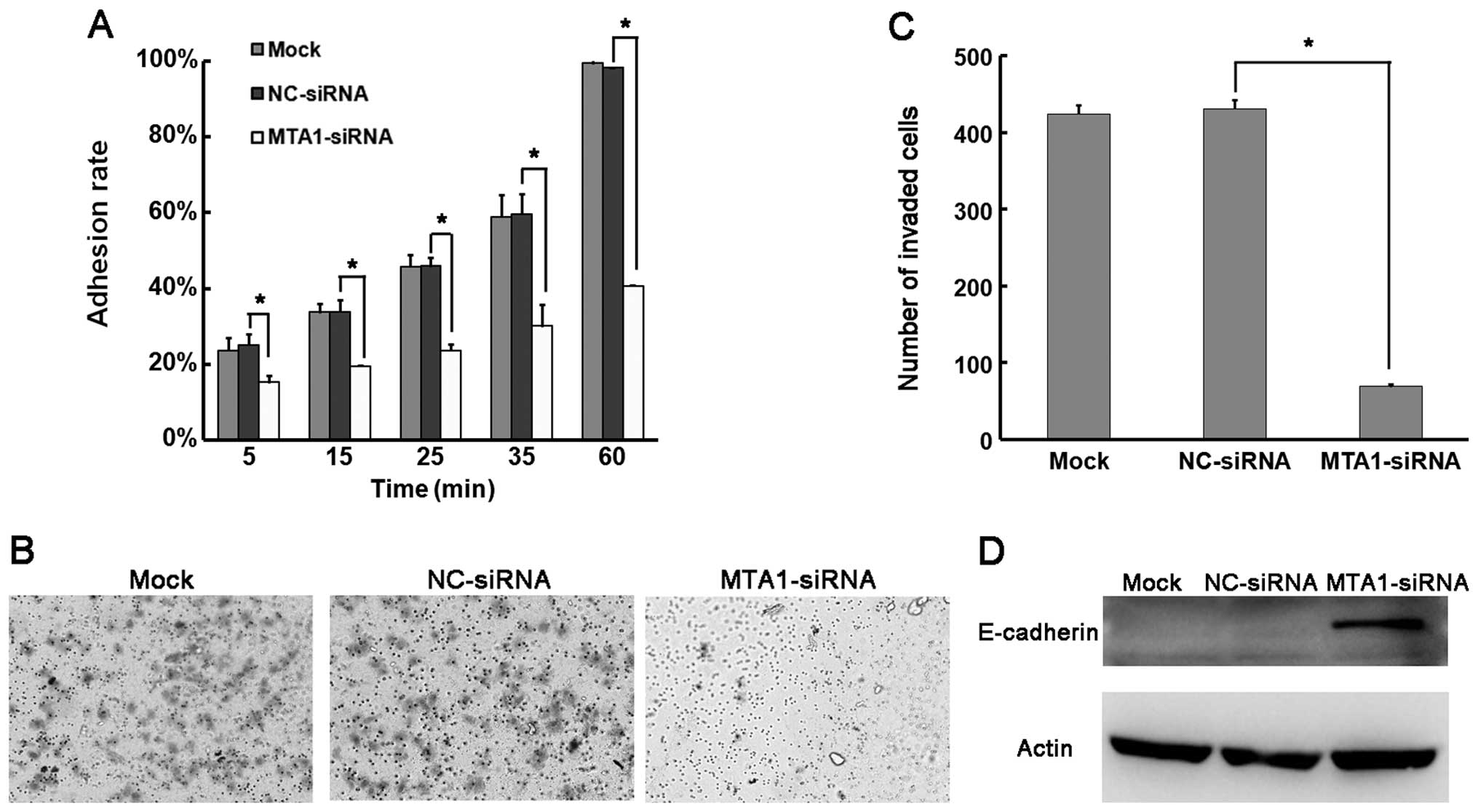
assays in NCI-H446 cells. First, we determine the adhesion ability
of NCI-H446 after MTA1 silencing. There was no significant
difference in the adhesion ability between the mock group and the
NC-siRNA-transfected group. However, the adhesion ability was
greatly impaired in the MTA1-siRNA-treated NCI-H446 cells (Fig. 3A). Next, we explored the invasive
potential of the NCI-H446 cells by the Transwell invasion model
after MTA1-siRNA and NC-siRNA transfection. We demonstrated that
the MTA1-siRNA cells had significantly reduced numbers of invaded
cells (69.5±3.5 vs. 424.5±11.5 in the control) (Fig. 3B and C). Adhesion and invasion are
two key steps during tumor cell metastasis. To further confirm that
MTA1 regulates SCLC metastasis at the molecular level, we detected
alteration in the adhesion-associated protein E-cadherin by MTA1
manipulation. As shown in Fig. 3D,
E-cadherin was much lower than that in the control group. These
results suggest that MTA1 has a positive role in promoting SCLC
metastasis.
MTA1 silencing reduces SCLC cell motility
in the wound healing assay
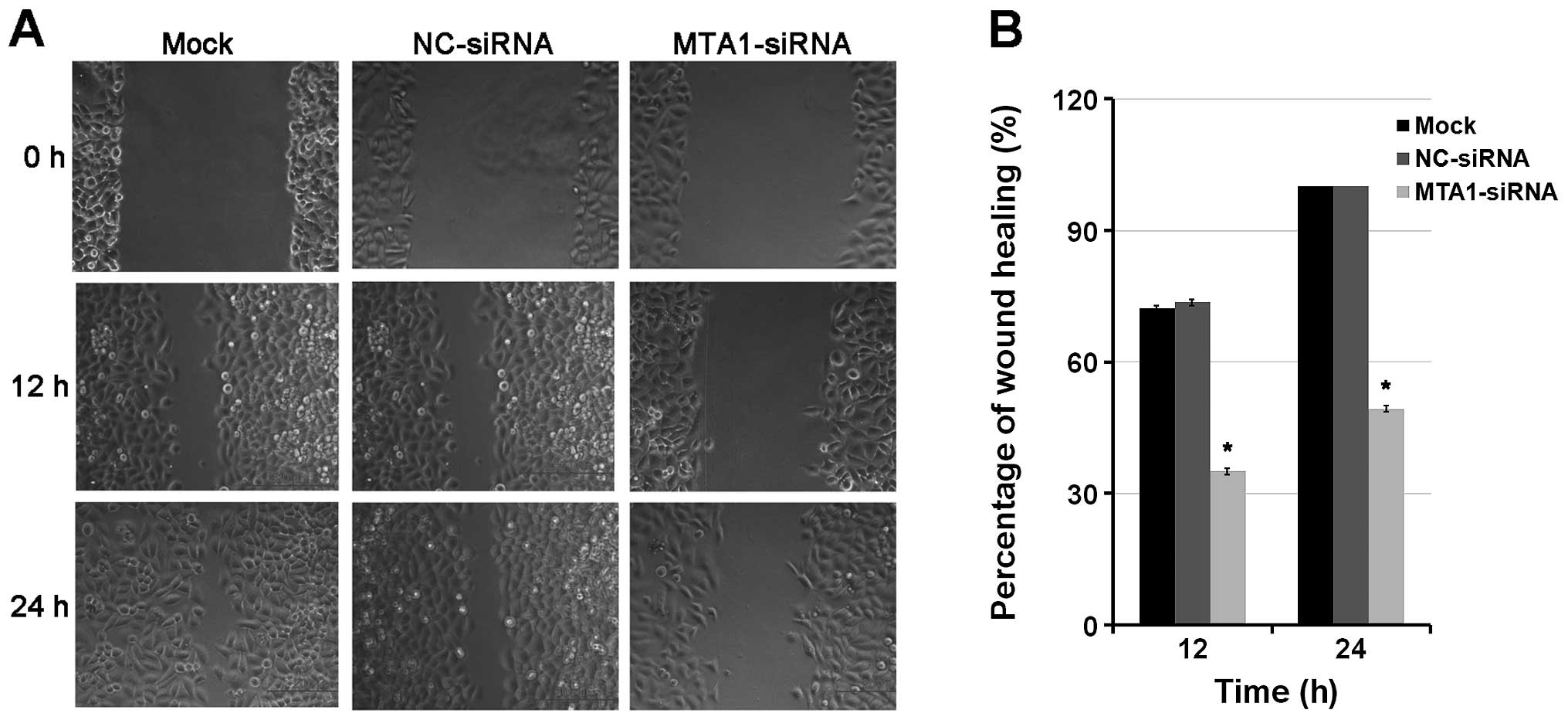
Next, we performed a wound healing assay in the
NCI-H446 cells to further understand the role of MTA1 in the
migration potential of SCLC cells. The results demonstrated that
the wound healing ability of the MTA1-siRNA group was significantly
reduced when compared to that of the mock group and negative
control group at 24 h after scratching. We measured the distance
between the migrating frontlines and calculated the speed of wound
healing. We found that the MTA1-siRNA-transfected group lagged
behind the mock and control group in the healing process (Fig. 4). These data demonstrated that MTA1
was effective in promoting SCLC migration which was significantly
inhibited by MTA1 silencing.
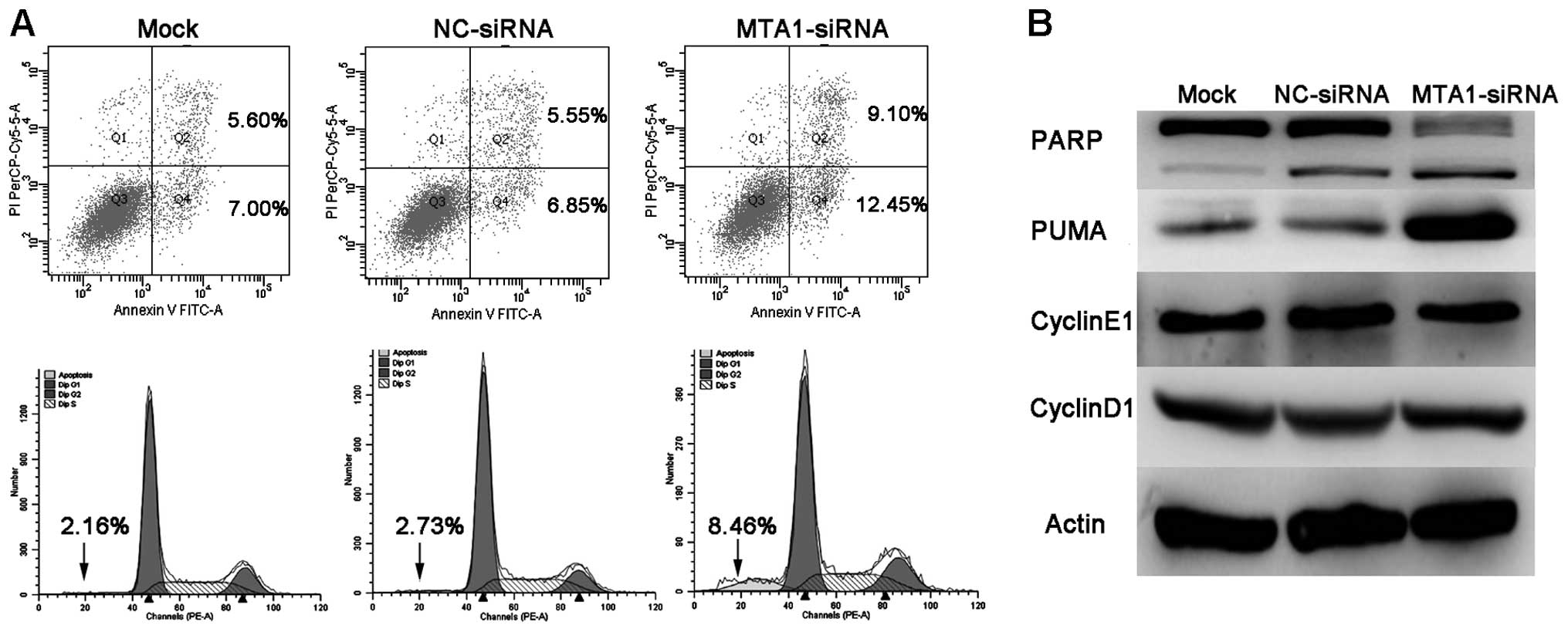
MTA1 knockdown affects the apoptosis but
not the cell cycle in the SCLC cell line
In order to explore whether MTA1 affects apoptosis
in SCLC cell lines, we performed Annexin V FITC assay by flow
cytometry. Data showed that MTA1 knockdown resulted in increased
percentage of cells in both early and particularly late apoptosis,
compared to the mock and negative groups (Fig. 5A). We further detected an alteration
in apoptosis-related proteins PARP and PUMA following MTA1
silencing. Results showed that MTA1-siRNA knockdown caused PUMA
upregulation and PARP cleavage. We did not observe a significant
change in the expression of cyclin E1 and cyclin D1, as consistent
with the cell cycle assay by flow cytometry (Fig. 5B).
Discussion
Despite decades of biological and clinical research,
there still remains a lack of consensus regarding the therapeutic
protocol of patients with small cell lung cancer (SCLC). An
understanding of the detailed molecular mechanisms are urgently
needed, and a series of molecules have been linked to SCLC
progression (19).
MTA1 is one of the molecules involved in
carcinogenesis and cancer progression that has been discovered in
the last few years. It was originally isolated from highly
metastatic rat mammary adenocarcinoma cell lines by differential
cDNA library screening (20). In
human cancer, it was first reported to be significantly correlated
with the malignant properties of gastrointestinal cancers in 1997
(21). Subsequent studies confirmed
a widespread upregulation of MTA1 in various human tumors (22), contributing to multiple malignant
properties of cancer, particularly invasion and metastasis.
The first evidence showing that MTA1 is upregulated
in advanced lung cancer came in 2002 by Sasaki et al
(23) via MTA1 mRNA detection in 74
non-small cell lung cancer (NSCLC) tissues. They indicated that
MTA1 upregulation was closely related to the invasion and
metastasis of NSCLC. Following that, several studies were performed
to investigate the role and mechanism of MTA1 in lung cancer
promotion (24–28). However, only NSCLC tissues or cell
lines were examined in these studies. Whether MTA1 is also
upregulated and tumor-promoting in SCLC is still unexplored to
date. To resolve this question, we first detected the expression of
MTA1 in SCLC by tissue microarray and the results showed that,
compared to the normal lung tissues, MTA1 was significantly
increased in SCLC, raising a probable role of MTA1 in SCLC
promotion.
Invasion and metastasis are typical hallmarks of
tumors. They are the main causes that makes it difficult to
eradicate all tumor cells from the body. Although the role of MTA1
in cancer metastasis is strongly indicated in a wide range of human
cancers, whether MTA1 is a potential target for cancer treatment
remains poorly studied, particularly in SCLC. Nicolson et al
(29) found that suppression of
MTA1 by antisense oligodeoxynucleotides greatly inhibited the
growth and metastasis of the MDA-MB-231 cancer cell line; Qian
et al (30) also found that
deletion of MTA1 by RNA interference suppressed the growth and
metastasis of B16F10 melanoma cells in vitro.
In the present study, we aimed to evaluate the
therapeutic value of MTA1 disruption in SCLC treatment. The
NCI-H446 cell line was selected because of its high expression of
MTA1 among the screened cell line models. Transfection of siRNA
specifically and significantly downregulated the MTA1 expression in
NCI-H446 cells, while there were no significant changes in the
NC-siRNA and mock groups, demonstrating the high specificity and
efficacy of the MTA1 siRNA. After MTA1 was downregulated by MTA1
siRNA, the cell growth and colony formation of NCI-H446 cells were
significantly decreased, which was consistent with the properties
in previous reports (29,31), indicating that MTA1 mediated the
regulation of the growth and proliferation of SCLC NCI-H446 cells
in vitro. The adhesion and invasion of NCI-H446 cells were
also significantly impaired by downregulation of MTA1. MTA1 is a
downstream effector of transforming growth factor-β1 (TGF-β1) which
mediates repression of epithelial-cadherin (E-cadherin) expression
(32). E-cadherin is essential for
maintaining cell-cell junctions which are important for tumor cell
invasion and metastasis (33).
Disruption of E-cadherin is correlated with tumor progression,
invasive growth, metastasis and poor prognosis (34). Notably, upregulation of E-cadherin
was observed after MTA1 downregulation, which coincided with the
adhesion and invasion results. Cell migration ability was
corroborated by a wound healing assay, and the results were further
confirmed. Collectively, these findings indicate that inhibition of
MTA1 significantly decreased the invasion and metastasis of SCLC
in vitro, showing promise as a target in cancer
treatment.
The MTA1/NuRD complex as a transcriptional
co-repressor of tumor suppressors is associated with deacetylated
p53 (35,36). p53 was downregulated in the SCLC
cell line following MTA1 overexpression, while depletion of MTA1
caused significant apoptosis as detected by flow cytometry and
western blotting for cleaved PARP, which is possibly associated
with upregulation of PUMA. Moreover, MTA1 also plays a crucial role
in the DNA repair process (37).
MTA1 is involved in DNA damage repair through a variety of ways,
among which the poly(ADP-ribose) polymerase (PARP)-dependent manner
was emphasized in human cancer cells (38). It was reported that overexpression
of MTA1 could lead to the activation of oncogenic cyclin D1
pathways (39), but no significant
change was observed when MTA1 was downregulated. This may be due to
the complex process as MTA1 has different regulatory effects on all
cycle-related proteins in different cancers. In summary, our
results showed that downregulation of MTA1 significantly reduced
the tumor malignant phenotype of SCLC in vitro, guaranteeing
further exploration of its application in SCLC treatment.
Our research collectively indicates that MTA1 is a
key regulator in SCLC. Reduction in MTA1 is an effective gene
therapeutic method by which to treat SCLC in vitro by
decreasing proliferation, invasion and metastasis abilities and
stimulating the apoptosis pathway. This may result in the
development of new treatments for SCLC.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (81372158, 81372159
and 81101518).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Owonikoko TK, Ragin CC, Belani CP, Oton
AB, Gooding WE, Taioli E and Ramalingam SS: Lung cancer in elderly
patients: an analysis of the surveillance, epidemiology, and end
results database. J Clin Oncol. 25:5570–5577. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jackman DM and Johnson BE: Small-cell lung
cancer. Lancet. 366:1385–1396. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Le Péchoux C, Dunant A, Senan S, Wolfson
A, Quoix E and Faivre-Finn C: Standard-dose versus higher-dose
prophylactic cranial irradiation (PCI) in patients with
limited-stage small-cell lung cancer in complete remission after
chemotherapy and thoracic radiotherapy (PCI 99-01, EORTC
22003-08004, RTOG 0212, and IFCT 99-01): a randomised clinical
trial. Lancet Oncol. 10:467–474. 2009. View Article : Google Scholar
|
|
5
|
Asai N, Ohkuni Y, Kaneko N, Yamaguchi E
and Kubo A: Relapsed small cell lung cancer: treatment options and
latest developments. Ther Adv Med Oncol. 6:69–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar R, Wang RA and Bagheri-Yarmand R:
Emerging roles of MTA family members in human cancers. Semin Oncol.
30:30–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xue Y, Wong J, Moreno GT, Young MK, Côté J
and Wang W: NURD, a novel complex with both ATP-dependent
chromatin-remodeling and histone deacetylase activities. Mol Cell.
2:851–861. 1998. View Article : Google Scholar
|
|
9
|
Jang KS, Paik SS, Chung H, Oh YH and Kong
G: MTA1 overexpression correlates significantly with tumor grade
and angiogenesis in human breast cancers. Cancer Sci. 97:374–379.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li SH, Wang Z and Liu XY:
Metastasis-associated protein 1 (MTA1) overexpression is closely
associated with shorter disease-free interval after complete
resection of histologically node-negative esophageal cancer. World
J Surg. 33:1876–1881. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawasaki G, Yanamoto S, Yoshitomi I,
Yamada S and Mizuno A: Overexpression of metastasis-associated MTA1
in oral squamous cell carcinomas: correlation with metastasis and
invasion. Int J Oral Maxillofac Surg. 37:1039–1046. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Higashijima J, Kurita N, Miyatani T,
Yoshikawa K, Morimoto S, Nishioka M, Iwata T and Shimada M:
Expression of histone deacetylase 1 and metastasis-associated
protein 1 as prognostic factors in colon cancer. Oncol Rep.
26:343–348. 2011.PubMed/NCBI
|
|
13
|
Zhu X, Guo Y, Li X, Ding Y and Chen L:
Metastasis-associated protein 1 nuclear expression is associated
with tumor progression and clinical outcome in patients with
non-small cell lung cancer. J Thorac Oncol. 5:1159–1166. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li DQ, Pakala SB, Nair SS, Eswaran J and
Kumar R: Metastasis-associated protein 1/nucleosome remodeling and
histone deacetylase complex in cancer. Cancer Res. 72:387–394.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian LL, Yue W, Zhu F, Li S and Li W:
Human mesenchymal stem cells play a dual role on tumor cell growth
in vitro and in vivo. J Cell Physiol. 226:1860–1867. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qian H, Lu N, Xue L, Liang X, Zhang X, Fu
M, et al: Reduced MTA1 expression by RNAi inhibits in vitro
invasion and migration of esophageal squamous cell carcinoma cell
line. Clin Exp Metastasis. 22:653–662. 2005. View Article : Google Scholar
|
|
17
|
Wang H, Zhang D, Wu W, Zhang J, Guo D,
Wang Q, et al: Overexpression and gender-specific differences of
SRC-3 (SRC-3/AIB1) immunoreactivity in human non-small cell lung
cancer: an in vivo study. J Histochem Cytochem. 58:1121–1127. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rudin CM, Durinck S, Stawiski EW, Poirier
JT, Modrusan Z, Shames DS, et al: Comprehensive genomic analysis
identifies SOX2 as a frequently amplified gene in small-cell lung
cancer. Nat Genet. 44:1111–1116. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sidransky D: Emerging molecular markers of
cancer. Nat Rev Cancer. 2:210–219. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Toh Y, Pencil SD and Nicolson GL: A novel
candidate metastasis-associated gene, mta1, differentially
expressed in highly metastatic mammary adenocarcinoma cell lines.
cDNA cloning, expression, and protein analyses. J Biol Chem.
269:22958–22963. 1994.PubMed/NCBI
|
|
21
|
Toh Y, Oki E, Oda S, Tokunaga E, Ohno S,
Maehara Y, Nicolson GL and Sugimachi K: Overexpression of the MTA1
gene in gastrointestinal carcinomas: correlation with invasion and
metastasis. Int J Cancer. 74:459–463. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Toh Y and Nicolson GL: The role of the MTA
family and their encoded proteins in human cancers: molecular
functions and clinical implications. Clin Exp Metastasis.
26:215–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sasaki H, Moriyama S, Nakashima Y,
Kobayashi Y, Yukiue H, Kaji M, Fukai I, Kiriyama M, Yamakawa Y and
Fujii Y: Expression of the MTA1 mRNA in advanced lung cancer. Lung
Cancer. 35:149–154. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu L, Mao XY, Fan CF and Zheng HC: MTA1
expression correlates significantly with cigarette smoke in
non-small cell lung cancer. Virchows Arch. 459:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu X, Zhang X, Wang H, Song Q, Zhang G,
Yang L, et al: MTA1 gene silencing inhibits invasion and alters the
microRNA expression profile of human lung cancer cells. Oncol Rep.
28:218–224. 2012.PubMed/NCBI
|
|
26
|
Xia Y, Chen Q, Zhong Z, Xu C, Wu C, Liu B
and Chen Y: Down-regulation of miR-30c promotes the invasion of
non-small cell lung cancer by targeting MTA1. Cell Physiol Biochem.
32:476–485. 2012. View Article : Google Scholar
|
|
27
|
Li S, Tian H, Yue W, Li L, Gao C, Si L, Li
W, Hu W, Qi L and Lu M: Down-regulation of MTA1 protein leads to
the inhibition of migration, invasion, and angiogenesis of
non-small-cell lung cancer cell line. Acta Biochim Biophys Sin
(Shanghai). 45:115–122. 2013. View Article : Google Scholar
|
|
28
|
Li Y, Chao Y, Fang Y, Wang J, Wang M,
Zhang H, Ying M, Zhu X and Wang H: MTA1 promotes the invasion and
migration of non-small cell lung cancer cells by downregulating
miR-125b. J Exp Clin Cancer Res. 32–33. 2003.
|
|
29
|
Nicolson GL, Nawa A, Toh Y, Taniguchi S,
Nishimori K and Moustafa A: Tumor metastasis-associated human MTA1
gene and its MTA1 protein product: role in epithelial cancer cell
invasion, proliferation and nuclear regulation. Clin Exp
Metastasis. 20:19–24. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qian H, Yu J, Li Y, Wang H, Song C, Zhang
X, Liang X, Fu M and Lin C: RNA interference of
metastasis-associated gene 1 inhibits metastasis of B16F10 melanoma
cells in a C57BL/6 mouse model. Biol Cell. 99:573–581. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Stephens LC and Kumar R:
Metastasis tumor antigen family proteins during breast cancer
progression and metastasis in a reliable mouse model for human
breast cancer. Clin Cancer Res. 12:1479–1486. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pakala SB, Singh K, Reddy SD, Ohshiro K,
Li DQ, Mishra L and Kumar R: TGF-β1 signaling targets
metastasis-associated protein 1, a new effector in epithelial
cells. Oncogene. 30:2230–2241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moon HE, Cheon H and Lee MS:
Metastasis-associated protein 1 inhibits p53-induced apoptosis.
Oncol Rep. 18:1311–1314. 2007.PubMed/NCBI
|
|
36
|
Kai L, Samuel SK and Levenson AS:
Resveratrol enhances p53 acetylation and apoptosis in prostate
cancer by inhibiting MTA1/ NuRD complex. Int J Cancer.
126:1538–1548. 2010.
|
|
37
|
Schmidt DR and Schreiber SL: Molecular
association between ATR and two components of the nucleosome
remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry.
38:14711–1477. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chou DM, Adamson B, Dephoure NE, Tan X,
Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP and Elledge SJ: A
chromatin localization screen reveals poly (ADP ribose)-regulated
recruitment of the repressive polycomb and NuRD complexes to sites
of DNA damage. Proc Natl Acad Sci USA. 107:18475–1880. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bagheri-Yarmand R, Talukder AH, Wang RA,
Vadlamudi RK and Kumar R: Metastasis-associated protein 1
deregulation causes inappropriate mammary gland development and
tumorigenesis. Development. 131:3469–3479. 2004. View Article : Google Scholar : PubMed/NCBI
|