Introduction
NF-κB is a family of proteins, including RelA, RelB,
c-Rel, NF-κBl, and NF-κB2, which play a critical role in a variety
of biological activities such as lymphocyte development, and
lymphoid tumor cell proliferation and apoptosis (1). The function of the NF-κB signaling
pathway in the proliferation and apoptosis of chronic lymphocytic
leukemia (CLL) cells is a hotspot in cancer research and is
therefore considered a promising therapeutic target for the
treatment of CLL (2). Findings of
recent studies have shown a frequency of only ~10% for NOTCH1
mutations in CLL at diagnosis, although this rate may reach 30%
with the disease progressing towards Richter’s transformation or
chemotherapy refractory CLL (3,4). It
remains unclear whether NOTCH1 mutations affect NF-κB signaling,
subsequently leading to drug resistance and disease progression.
Therefore, we assessed 3 patients with features of CLL carrying
NOTCH1 mutations, and investigated in detail the NOTCH1
mutation-induced constitutive activation of NF-κB signaling.
Materials and methods
Cells
The primary CLL cells were derived from samples
obtained from naive CLL patients diagnosed at Union hospital of
Fujian Medical University (Fuzhou, China). Normal bone marrow
controls were collected from healthy donors. Written informed
consent was provided by each participant. The study was approved by
the institutional review board and conducted in accordance with the
Declaration of Helsinki.
Reagents and devices
Anti-human-CD19, CD5 and CD20 antibodies were
purchased from BD Biosciences (San Diego, CA, USA). The human
CD19+ magnetic bead sorting kit was obtained from
Miltenyi Biotec (Shanghai, China). GSI and PDTC were purchased from
Sigma-Aldrich Corp. (Shanghai, China). Flow cytometry kits were
provided by BD Biosciences, and the ABI PCR instrument was
purchased from ABI Biosystems (Shanghai, China).
Isolation of CLL- and normal
CD19+ B cells
Mononuclear cells (MNCs) were isolated from 5 ml of
anti-coagulated bone marrow samples from CLL patients by density
gradient centrifugation using the lymphocyte separation medium.
Subsequently, CD19+ cells were sorted using
CD19+ magnetic beads and stained with anti-CD19, CD5 and
CD20 antibodies. Flow cytometric analysis showed a purity of ≥95%
for CD5+CD19+ cells. As control samples,
CD19+ cells from 10 ml of bone marrow samples from 5
healthy individuals were isolated with a final purity of ≥90%.
Chromosomal karyotype analysis
Bone marrow cells were briefly cultured and
processed according to conventional R-banding techniques. The
probes used in FISH included the centromeric (CSP12), 11q22-23
deletion (ATM), 17p13 deletion (P53), and 13q14 deletion (RB1 and
D13S25) probes obtained from Abbott (Beijing, China).
Gene sequencing
Gene mutation analysis was performed by
amplification of the IgVH and NOTCH1 fragments by RT-PCR. After
purification, the PCR products underwent direct sequencing. The
sequencing results were analyzed with the V-QUEST software, and
IgVH mutations were validated with >2% base alteration as
compared to the germline gene.
Immunophenotyping and detection of Zap70
and CD38 by flow cytometry
Cells (1×106) were incubated with the
corresponding antibodies for 5 min at room temperature after
washing with ice-cold FACS buffer. The cells were then resuspended
to obtain single-cell suspensions after routine washes and examined
by flow cytometry (BD Biosciences). The cells were designated as
Zap70+ and CD38+ with >20% cells staining
positive for these markers, respectively.
Immunofluorescence analysis of ICN
expression
After fixation with 4% formaldehyde and blocking (3%
BSA-PBS), the cells were incubated with primary antibodies and
fluorescence-conjugated secondary antibodies. After DAPI nuclear
counterstaining, the cells were observed by fluorescence
microscopy.
Western blot detection of ICN
expression
The cells were collected, lysed and boiled for 8 min
in loading buffer. The resulting protein samples were subjected to
polyacrylamide gel electrophoresis followed by membrane transfer.
The membrane was sequentially incubated with anti-ICN primary- and
horseradish peroxidase-conjugated secondary antibodies
(eBioscience) prior to development.
RT-PCR
One-step TRIzol extraction of total RNA was
performed. cDNA was synthesized using primers designed with the
Blast-Primer software. The nucleotide sequences of the forward
primers of RelA and RelB were 5′-GACCGCT GCATCCACAGTTT-3′ and
5′-CCCCTACAATGCTGGCTCC CTGAA-3′ respectively. RT-PCR was conducted
on an ABI PCR instrument using β-actin as an internal control.
Measurement of NF-κB activity
The nuclear proteins were isolated and the DNA
binding activity of NF-κB was measured using a TransAM™ ELISA
detection kit (Shenzhen, China) according to the manufacturer’s
instructions. Briefly, nuclear proteins were incubated with a
biotin-labeled κB oligonucleo-tide probe to form a binding complex,
which was then isolated by non-denaturing acrylamide gel shift,
followed by infrared imaging to assess NF-κB activity.
Assessment of apoptosis by flow
cytometry
Cells (1×106) were collected at different
time points after drug treatment and incubated with Annexin V in
combination with other antibodies for 5 min at room temperature.
After washing, the cells were resuspended to obtain single-cell
suspensions, which were analyzed via flow cytometry. The apoptotic
rate was calculated as: (drug-induced apoptotic cell number -
spontaneous apoptotic cell number) ×100/(100 − control group).
Statistical analysis
The experiments were repeated for a total of three
times. The Student’s t-test was used to compare the means of two
independent samples with the SPSS16.0 software (SPSS Inc., Armonk,
NY, USA). P<0.05 was considered statistically significant.
Results
The gene sequencing results for samples from the 3
naive CLL patients are shown in Table
I. All patients presented a CT frameshift deletion in the PEST
domain of the NOTCH1 gene, which was associated with an
abnormal chromosomal karyotype such as trisomy 12 and 13q deletion.
The patients carrying NOTCH1 mutations had a poor prognosis. One of
the patients experienced disease progression characterized by fever
and lymph node enlargement during the 4th course of FCR therapy.
Lymph node biopsy suggested diffuse large B-cell lymphoma
transformation.
 | Table IClinical factors and therapeutic
outcomes of the three naive CLL patients. |
Table I
Clinical factors and therapeutic
outcomes of the three naive CLL patients.
| Characteristics | Patient 1 | Patient 2 | Patient 3 |
|---|
| Age (years) | 56 | 76 | 61 |
| Gender | F | M | M |
| ECOG | 0 | 1 | 2 |
| Binet stage | B | C | B |
| β2-MG | 3.5 mg/l | 3.8 mg/l | 3.2 mg/l |
| Zap-70 | + | + | − |
| CD38 | − | − | − |
| Karyotype | 47,XX,+12[19]
/46XX[1] | 46,XY,del(13)(q14q21)
[8]/46,XY[12] | 46,XY,+12,del(13)
(q13q21)[5]/46,XY[15] |
| IgVH | Mutated | Unmutated | Mutated |
| NOTCH1
nucleotide |
c7541_7542het_delCT |
c7541_7542het_delCT |
c7541_7542het_delCT |
| Treatment and
outcome | FCR 4 courses Richter
transformation | FCR 4 courses +
R-CHOP 2 courses PR | FCR 3 courses died
due to cerebral hemorrhage |
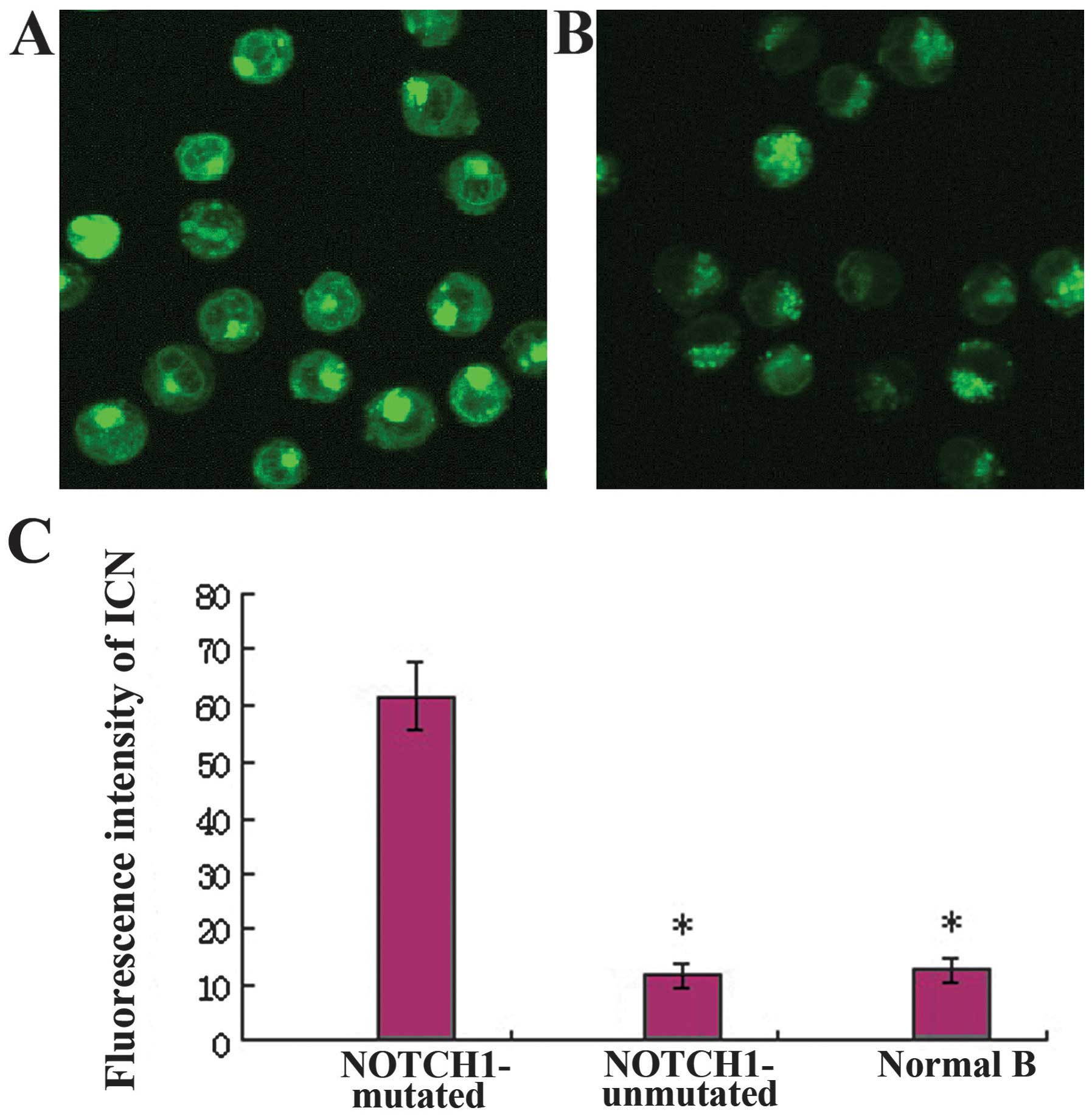
In CLL cells carrying NOTCH1 mutations, the ICN
protein was abundantly expressed in the cytoplasm and nuclei, at
significantly higher levels than that observed in unmutated and
normal CD19+ B cells by immunofluorescence (P<0.01)
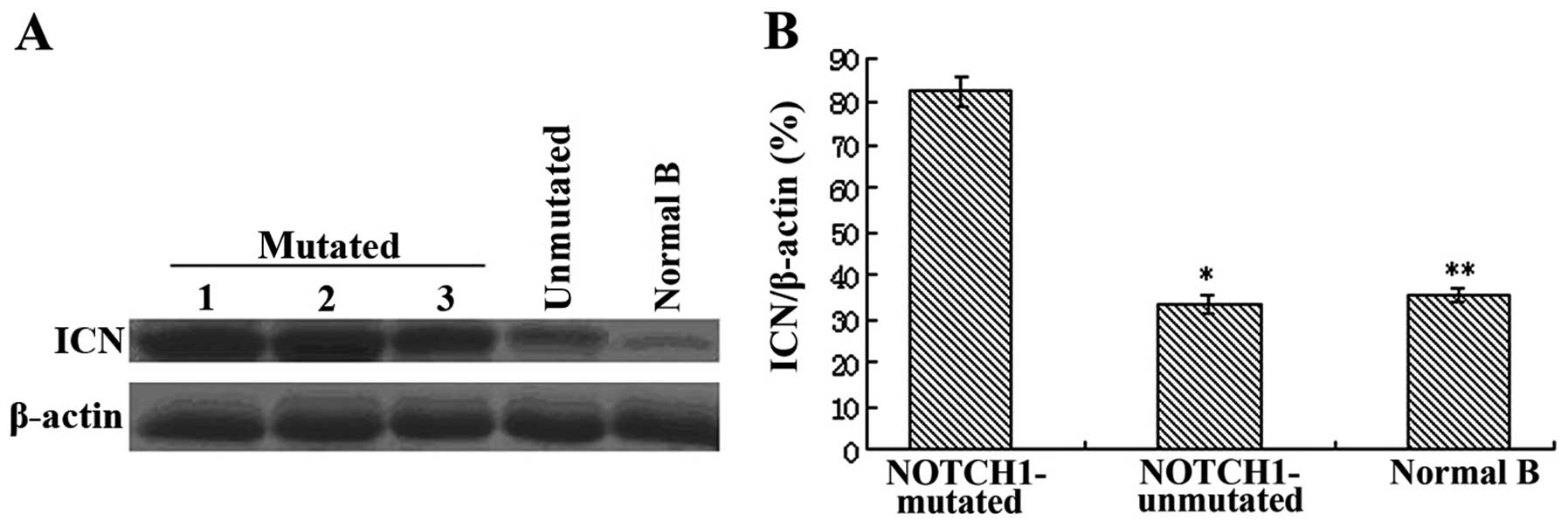
(Fig. 1). Western blot analysis
revealed similar results, with markedly reduced ICN protein levels
in unmutated- and normal B cells compared with NOTCH1-mutated CLL
cells (P<0.01 and P<0.001) (Fig.
2).
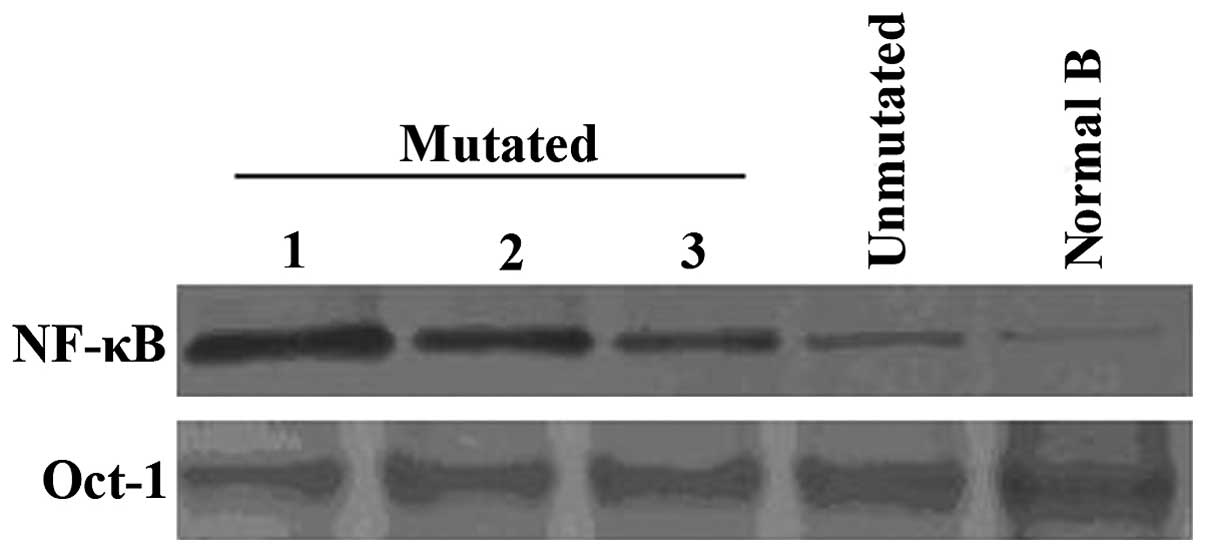
NF-κB activity assessment showed robust DNA-binding
activity of the NF-κB nuclear protein in NOTCH1-mutated CLL cells,
whereas only weak binding activity was detected in unmutated- and
normal B lymphocytes (Fig. 3). In
addition, ELISA showed markedly increased RelA and RelB activities
in mutated CLL cells (0.56±0.03 and 0.28±0.05, respectively)
compared with unmutated CLL cells (0.19±0.01 and 0.08±0.00,
respectively, P<0.01) and normal B-cells (0.01±0.0 and
0.007±0.00, respectively, P<0.01).
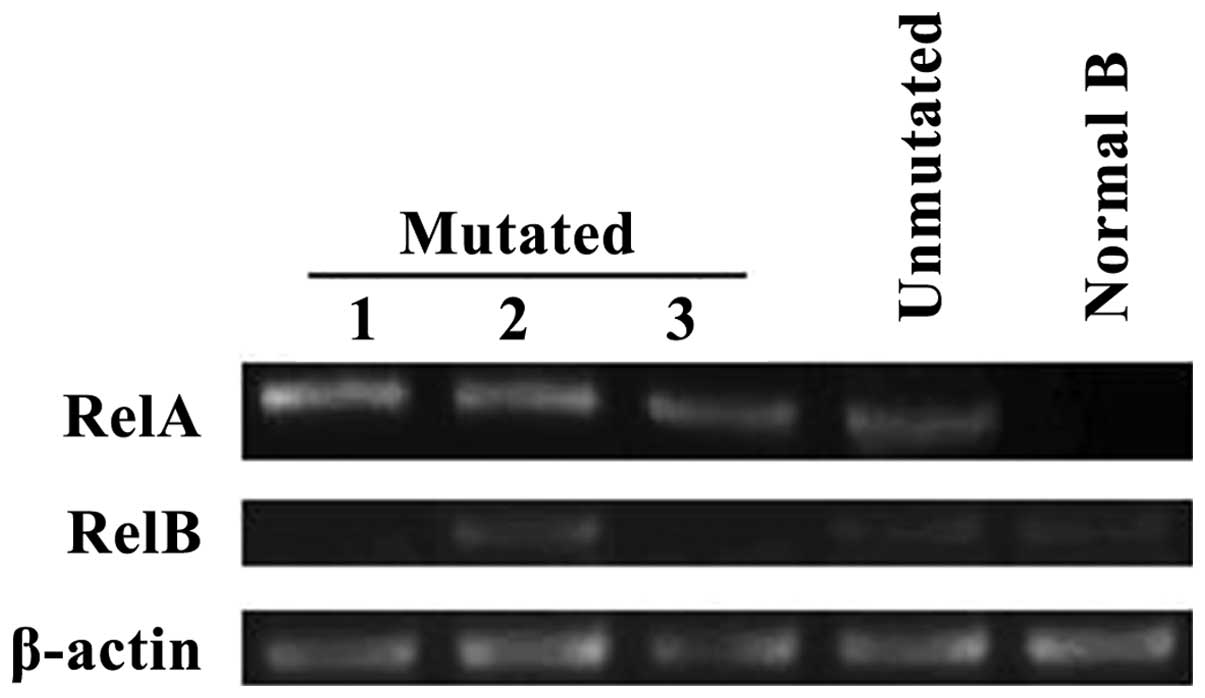
The mRNA levels of RelA and RelB in CLL cells were
examined by RT-PCR. As shown in Fig.
4, the expression of RelA and RelB mRNA in CLL cells varied
considerably. Regardless of the presence or absence of NOTCH1
mutations, RelA mRNA was always detected in CLL cells. However,
RelB mRNA was only found in the second patient carrying NOTCH1
mutations.
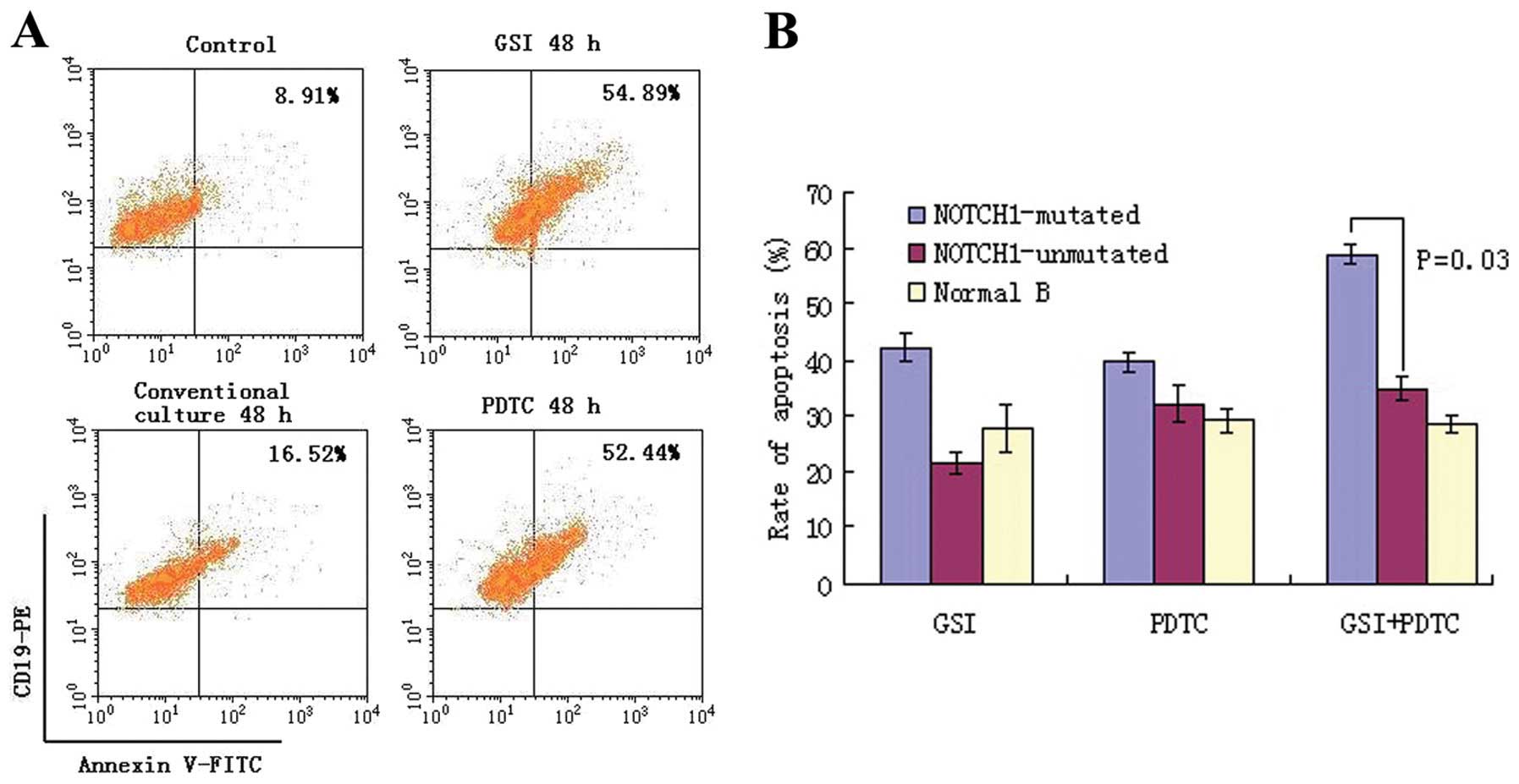
We hypothesized that NF-κB inhibition was able to
induce apoptosis in CLL cells. As shown in Fig. 5, the ratios of apoptotic cells in
NOTCH1-mutated CLL cells were significantly increased following
treatment with GSI (ICN-specific inhibitor) and PDTC
(NF-κB-specific inhibitor) compared with those obtained in
unmutated- and normal B cells, and this effect was even greater
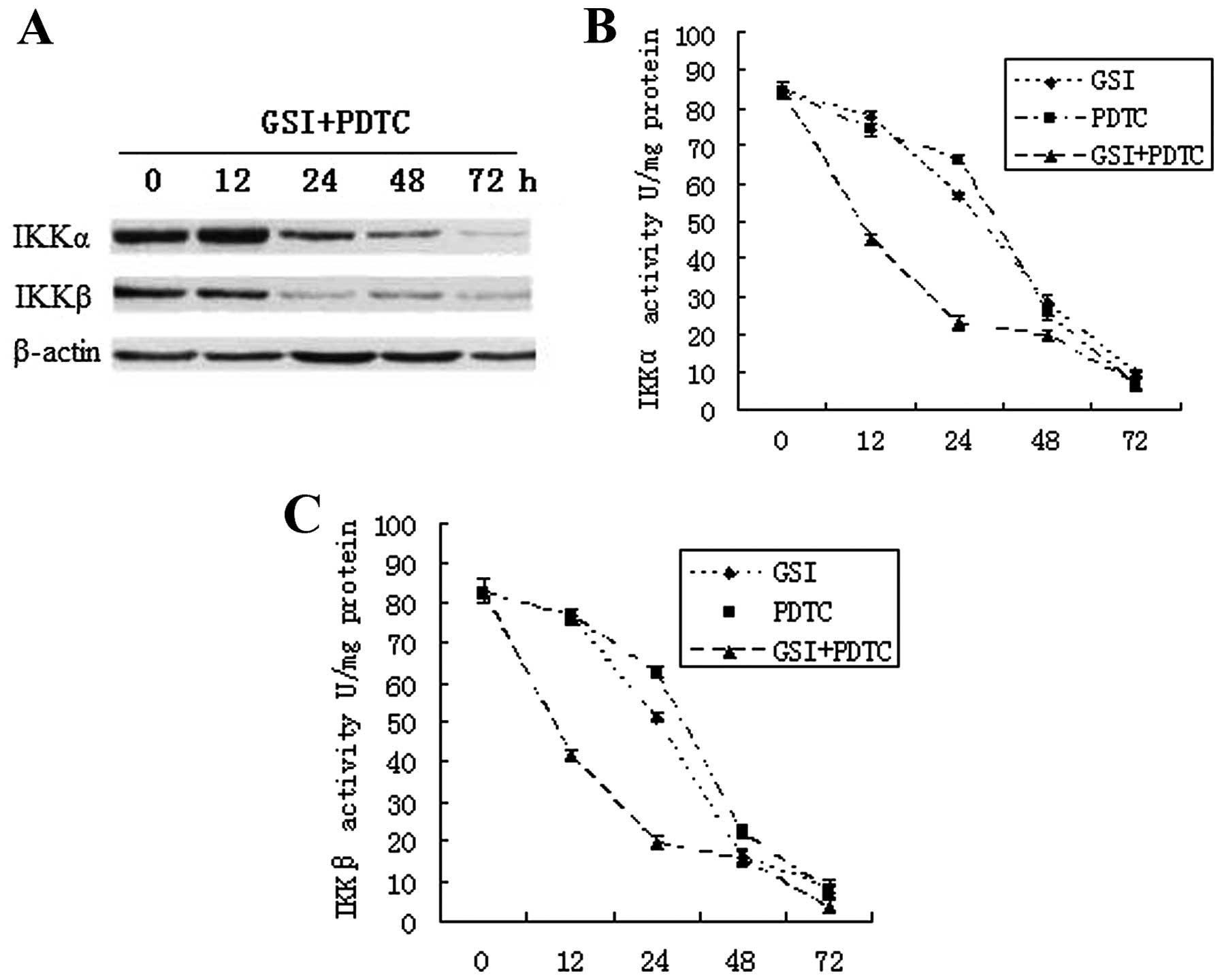
when the two drugs were used (P=0.03). Western blotting confirmed
suppression of the active IKKα and IKKβ together with the prolonged
treatment of GSI in combination with PDTC (Fig. 6).
Discussion
Results of the present study are in agreement with
those of previous reports, which indicated that the clinical
factors, biological characteristics, treatment response and
prognosis of CLL patients in China vary from those of other
countries (5,6). The median survival time of CLL
patients was ~67 months, with many individuals developing drug
resistance or progressing to aggressive lymphoma, or Richter’s
syndrome, during the therapeutic procedure. In this case, the
patients showed poorer prognosis with survival for only 5–8 months.
Thus, it is important to further assess the mechanisms involved in
the malignant transformation of CLL and explore novel therapeutic
targets for its treatment.
The NF-κB signaling pathway has been shown to play a
critical role in the promotion of proliferation and anti-apoptosis
of B-lymphoid tumor cells (1,7,8).
Unlike other lymphoid tumors, IKK kinase activation is
indispensable to the NF-κB signaling pathway in CLL cells (9). Ubiquitination by the CYLD enzyme K63,
the adapter protein of IKK kinase, can effectively regulate NF-κB
signaling. Other regulatory factors such as NOTCH1 and Bcl-10 also
have crucial functions in this pathway. For instance, through
direct or indirect effects on CYLD ubiquitination, NOTCH1
constitutively activates NF-κB signaling in CLL cells, which
results in resistance to apoptosis and consequently contributes to
the development and progression of CLL (10,11).
Recent studies have also shown that mutations of
NOTCH1 promoted the release and stabilization of ICN, which in turn
excessively inhibited CLL cell apoptosis and to a greater extent,
led to aggressive progression to Richter’s syndrome (12,13).
This may be a consequence of the direct activation of NF-κB
signaling by ICN following NOTCH1 mutation (3). Although no exact mechanisms were
revealed for ICN activation of NF-κB, these findings demonstrated
that ICN, a dominant active form of NOTCH1, play an indispensable
role in the NF-κB signaling pathway, particularly following NOTCH1
mutation. Therefore, NOTCH1 mutation is considered a critical
indicator for unfavorable prognosis in CLL (14–16).
Nevertheless, NOTCH1 mutation is not a common event
in CLL. Shen et al (17),
Giulia et al (3), and
Shedden et al (4) performed
second-generation sequencing and found an incidence of ~10% for
NOTCH1 mutations in CLL patients at diagnosis. However, this
incidence may reach 30% following aggressive transformation of the
disease or occurrence of chemotherapy resistance, particularly in
association with cytogenetic abnormalities including 17p- and
trisomy 12 (18). In this case,
poor prognosis is expected for patients. It is conceivable that
certain connections exist between NOTCH1 mutations, NF-κB signal
aberrations and malignant transformation of the disease. In this
study, all three CLL patients had additional abnormal karyotype of
trisomy 12 or 13q-deletion, together with unfavorable prognosis:
one of these patients rapidly progressed to disease transformation
into diffuse large B-cell lymphoma (Richter’s syndrome).
Further sequencing analysis revealed dinucleotide
frame-shift deletion in the PEST domain of NOTCH1. Previously, such
mutations identified in NOTCH1 exon 34 resulted in truncated
C-terminal PEST domain. Consequently, ICN, the protein product of
the truncated PEST domain mediated the sustained activation of the
NF-κB signaling pathway (4).
Additional studies have indicated enriched ICN proteins in the
cytoplasm and nuclei of mutated CLL cells exhibiting strong
NF-κB-dependent anti-apoptotic properties. This may constitute a
critical molecular basis for the aggressive transformation of CLL
cells. An important issue is how ICN proteins affect NF-κB
signaling. Different findings have been provided from
investigations on assorted lymphoid tumors. For instance, Schwarzer
et al found that ICN directly activates IKK kinase in
Hodgkin lymphoma cells, thereby activating the NF-κB signaling
pathway (19).
D’Altri et al demonstrated that ICN
constitutively activates NF-κB through CYLD inhibition and NF-κB
de-ubiquitination in T-ALL cells (20). Following treatment of NOTCH1-mutated
cells with specific inhibitors, we observed a marked inhibition of
IKKα and IKKβ, accompanied by prolonged GSI and PDTC effects. These
results suggest that NOTCH1 mutations in CLL lead to constitutive
activation of NF-κB signaling, likely due to ICN overexpression.
Nevertheless, it remains to be elucidated how ICN activates the
NF-κB signaling pathway following NOTCH1 mutation, which is
possibly a key mechanism involved in the malignant transformation
of CLL, and requires further investigation.
Acknowledgements
This study was supported by National and Fujian
Provincial Key Clinical Specialty Discipline Construction Program
PRC and by a grant of New Century Talent in Fujian (No. JA10128) to
Z.-S.X.
References
|
1
|
Davis RE, Brown KD, Siebenlist U and
Staudt LM: Constitutive nuclear factor κB activity is required for
survival of activated B cell-like diffuse large B cell lymphoma
cells. J Exp Med. 194:1861–1874. 2010. View Article : Google Scholar
|
|
2
|
Lopez-Guerra M and Colomer D: NF-kappaB as
a therapeutic target in chronic lymphocytic leukemia. Expert Opin
Ther Targets. 14:275–288. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giulia F, Silvia R, Davide R, et al:
Analysis of the chronic lymphocytic leukemia coding genome: role of
NOTCH1 mutational activation. J Exp Med. 208:1389–1401. 2011.
View Article : Google Scholar
|
|
4
|
Shedden K, Li Y, Ouillette P and Malek SN:
Characteristics of chronic lymphocytic leukemia with somatically
acquired mutations in NOTCH1 exon 34. Leukemia. 26:1108–1110. 2012.
View Article : Google Scholar
|
|
5
|
Xu Z, Zhang J, Wu S, et al: Younger
patients with chronic lymphocytic leukemia benefit from rituximab
treatment: A single center study in China. Oncol Lett. 5:1266–1272.
2013.PubMed/NCBI
|
|
6
|
Chen L, Zhang Y, Zheng W, et al:
Distinctive IgVH gene segments usage and mutation status in Chinese
patients with chronic lymphocytic leukemia. Leuk Res. 32:1491–1498.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang S, Wang S, Yang T, et al:
CD40L-mediated inhibition of NF-kappaB in CA46 Burkitt lymphoma
cells promotes apoptosis. Leuk Lymphoma. 49:1792–1799. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Capaccione KM and Pine SR: The Notch
signaling pathway as a mediator of tumor survival. Carcinogenesis.
34:1420–1430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lutzny G, Kocher T, Schmidt-Supprian M, et
al: Protein kinase c-β-dependent activation of NF-κB in stromal
cells is indispensable for the survival of chronic lymphocytic
leukemia B cells in vivo. Cancer Cell. 23:77–92. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baldoni S, Sportoletti P, Del Papa B, et
al: NOTCH and NF-κB interplay in chronic lymphocytic leukemia is
independent of genetic lesion. Int J Hematol. 98:153–157. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rosati E, Sabatini R, Rampino G, et al:
Constitutively activated Notch signaling is involved in survival
and apoptosis resistance of B-CLL cells. Blood. 113:856–865. 2009.
View Article : Google Scholar
|
|
12
|
Rossi D, Rasi S, Spina V, et al: Different
impact of NOTCH1 and SF3B1 mutations on the risk of chronic
lymphocytic leukemia transformation to Richter syndrome. Br J
Haematol. 158:426–429. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Puente XS, Pinyol M, Quesada V, et al:
Whole-genome sequencing identifies recurrent mutations in chronic
lymphocytic leukaemia. Nature. 475:101–105. 2012. View Article : Google Scholar :
|
|
14
|
Del Giudice I, Rossi D, Chiaretti S, et
al: NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL)
confer an unfavorable prognosis, induce a distinctive
transcriptional profiling and refine the intermediate prognosis of
+12 CLL. Haematologica. 97:437–441. 2012. View Article : Google Scholar :
|
|
15
|
Villamor N, Conde L, Martínez-Trillos A,
et al: NOTCH1 mutations identify a genetic subgroup of chronic
lymphocytic leukemia patients with high risk of transformation and
poor outcome. Leukemia. 27:1100–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mansouri L, Cahill1 N, Gunnarsson R, et
al: NOTCH1 and SF3B1 mutations can be added to the hierarchical
prognostic classification in chronic lymphocytic leukemia.
Leukemia. 27:512–514. 2013. View Article : Google Scholar
|
|
17
|
Shen W, Qiao C, Xu W, et al: Investigation
of NOTCH1 mutation in Chinese patients with B-cell chronic
lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 11(Suppl 2):
2222011. View Article : Google Scholar
|
|
18
|
López C, Delgado J, Costa D, et al:
Different distribution of NOTCH1 mutations in chronic lymphocytic
leukemia with isolated trisomy 12 or associated with other
chromosomal alterations. Genes Chromosomes Cancer. 51:881–889.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwarzer R, Dörken B and Jundt F: Notch
is an essential upstream regulator of NF-κB and is relevant for
survival of Hodgkin and Reed-Sternberg cells. Leukemia. 26:806–813.
2012. View Article : Google Scholar
|
|
20
|
D’Altri T, Gonzalez J, Aifantis I,
Espinosa L and Bigas A: Hes1 expression and CYLD repression are
essential events downstream of NOTCH1 in T-cell leukemia. Cell
Cycle. 10:1031–1036. 2011. View Article : Google Scholar
|